Gold Nanoparticles in Conjunction with Nucleic Acids as a Modern Molecular System for Cellular Delivery
Abstract
1. Introduction
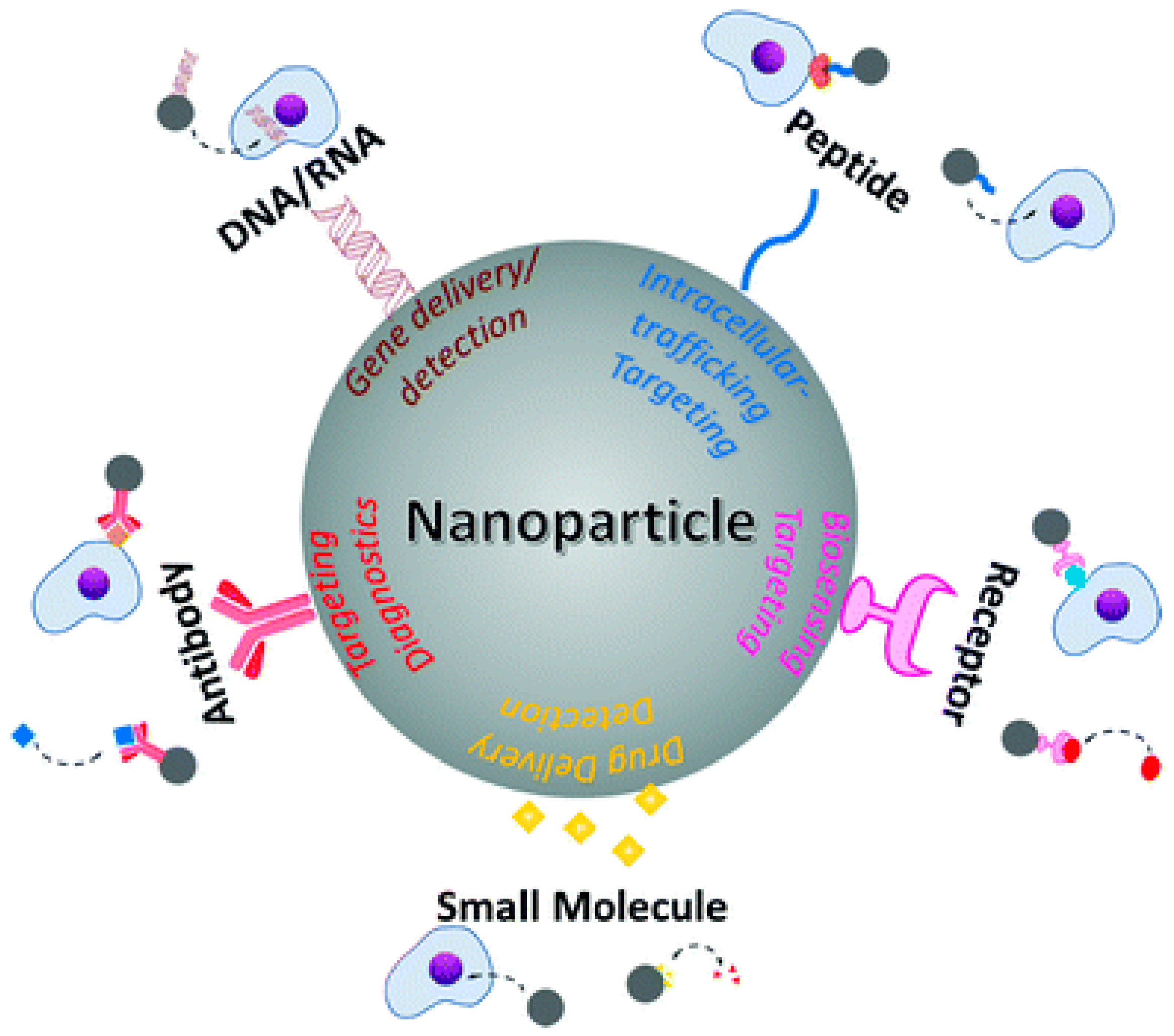
2. Metal Nanoparticles in Medicine
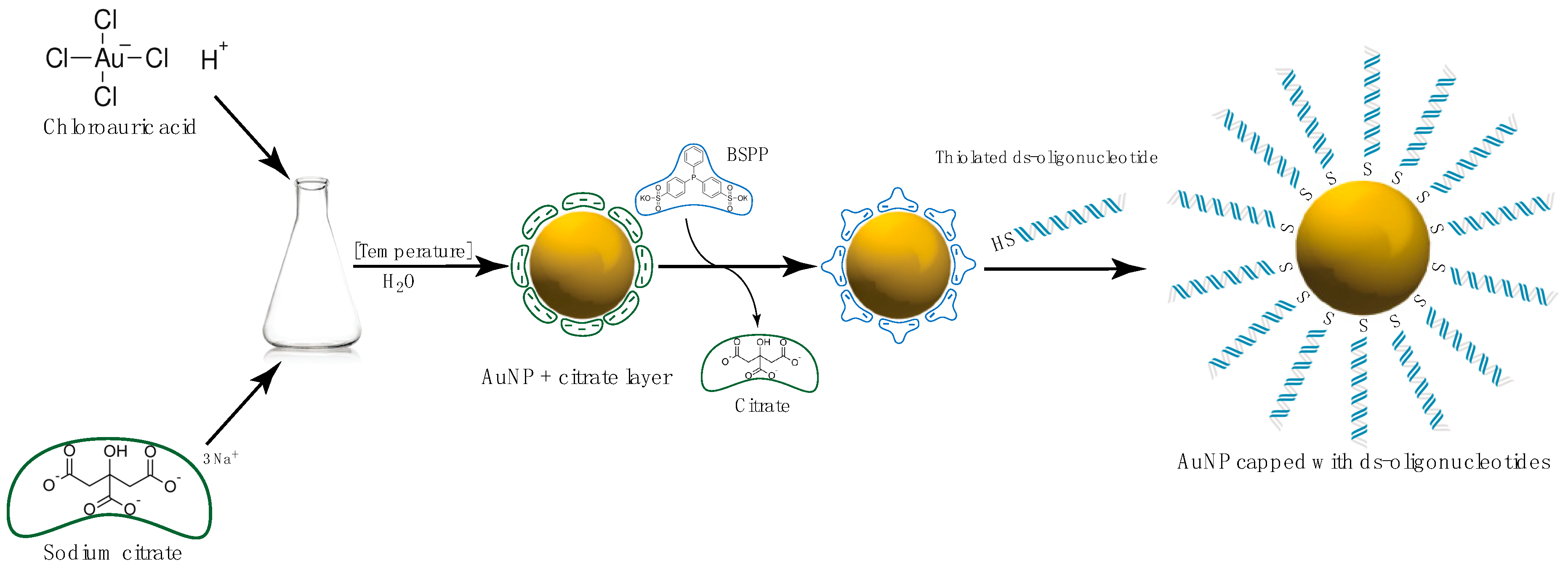
Gold Nanoparticles
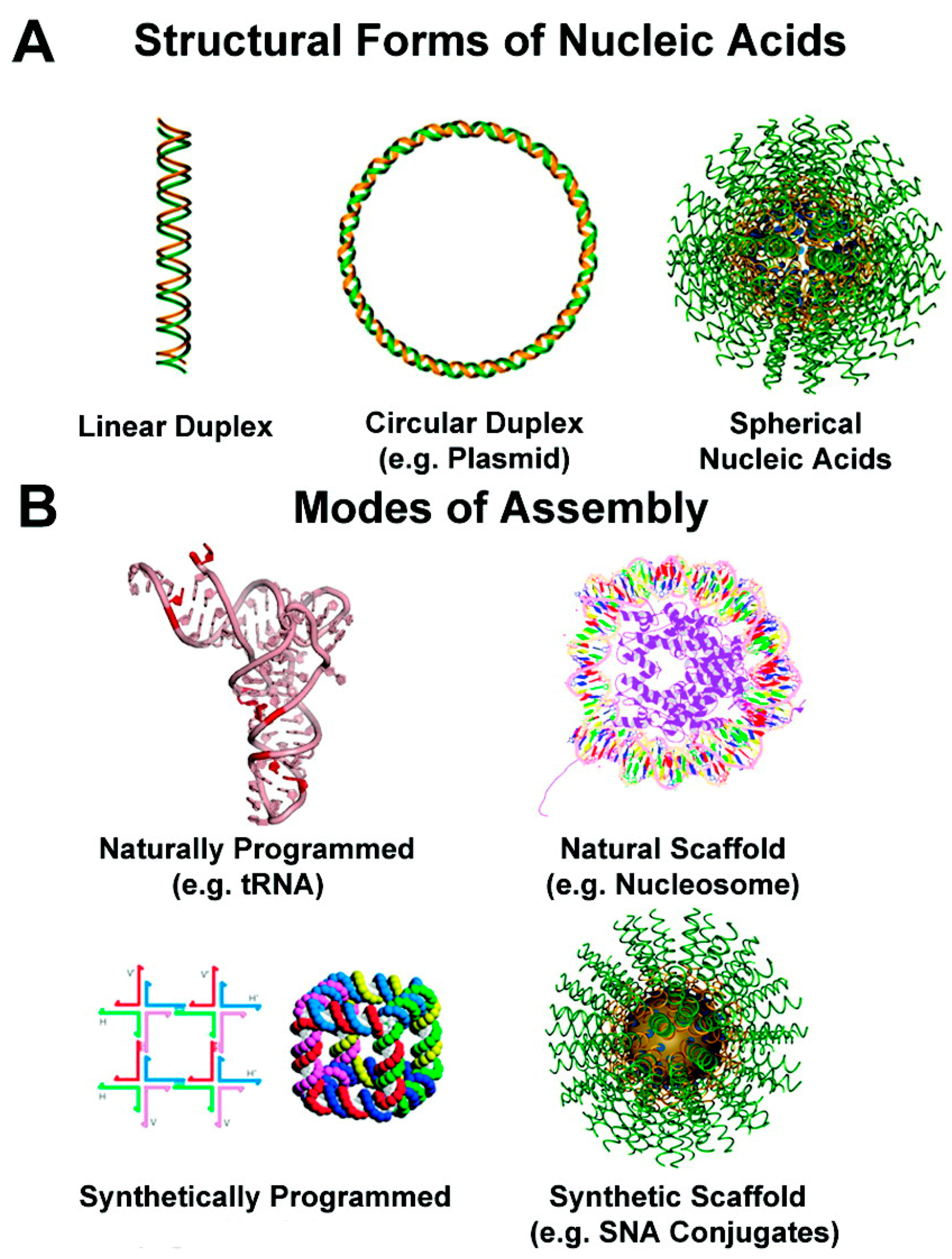
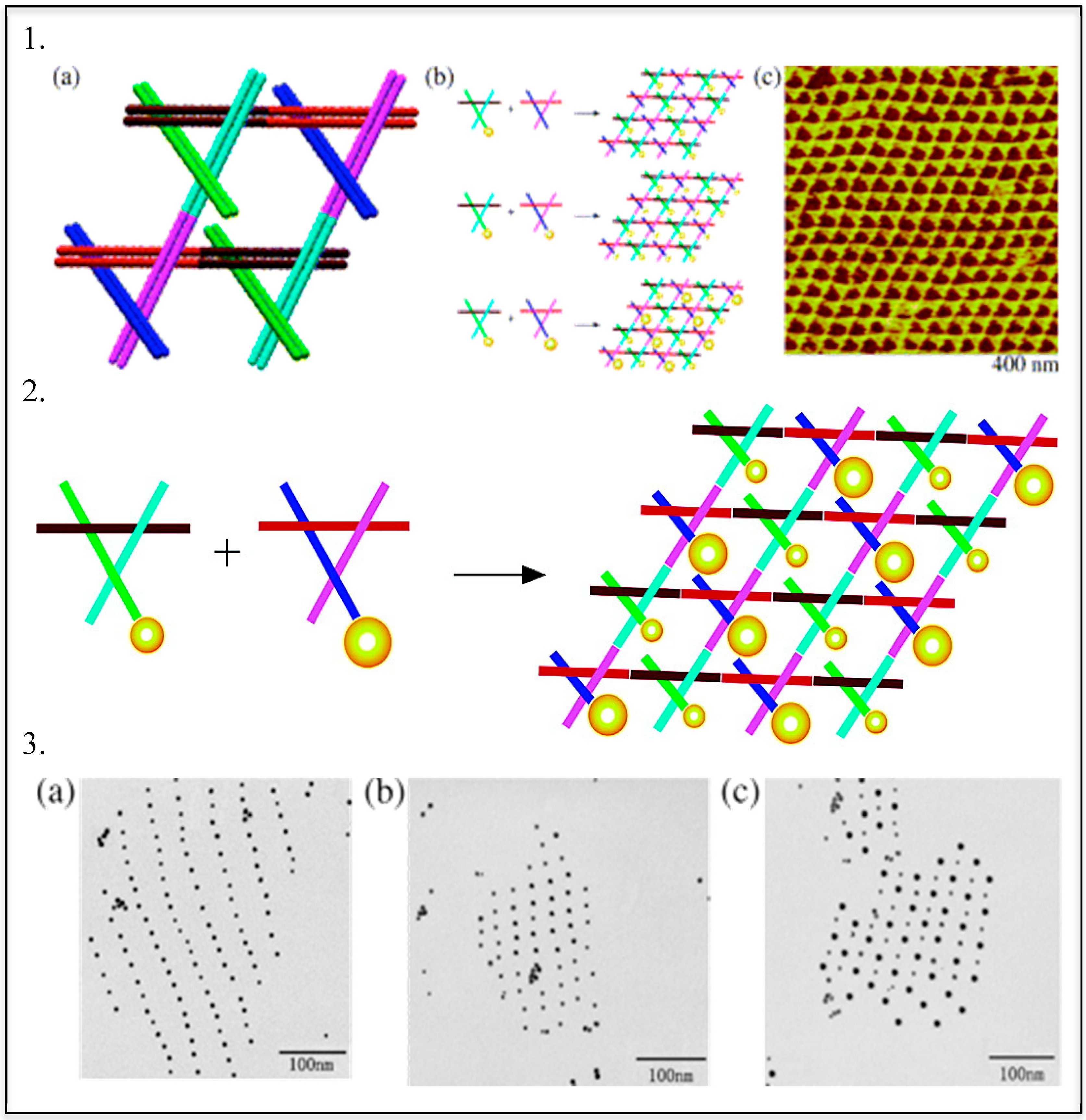
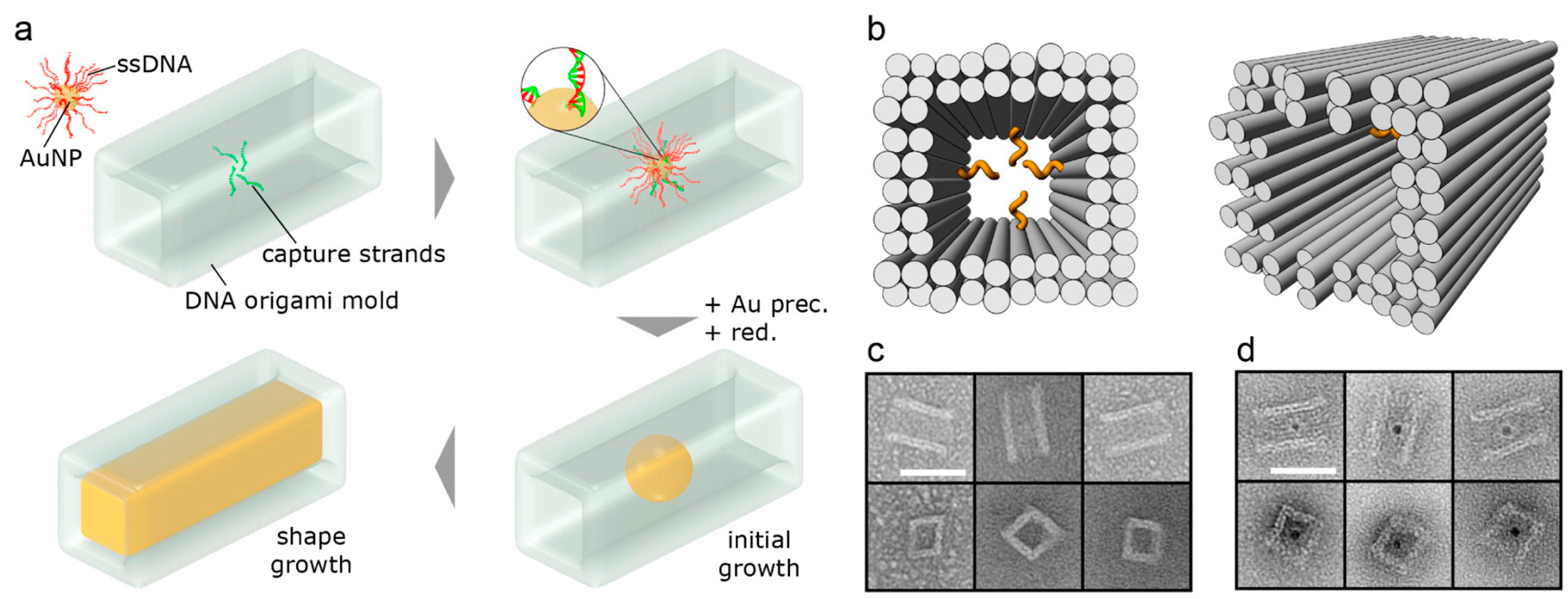
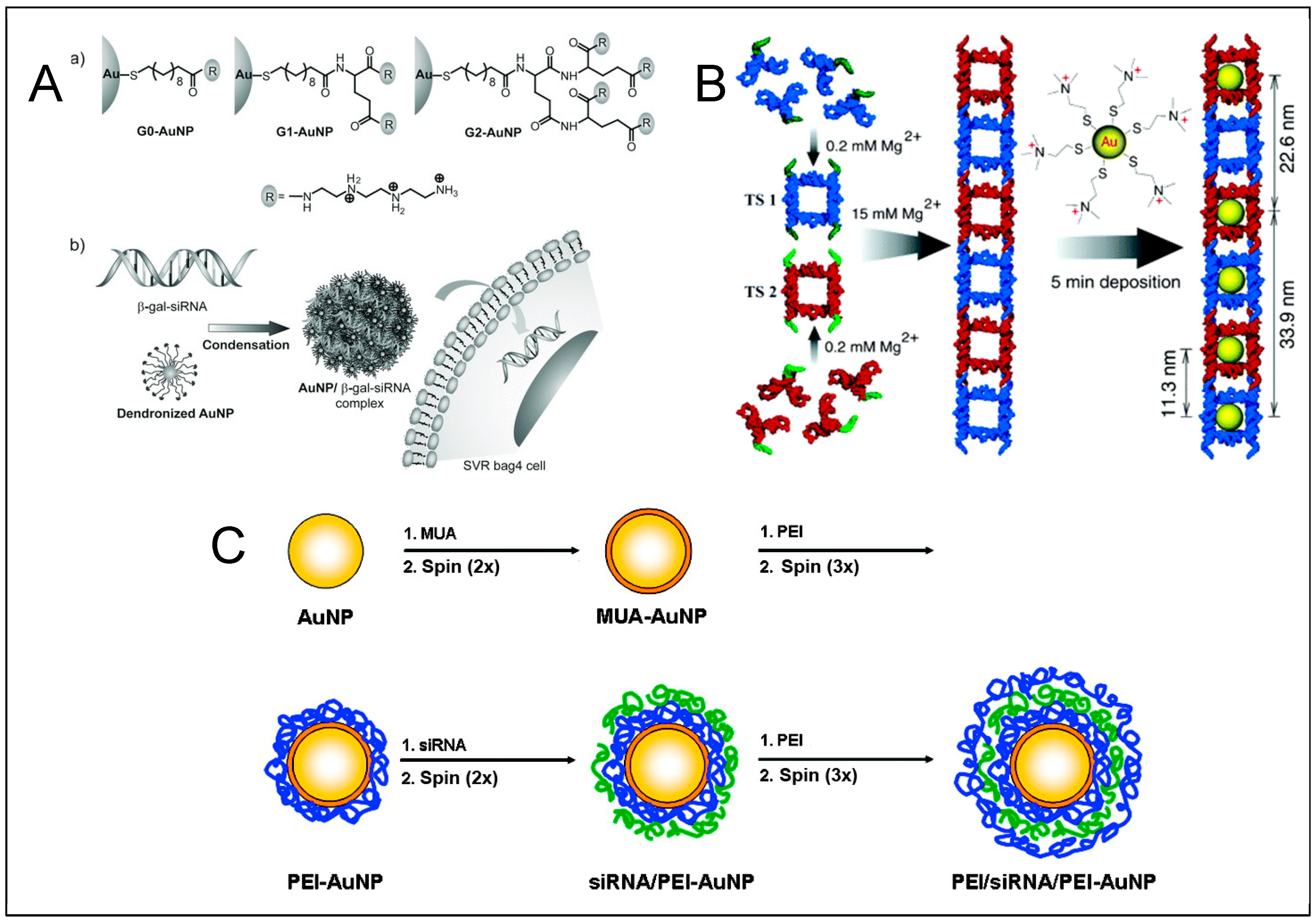
3. Templated DNA Structures—DNA Nanotechnology
DNA—Metal Nanoparticles Conjugates
4. RNA—Gold Nanoparticles (AuNP) Conjugates
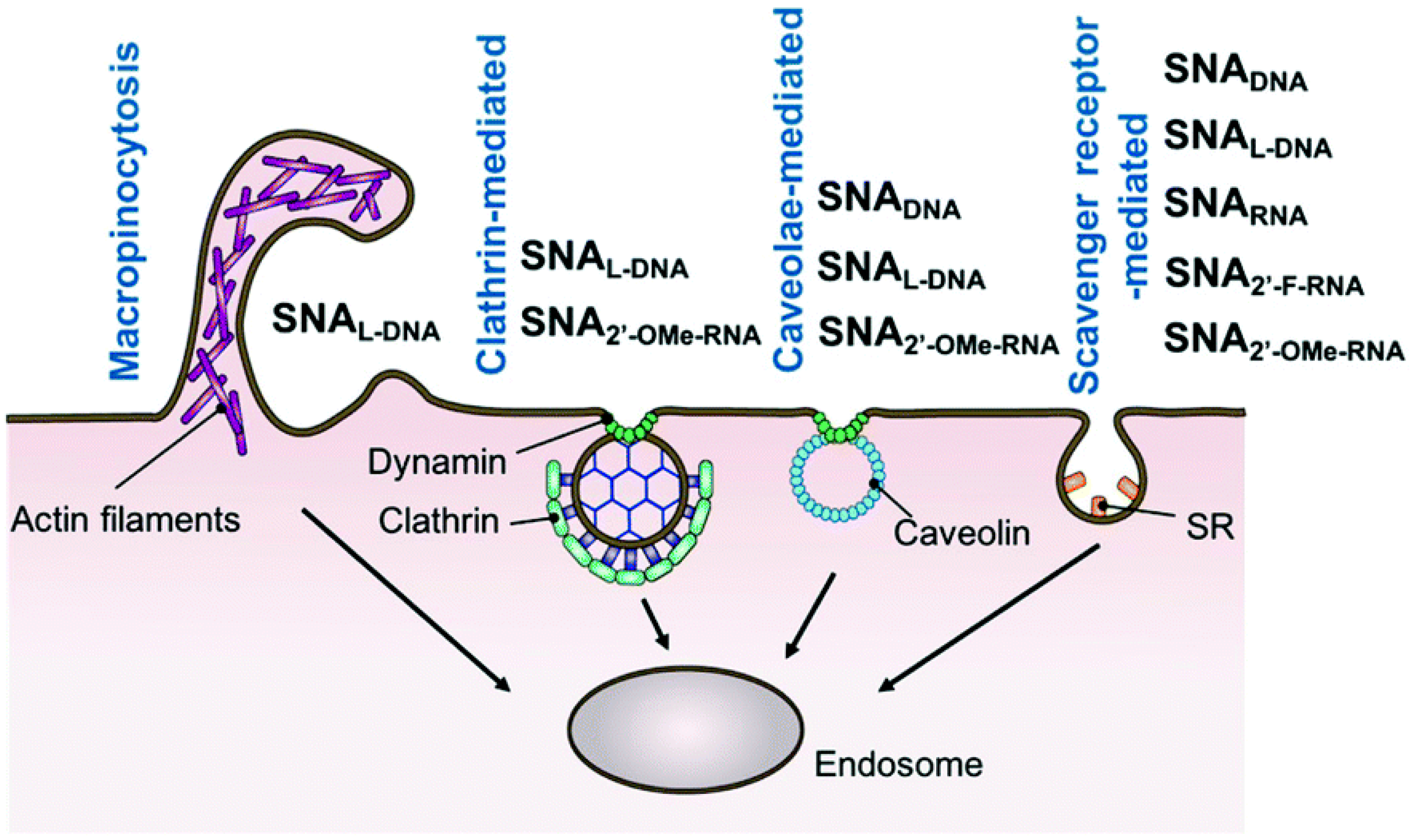
5. Transport of DNA and RNA Conjugated Nanoparticles inside a Cell
- Cell surface binding,
- Translocation across the plasma membrane and thus penetration inside the cell (include membrane invagination and sorting into early endosomes)
- Escape from endosomes or lysosomes
5.1. Intracellular Trafficking Organelle Distribution and Processing of Nanostructures
- Low-motility particles (adhered to the membrane or bound to receptors)
- High-motility particles (particles wrapped in early endosomal vesicles)
- Low-motility particles (wrapped in late endosomes or lysosomes in the perinuclear region of the cell)
- The fast-moving single particle chases a slow-moving one to merge (this type of motion is associated with vesicular fusion of early endosomes)
- The single nanoparticle connects to small cluster on a different track, then rapidly separates and both molecules continue the moving along original tracks.
- The small cluster moves rapidly towards another one, which is static and suddenly reverses its moving direction. This motion is similar to a dynein- and kinesin-based bidirectional cargo transport along the microtubule.
- The two small clusters move along two tracks in the cross-section of the microtubules and one of them looses its mobility and becomes static while the second one departs with high speed.
5.2. Physico-Chemical Properties of Nanoparticles and Their Cellular Uptake and Transport
6. Summary
Funding
Acknowledgments
Conflicts of Interest
References
- Jeevanandam, J.; Barhoum, A.; Chan, Y.S.; Dufresne, A.; Danquah, M.K. Review on nanoparticles and nanostructured materials: History, sources, toxicity and regulations. Beilstein J. Nanotechnol. 2018, 9, 1050–1074. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.; Saeed, K.; Khan, I. Nanoparticles: Properties, applications and toxicities. Arab. J. Chem. 2019, 12, 908–931. [Google Scholar] [CrossRef]
- Iravani, S. Green synthesis of metal nanoparticles using plants. Green Chem. 2011, 13, 2638–2650. [Google Scholar] [CrossRef]
- Khan, Z.U.; Khan, A.; Chen, Y.M.; Shah, N.S.; Muhammad, N.; Khan, A.U.; Tahir, K.; Khan, F.U.; Murtaza, B.; Ul Hassan, S.; et al. Biomedical applications of green synthesized Nobel metal nanoparticles. J. Photochem. Photobiol. B 2017, 173, 150–164. [Google Scholar] [CrossRef]
- Meziani, M.J.; Sun, Y.P. Protein-conjugated nanoparticles from rapid expansion of supercritical fluid solution into aqueous solution. J. Am. Chem. Soc. 2003, 125, 8015–8018. [Google Scholar] [CrossRef]
- Piella, J.; Bastus, N.G.; Puntes, V. Size-Dependent Protein-Nanoparticle Interactions in Citrate-Stabilized Gold Nanoparticles: The Emergence of the Protein Corona. Bioconjug. Chem. 2017, 28, 88–97. [Google Scholar] [CrossRef]
- Draz, M.S.; Fang, B.A.; Zhang, P.F.; Hu, Z.; Gu, S.D.; Weng, K.C.; Gray, J.W.; Chen, F.F. Nanoparticle-Mediated Systemic Delivery of siRNA for Treatment of Cancers and Viral Infections. Theranostics 2014, 4, 872–892. [Google Scholar] [CrossRef]
- Brigger, I.; Dubernet, C.; Couvreur, P. Nanoparticles in cancer therapy and diagnosis. Adv. Drug Deliv. Rev. 2002, 54, 631–651. [Google Scholar] [CrossRef]
- Azharuddin, M.; Zhu, G.H.; Das, D.; Ozgur, E.; Uzun, L.; Turner, A.P.F.; Patra, H.K. A repertoire of biomedical applications of noble metal nanoparticles. Chem. Commun. 2019, 55, 6964–6996. [Google Scholar] [CrossRef]
- Mirkin, C.A.; Letsinger, R.L.; Mucic, R.C.; Storhoff, J.J. A DNA-based method for rationally assembling nanoparticles into macroscopic materials. Nature 1996, 382, 607–609. [Google Scholar] [CrossRef]
- Exicure. Available online: http://www.exicuretx.com/index.php (accessed on 27 November 2019).
- Cutler, J.I.; Auyeung, E.; Mirkin, C.A. Spherical Nucleic Acids. J. Am. Chem. Soc. 2012, 134, 1376–1391. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhang, B.; Lu, X.; Tan, X.; Jia, F.; Xiao, Y.; Cheng, Z.; Li, Y.; Silva, D.O.; Schrekker, H.S.; et al. Molecular spherical nucleic acids. Proc. Natl. Acad. Sci. USA 2018, 115, 4340–4344. [Google Scholar] [CrossRef] [PubMed]
- Cutler, J.I.; Zhang, K.; Zheng, D.; Auyeung, E.; Prigodich, A.E.; Mirkin, C.A. Polyvalent Nucleic Acid Nanostructures. J. Am. Chem. Soc. 2011, 133, 9254–9257. [Google Scholar] [CrossRef] [PubMed]
- Connor, E.E.; Mwamuka, J.; Gole, A.; Murphy, C.J.; Wyatt, M.D. Gold nanoparticles are taken up by human cells but do not cause acute cytotoxicity. Small 2005, 1, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Yeh, Y.C.; Creran, B.; Rotello, V.M. Gold nanoparticles: Preparation, properties, and applications in bionanotechnology. Nanoscale 2012, 4, 1871–1880. [Google Scholar] [CrossRef] [PubMed]
- Amendola, V.; MeneghettI, M.; Stener, M.; Guo, Y.; Chen, S.; Crespo, P.; Garcia, M.A.; Hernando, A.; Pengo, P.; Pasquato, L. Physico-Chemical Characteristics of Gold Nanoparticles. In Gold Nanoparticles in Analytical Chemistry; Elsevier: Amsterdam, The Netherlands, 2014; Volume 66, pp. 81–152. [Google Scholar]
- Amendola, V.; Pilot, R.; Frasconi, M.; Marago, O.M.; Iati, M.A. Surface plasmon resonance in gold nanoparticles: A review. J. Phys. Condens. Matter 2017, 29. [Google Scholar] [CrossRef]
- Das, M.; Shim, K.H.; An, S.S.A.; Yi, D.K. Review on gold nanoparticles and their applications. Toxicol. Environ. Health Sci. 2011, 3, 193–205. [Google Scholar] [CrossRef]
- Han, G.; You, C.C.; Kim, B.J.; Turingan, R.S.; Forbes, N.S.; Martin, C.T.; Rotello, V.M. Light-regulated release of DNA and its delivery to nuclei by means of photolabile gold nanoparticles. Angew. Chem. Int. Ed. 2006, 45, 3165–3169. [Google Scholar] [CrossRef]
- Weintraub, K. The new gold standard. Nature 2013, 495, S14–S16. [Google Scholar] [CrossRef]
- Hong, R.; Han, G.; Fernandez, J.M.; Kim, B.J.; Forbes, N.S.; Rotello, V.M. Glutathione-mediated delivery and release using monolayer protected nanoparticle carriers. J. Am. Chem. Soc. 2006, 128, 1078–1079. [Google Scholar] [CrossRef]
- Weissleder, R. A clearer vision for in vivo imaging. Nat. Biotechnol. 2001, 19, 316–317. [Google Scholar] [CrossRef] [PubMed]
- Sotnikov, D.V.; Berlina, A.N.; Ivanov, V.S.; Zherdev, A.V.; Dzantiev, B.B. Adsorption of proteins on gold nanoparticles: One or more layers? Colloids Surf. B Biointerfaces 2019, 173, 557–563. [Google Scholar] [CrossRef] [PubMed]
- Khashayar, P.; Amoabediny, G.; Larijani, B.; Hosseini, M.; Vanfleteren, J. Fabrication and Verification of Conjugated AuNP-Antibody Nanoprobe for Sensitivity Improvement in Electrochemical Biosensors. Sci. Rep. 2017, 7, 16070. [Google Scholar] [CrossRef] [PubMed]
- Mustafaoglu, N.; Kiziltepe, T.; Bilgicer, B. Site-specific conjugation of an antibody on a gold nanoparticle surface for one-step diagnosis of prostate specific antigen with dynamic light scattering. Nanoscale 2017, 9, 8684–8694. [Google Scholar] [CrossRef] [PubMed]
- Pissuwan, D.; Niidome, T.; Cortie, M.B. The forthcoming applications of gold nanoparticles in drug and gene delivery systems. J. Control. Release 2011, 149, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Camerin, M.; Magaraggia, M.; Soncin, M.; Jori, G.; Moreno, M.; Chambrier, I.; Cook, M.J.; Russell, D.A. The in vivo efficacy of phthalocyanine-nanoparticle conjugates for the photodynamic therapy of amelanotic melanoma. Eur. J. Cancer 2010, 46, 1910–1918. [Google Scholar] [CrossRef]
- Labala, S.; Jose, A.; Venuganti, V.V. Transcutaneous iontophoretic delivery of STAT3 siRNA using layer-by-layer chitosan coated gold nanoparticles to treat melanoma. Colloids Surf. B Biointerfaces 2016, 146, 188–197. [Google Scholar] [CrossRef]
- Mohammadi, Z.; Sazgarnia, A.; Rajabi, O.; Seilanian Toosi, M. Comparative study of X-ray treatment and photodynamic therapy by using 5-aminolevulinic acid conjugated gold nanoparticles in a melanoma cell line. Artif. Cells Nanomed. Biotechnol. 2017, 45, 467–473. [Google Scholar] [CrossRef]
- Huang, X.; Jain, P.K.; El-Sayed, I.H.; El-Sayed, M.A. Plasmonic photothermal therapy (PPTT) using gold nanoparticles. Lasers Med. Sci. 2008, 23, 217–228. [Google Scholar] [CrossRef]
- Kennedy, L.C.; Bickford, L.R.; Lewinski, N.A.; Coughlin, A.J.; Hu, Y.; Day, E.S.; West, J.L.; Drezek, R.A. A new era for cancer treatment: Gold-nanoparticle-mediated thermal therapies. Small 2011, 7, 169–183. [Google Scholar] [CrossRef]
- Day, E.S.; Zhang, L.; Thompson, P.A.; Zawaski, J.A.; Kaffes, C.C.; Gaber, M.W.; Blaney, S.M.; West, J.L. Vascular-targeted photothermal therapy of an orthotopic murine glioma model. Nanomedicine 2012, 7, 1133–1148. [Google Scholar] [CrossRef]
- Vines, J.B.; Yoon, J.H.; Ryu, N.E.; Lim, D.J.; Park, H. Gold Nanoparticles for Photothermal Cancer Therapy. Front. Chem. 2019, 7, 167. [Google Scholar] [CrossRef] [PubMed]
- D’Acunto, M. Detection of Intracellular Gold Nanoparticles: An Overview. Materials 2018, 11, 882. [Google Scholar] [CrossRef] [PubMed]
- Huefner, A.; Septiadi, D.; Wilts, B.D.; Patel, I.I.; Kuan, W.L.; Fragniere, A.; Barker, R.A.; Mahajan, S. Gold nanoparticles explore cells: Cellular uptake and their use as intracellular probes. Methods 2014, 68, 354–363. [Google Scholar] [CrossRef] [PubMed]
- Song, K.H.; Kim, C.; Maslov, K.; Wang, L.V. Noninvasive in vivo spectroscopic nanorod-contrast photoacoustic mapping of sentinel lymph nodes. Eur. J. Radiol. 2009, 70, 227–231. [Google Scholar] [CrossRef] [PubMed]
- Rogers, N.J.; Jeffery, H.C.; Claire, S.; Lewis, D.J.; Zikeli, G.; Hodges, N.J.; Egginton, S.; Nash, G.B.; Pikramenou, Z. Tailoring iridium luminescence and gold nanoparticle size for imaging of microvascular blood flow. Nanomedicine 2017, 12, 2725–2740. [Google Scholar] [CrossRef] [PubMed]
- Bagheri, S.; Yasemi, M.; Safaie-Qamsari, E.; Rashidiani, J.; Abkar, M.; Hassani, M.; Mirhosseini, S.A.; Kooshki, H. Using gold nanoparticles in diagnosis and treatment of melanoma cancer. Artif. Cells Nanomed. Biotechnol. 2018, 46, 462–471. [Google Scholar] [CrossRef] [PubMed]
- Meola, A.; Rao, J.; Chaudhary, N.; Sharma, M.; Chang, S.D. Gold Nanoparticles for Brain Tumor Imaging: A Systematic Review. Front. Neurol. 2018, 9, 328. [Google Scholar] [CrossRef]
- Cruz, L.J.; Tacken, P.J.; Rueda, F.; Domingo, J.C.; Albericio, F.; Figdor, C.G. Targeting nanoparticles to dendritic cells for immunotherapy. Methods Enzymol. 2012, 509, 143–163. [Google Scholar] [CrossRef]
- Jia, J.; Zhang, Y.; Xin, Y.; Jiang, C.; Yan, B.; Zhai, S. Interactions Between Nanoparticles and Dendritic Cells: From the Perspective of Cancer Immunotherapy. Front. Oncol. 2018, 8, 404. [Google Scholar] [CrossRef]
- Surendran, S.P.; Moon, M.J.; Park, R.; Jeong, Y.Y. Bioactive Nanoparticles for Cancer Immunotherapy. Int. J. Mol. Sci. 2018, 19, 3877. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Wu, T.; Qin, X.; Qiao, Q.; Shang, L.; Song, Q.; Yang, C.; Zhang, Z. Intracellularly Generated Immunological Gold Nanoparticles for Combinatorial Photothermal Therapy and Immunotherapy against Tumor. Nano Lett. 2019, 19, 6635–6646. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Jiang, Z.; Saha, K.; Kim, C.S.; Kim, S.T.; Landis, R.F.; Rotello, V.M. Gold nanoparticles for nucleic acid delivery. Mol. Ther. 2014, 22, 1075–1083. [Google Scholar] [CrossRef] [PubMed]
- Seeman, N.C. Nucleic acid junctions and lattices. J. Theor. Biol. 1982, 99, 237–247. [Google Scholar] [CrossRef]
- Fu, T.J.; Seeman, N.C. DNA double-crossover molecules. Biochemistry 1993, 32, 3211–3220. [Google Scholar] [CrossRef]
- Sa-Ardyen, P.; Vologodskii, A.V.; Seeman, N.C. The flexibility of DNA double crossover molecules. Biophys. J. 2003, 84, 3829–3837. [Google Scholar] [CrossRef]
- Ding, B.Q.; Sha, R.J.; Seeman, N.C. Pseudohexagonal 2D DNA crystals from double crossover cohesion. J. Am. Chem. Soc. 2004, 126, 10230–10231. [Google Scholar] [CrossRef]
- Wang, W.; Lin, T.; Zhang, S.Y.; Bai, T.X.; Mi, Y.L.; Wei, B.Y. Self-assembly of fully addressable DNA nanostructures from double crossover tiles. Nucleic Acids Res. 2016, 44, 7989–7996. [Google Scholar] [CrossRef]
- LaBean, T.H.; Yan, H.; Kopatsch, J.; Liu, F.R.; Winfree, E.; Reif, J.H.; Seeman, N.C. Construction, analysis, ligation, and self-assembly of DNA triple crossover complexes. J. Am. Chem. Soc. 2000, 122, 1848–1860. [Google Scholar] [CrossRef]
- Liu, D.; Park, S.H.; Reif, J.H.; LaBean, T.H. DNA nanotubes self-assembled from triple-crossover tiles as templates for conductive nanowires. Proc. Natl. Acad. Sci. USA 2004, 101, 717–722. [Google Scholar] [CrossRef]
- Wei, B.; Mi, Y.L. A new triple crossover triangle (TXT) motif for DNA self-assembly. Biomacromolecules 2005, 6, 2528–2532. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.Y.; Yan, H.; Wang, T.; Seeman, N.C. Paranemic crossover DNA: A generalized Holliday structure with applications in nanotechnology. J. Am. Chem. Soc. 2004, 126, 1666–1674. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.Y.; Wang, X.; Wang, T.; Sha, R.J.; Seeman, N.C. PX DNA triangle oligomerized using a novel three-domain motif. Nano Lett. 2008, 8, 317–322. [Google Scholar] [CrossRef] [PubMed]
- Winfree, E.; Liu, F.R.; Wenzler, L.A.; Seeman, N.C. Design and self-assembly of two-dimensional DNA crystals. Nature 1998, 394, 539–544. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Park, S.H.; Finkelstein, G.; Reif, J.H.; LaBean, T.H. DNA-templated self-assembly of protein arrays and highly conductive nanowires. Science 2003, 301, 1882–1884. [Google Scholar] [CrossRef]
- Chen, J.; Seeman, N.C. The Synthesis from DNA of a Molecule with the Connectivity of a Cube. Nature 1991, 350, 631–633. [Google Scholar] [CrossRef]
- Shih, W.M.; Quispe, J.D.; Joyce, G.F. A 1.7-kilobase single-stranded DNA that folds into a nanoscale octahedron. Nature 2004, 427, 618–621. [Google Scholar] [CrossRef]
- Rothemund, P.W. Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440, 297–302. [Google Scholar] [CrossRef]
- Mao, C.D.; Sun, W.Q.; Shen, Z.Y.; Seeman, N.C. A nanomechanical device based on the B-Z transition of DNA. Nature 1999, 397, 144–146. [Google Scholar] [CrossRef]
- Modi, S.; Swetha, M.G.; Goswami, D.; Gupta, G.D.; Mayor, S.; Krishnan, Y. A DNA nanomachine that maps spatial and temporal pH changes inside living cells. Nat. Nanotechnol. 2009, 4, 325–330. [Google Scholar] [CrossRef]
- Modi, S.; Nizak, C.; Surana, S.; Halder, S.; Krishnan, Y. Two DNA nanomachines map pH changes along intersecting endocytic pathways inside the same cell. Nat. Nanotechnol. 2013, 8, 459–467. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.G.; Liang, X.G.; Mochizuki, T.; Asanuma, H. A Light-Driven DNA Nanomachine for the Efficient Photoswitching of RNA Digestion. Angew. Chem. Int. Ed. 2010, 49, 2167–2170. [Google Scholar] [CrossRef] [PubMed]
- Lund, K.; Manzo, A.J.; Dabby, N.; Michelotti, N.; Johnson-Buck, A.; Nangreave, J.; Taylor, S.; Pei, R.J.; Stojanovic, M.N.; Walter, N.G.; et al. Molecular robots guided by prescriptive landscapes. Nature 2010, 465, 206–210. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, M.S.; Mao, C.D. An autonomous DNA nanomotor powered by a DNA enzyme. Angew. Chem. Int. Ed. 2004, 43, 3554–3557. [Google Scholar] [CrossRef] [PubMed]
- Yin, P.; Yan, H.; Daniell, X.G.; Turberfield, A.J.; Reif, J.H. A unidirectional DNA walker that moves autonomously along a track. Angew. Chem. Int. Ed. 2004, 43, 4906–4911. [Google Scholar] [CrossRef] [PubMed]
- Bath, J.; Green, S.J.; Turberfield, A.J. A free-running DNA motor powered by a nicking enzyme. Angew. Chem. Int. Ed. 2005, 44, 4358–4361. [Google Scholar] [CrossRef]
- Yurke, B.; Turberfield, A.J.; Mills, A.P.; Simmel, F.C.; Neumann, J.L. A DNA-fuelled molecular machine made of DNA. Nature 2000, 406, 605–608. [Google Scholar] [CrossRef]
- Shin, J.S.; Pierce, N.A. A synthetic DNA walker for molecular transport. J. Am. Chem. Soc. 2004, 126, 10834–10835. [Google Scholar] [CrossRef]
- Tian, Y.; Mao, C.D. Molecular gears: A pair of DNA circles continuously rolls against each other. J. Am. Chem. Soc. 2004, 126, 11410–11411. [Google Scholar] [CrossRef]
- Douglas, S.M.; Bachelet, I.; Church, G.M. A Logic-Gated Nanorobot for Targeted Transport of Molecular Payloads. Science 2012, 335, 831–834. [Google Scholar] [CrossRef]
- Andersen, E.S.; Dong, M.; Nielsen, M.M.; Jahn, K.; Subramani, R.; Mamdouh, W.; Golas, M.M.; Sander, B.; Stark, H.; Oliveira, C.L.P.; et al. Self-assembly of a nanoscale DNA box with a controllable lid. Nature 2009, 459, 73–76. [Google Scholar] [CrossRef] [PubMed]
- Bujold, K.E.; Fakhoury, J.; Edwardson, T.G.W.; Carneiro, K.M.M.; Briard, J.N.; Godin, A.G.; Amrein, L.; Hamblin, G.D.; Panasci, L.C.; Wiseman, P.W.; et al. Sequence-responsive unzipping DNA cubes with tunable cellular uptake profiles. Chem. Sci. 2014, 5, 2449–2455. [Google Scholar] [CrossRef]
- Liu, M.; Li, Q.; Liang, L.; Li, J.; Wang, K.; Li, J.; Lv, M.; Chen, N.; Song, H.; Lee, J.; et al. Real-time visualization of clustering and intracellular transport of gold nanoparticles by correlative imaging. Nat. Commun. 2017, 8, 15646. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.Y.; Li, X.F.; Zhang, H.Q.; Le, X.C. A microRNA-initiated DNAzyme motor operating in living cells. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef]
- Rajendran, A.; Nakata, E.; Nakano, S.; Morii, T. Nucleic-Acid-Templated Enzyme Cascades. ChemBioChem 2017, 18, 696–716. [Google Scholar] [CrossRef]
- Gelinas, A.D.; Davies, D.R.; Janjic, N. Embracing proteins: Structural themes in aptamer-protein complexes. Curr. Opin. Struct. Biol. 2016, 36, 122–132. [Google Scholar] [CrossRef]
- Sacca, B.; Niemeyer, C.M. Functionalization of DNA nanostructures with proteins. Chem. Soc. Rev. 2011, 40, 5910–5921. [Google Scholar] [CrossRef]
- Niemeyer, C.M.; Sano, T.; Smith, C.L.; Cantor, C.R. Oligonucleotide-Directed Self-Assembly of Proteins—Semisynthetic DNA Streptavidin Hybrid Molecules as Connectors for the Generation of Macroscopic Arrays and the Construction of Supramolecular Bioconjugates. Nucleic Acids Res. 1994, 22, 5530–5539. [Google Scholar] [CrossRef]
- Niemeyer, C.M.; Adler, M.; Gao, S.; Chi, L.F. Supramolecular nanocircles consisting of streptavidin and DNA. Angew. Chem. Int. Ed. 2000, 39, 3055–3059. [Google Scholar] [CrossRef]
- Li, H.Y.; Park, S.H.; Reif, J.H.; LaBean, T.H.; Yan, H. DNA-templated self-assembly of protein and nanoparticle linear arrays. J. Am. Chem. Soc. 2004, 126, 418–419. [Google Scholar] [CrossRef]
- Park, S.H.; Yin, P.; Liu, Y.; Reif, J.H.; LaBean, T.H.; Yan, H. Programmable DNA self-assemblies for nanoscale organization of ligands and proteins. Nano Lett. 2005, 5, 729–733. [Google Scholar] [CrossRef] [PubMed]
- Koyfman, A.Y.; Braun, G.B.; Reich, N.O. Cell-Targeted Self-Assembled DNA Nanostructures. J. Am. Chem. Soc. 2009, 131, 14237–14239. [Google Scholar] [CrossRef] [PubMed]
- Malo, J.; Mitchell, J.C.; Venien-Bryan, C.; Harris, J.R.; Wille, H.; Sherratt, D.J.; Turberfield, A.J. Engineering a 2D protein-DNA crystal. Angew. Chem. Int. Ed. 2005, 44, 3057–3061. [Google Scholar] [CrossRef] [PubMed]
- Aptagen. Available online: https://www.aptagen.com/apta-index/ (accessed on 27 November 2019).
- Liu, Y.; Lin, C.X.; Li, H.Y.; Yan, H. Protein nanoarrays—Aptamer-directed self-assembly of protein arrays on a DNA nanostructure. Angew. Chem. Int. Ed. 2005, 44, 4333–4338. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.X.; Katilius, E.; Liu, Y.; Zhang, J.P.; Yan, H. Self-assembled signaling aptamer DNA arrays for protein detection. Angew. Chem. Int. Ed. 2006, 45, 5296–5301. [Google Scholar] [CrossRef] [PubMed]
- Niemeyer, C.M. Bioorganic applications of semisynthetic DNA-protein conjugates. Chem. A Eur. J. 2001, 7, 3189–3195. [Google Scholar] [CrossRef]
- Niemeyer, C.M. The developments of semisynthetic DNA-protein conjugates. Trends Biotechnol. 2002, 20, 395–401. [Google Scholar] [CrossRef]
- Li, H.Y.; LaBean, T.H.; Leong, K.W. Nucleic acid-based nanoengineering: Novel structures for biomedical applications. Interface Focus 2011, 1, 702–724. [Google Scholar] [CrossRef]
- Hu, Q.; Li, H.; Wang, L.; Gu, H.; Fan, C. DNA Nanotechnology-Enabled Drug Delivery Systems. Chem. Rev. 2019, 119, 6459–6506. [Google Scholar] [CrossRef]
- Yang, D.Y.; Campolongo, M.J.; Tran, T.N.N.; Ruiz, R.C.H.; Kahn, J.S.; Luo, D. Novel DNA materials and their applications. Wires Nanomed. Nanobiotechnol. 2010, 2, 648–669. [Google Scholar] [CrossRef]
- Seeman, N.; Sleiman, H. DNA nanotechnology. Nat. Rev. Mater. 2017, 3. [Google Scholar] [CrossRef]
- Bath, J.; Turberfield, A.J. DNA nanomachines. Nat. Nanotechnol 2007, 2, 275–284. [Google Scholar] [CrossRef] [PubMed]
- Liedl, T.; Sobey, T.L.; Simmel, F.C. DNA-based nanodevices. Nano Today 2007, 2, 36–41. [Google Scholar] [CrossRef]
- Niemeyer, C.M. Functional devices from DNA and proteins. Nano Today 2007, 2, 42–52. [Google Scholar] [CrossRef]
- Kwon, Y.W.; Lee, C.H.; Choi, D.H.; Jin, J.I. Materials science of DNA. J. Mater. Chem. 2009, 19, 1353–1380. [Google Scholar] [CrossRef]
- Carter, J.D.; LaBean, T.H. Organization of Inorganic Nanomaterials via Programmable DNA Self-Assembly and Peptide Molecular Recognition. ACS Nano 2011, 5, 2200–2205. [Google Scholar] [CrossRef] [PubMed]
- Torimoto, T.; Yamashita, M.; Kuwabata, S.; Sakata, T.; Mori, H.; Yoneyama, H. Fabrication of CdS nanoparticle chains along DNA double strands. J. Phys. Chem. B 1999, 103, 8799–8803. [Google Scholar] [CrossRef]
- Richter, J.; Seidel, R.; Kirsch, R.; Mertig, M.; Pompe, W.; Plaschke, J.; Schackert, H.K. Nanoscale palladium metallization of DNA. Adv. Mater. 2000, 12, 507–510. [Google Scholar] [CrossRef]
- Ford, W.E.; Harnack, O.; Yasuda, A.; Wessels, J.M. Platinated DNA as precursors to templated chains of metal nanoparticles. Adv. Mater. 2001, 13, 1793–1797. [Google Scholar] [CrossRef]
- Patolsky, F.; Weizmann, Y.; Lioubashevski, O.; Willner, I. Au-nanoparticle nanowires based on DNA and polylysine templates. Angew. Chem. Int. Ed. 2002, 41, 2323–2327. [Google Scholar] [CrossRef]
- Stoltenberg, R.M.; Woolley, A.T. DNA-templated nanowire fabrication. Biomed. Microdevices 2004, 6, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Alivisatos, A.P.; Johnsson, K.P.; Peng, X.G.; Wilson, T.E.; Loweth, C.J.; Bruchez, M.P.; Schultz, P.G. Organization of ‘nanocrystal molecules’ using DNA. Nature 1996, 382, 609–611. [Google Scholar] [CrossRef] [PubMed]
- Le, J.D.; Pinto, Y.; Seeman, N.C.; Musier-Forsyth, K.; Taton, T.A.; Kiehl, R.A. DNA-templated self-assembly of metallic nanocomponent arrays on a surface. Nano Lett. 2004, 4, 2343–2347. [Google Scholar] [CrossRef]
- Pinto, Y.Y.; Le, J.D.; Seeman, N.C.; Musier-Forsyth, K.; Taton, T.A.; Kiehl, R.A. Sequence-encoded self-assembly of multiple-nanocomponent arrays by 2D DNA scaffolding. Nano Lett. 2005, 5, 2399–2402. [Google Scholar] [CrossRef]
- Sharma, J.; Chhabra, R.; Liu, Y.; Ke, Y.G.; Yan, H. DNA-templated self-assembly of two-dimensional and periodical gold nanoparticle arrays. Angew. Chem. Int. Ed. 2006, 45, 730–735. [Google Scholar] [CrossRef]
- Zhang, J.P.; Liu, Y.; Ke, Y.G.; Yan, H. Periodic square-like gold nanoparticle arrays templated by self-assembled 2D DNA nanogrids on a surface. Nano Lett. 2006, 6, 248–251. [Google Scholar] [CrossRef]
- Zheng, J.W.; Constantinou, P.E.; Micheel, C.; Alivisatos, A.P.; Kiehl, R.A.; Seeman, N.C. Two-dimensional nanoparticle arrays show the organizational power of robust DNA motifs. Nano Lett. 2006, 6, 1502–1504. [Google Scholar] [CrossRef]
- Nykypanchuk, D.; Maye, M.M.; van der Lelie, D.; Gang, O. DNA-guided crystallization of colloidal nanoparticles. Nature 2008, 451, 549–552. [Google Scholar] [CrossRef]
- Park, S.Y.; Lytton-Jean, A.K.R.; Lee, B.; Weigand, S.; Schatz, G.C.; Mirkin, C.A. DNA-programmable nanoparticle crystallization. Nature 2008, 451, 553–556. [Google Scholar] [CrossRef]
- Julin, S.; Nummelin, S.; Kostiainen, M.A.; Linko, V. DNA nanostructure-directed assembly of metal nanoparticle superlattices. J. Nanoparticle Res. 2018, 20, 119. [Google Scholar] [CrossRef]
- Mastroianni, A.J.; Claridge, S.A.; Alivisatos, A.P. Pyramidal and Chiral Groupings of Gold Nanocrystals Assembled Using DNA Scaffolds. J. Am. Chem. Soc. 2009, 131, 8455–8459. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, N.; Schlapak, R.; Kastner, M.; Armitage, D.; Chrzanowski, W.; Riener, J.; Hinterdorfer, P.; Ebner, A.; Howorka, S. A DNA Nanostructure for the Functional Assembly of Chemical Groups with Tunable Stoichiometry and Defined Nanoscale Geometry. Angew. Chem. Int. Ed. 2009, 48, 525–527. [Google Scholar] [CrossRef] [PubMed]
- Sharma, J.; Chhabra, R.; Andersen, C.S.; Gothelf, K.V.; Yan, H.; Liu, Y. Toward reliable gold nanoparticle patterning on self-assembled DNA nanoscaffold. J. Am. Chem. Soc. 2008, 130, 7820–7821. [Google Scholar] [CrossRef] [PubMed]
- Ding, B.Q.; Deng, Z.T.; Yan, H.; Cabrini, S.; Zuckermann, R.N.; Bokor, J. Gold Nanoparticle Self-Similar Chain Structure Organized by DNA Origami. J. Am. Chem. Soc. 2010, 132, 3248–3249. [Google Scholar] [CrossRef] [PubMed]
- Pal, S.; Deng, Z.T.; Ding, B.Q.; Yan, H.; Liu, Y. DNA-Origami-Directed Self-Assembly of Discrete Silver-Nanoparticle Architectures. Angew. Chem. Int. Ed. 2010, 49, 2700–2704. [Google Scholar] [CrossRef]
- Helmi, S.; Ziegler, C.; Kauert, D.J.; Seidel, R. Shape-Controlled Synthesis of Gold Nanostructures Using DNA Origami Molds. Nano Lett. 2014, 14, 6693–6698. [Google Scholar] [CrossRef]
- Sun, W.; Boulais, E.; Hakobyan, Y.; Wang, W.L.; Guan, A.; Bathe, M.; Yin, P. Casting inorganic structures with DNA molds. Science 2014, 346, 1258361. [Google Scholar] [CrossRef]
- Yan, H.; Zhang, X.P.; Shen, Z.Y.; Seeman, N.C. A robust DNA mechanical device controlled by hybridization topology. Nature 2002, 415, 62–65. [Google Scholar] [CrossRef]
- Mao, C.D.; LaBean, T.H.; Reif, J.H.; Seeman, N. Logical computation using algorithmic self-assembly of DNA triple-crossover molecules (vol 407, pg 493, 2000). Nature 2000, 408, 750. [Google Scholar] [CrossRef]
- Benenson, Y.; Paz-Elizur, T.; Adar, R.; Keinan, E.; Livneh, Z.; Shapiro, E. Programmable and autonomous computing machine made of biomolecules. Nature 2001, 414, 430–434. [Google Scholar] [CrossRef]
- Fujibayashi, K.; Hariadi, R.; Park, S.H.; Winfree, E.; Murata, S. Toward reliable algorithmic self-assembly of DNA tiles: A fixed-width cellular automaton pattern. Nano Lett. 2008, 8, 1791–1797. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Xie, Y.J.; Zhou, S.H.; Zhou, C.J.; Zheng, X.D. Reversible Data Hiding Based on DNA Computing. Comput. Intell. Neurosci. 2017. [Google Scholar] [CrossRef] [PubMed]
- Green, S.J.; Lubrich, D.; Turberfield, A.J. DNA hairpins: Fuel for autonomous DNA devices. Biophys. J. 2006, 91, 2966–2975. [Google Scholar] [CrossRef] [PubMed]
- Venkataraman, S.; Dirks, R.M.; Rothemund, P.W.K.; Winfree, E.; Pierce, N.A. An autonomous polymerization motor powered by DNA hybridization. Nat. Nanotechnol. 2007, 2, 490–494. [Google Scholar] [CrossRef]
- Lubrich, D.; Lin, J.; Yan, J. A contractile DNA machine. Angew. Chem. Int. Ed. 2008, 47, 7026–7028. [Google Scholar] [CrossRef]
- Deng, Z.X.; Mao, C.D. Molecular lithography with DNA nanostructures. Angew. Chem. Int. Ed. 2004, 43, 4068–4070. [Google Scholar] [CrossRef]
- Edwardson, T.G.W.; Lau, K.L.; Bousmail, D.; Serpell, C.J.; Sleiman, H.F. Transfer of molecular recognition information from DNA nanostructures to gold nanoparticles. Nat. Chem. 2016, 8, 162–170. [Google Scholar] [CrossRef]
- Kuzyk, A.; Schreiber, R.; Fan, Z.Y.; Pardatscher, G.; Roller, E.M.; Hogele, A.; Simmel, F.C.; Govorov, A.O.; Liedl, T. DNA-based self-assembly of chiral plasmonic nanostructures with tailored optical response. Nature 2012, 483, 311–314. [Google Scholar] [CrossRef]
- Wang, X.; Sha, R.J.; Kristiansen, M.; Hernandez, C.; Hao, Y.D.; Mao, C.D.; Canary, J.W.; Seeman, N.C. An Organic Semiconductor Organized into 3D DNA Arrays by “Bottom-up” Rational Design. Angew. Chem. Int. Ed. 2017, 56, 6445–6448. [Google Scholar] [CrossRef]
- Tyagi, S.; Kramer, F.R. Molecular beacons: Probes that fluoresce upon hybridization. Nat. Biotechnol. 1996, 14, 303–308. [Google Scholar] [CrossRef]
- Drake, T.J.; Tan, W.H. Molecular beacon DNA probes and their bioanalytical applications. Appl. Spectrosc. 2004, 58, 269A–280A. [Google Scholar] [CrossRef] [PubMed]
- Mirkin, C.A. Programming the assembly of two- and three-dimensional architectures with DNA and nanoscale inorganic building blocks. Inorg. Chem. 2000, 39, 2258–2272. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; McLaughlin, C.K.; Aldaye, F.A.; Hamblin, G.D.; Rys, A.Z.; Rouiller, I.; Sleiman, H.F. Metal-nucleic acid cages. Nat. Chem. 2009, 1, 390–396. [Google Scholar] [CrossRef] [PubMed]
- Nam, J.M.; Thaxton, C.S.; Mirkin, C.A. Nanoparticle-based bio-bar codes for the ultrasensitive detection of proteins. Science 2003, 301, 1884–1886. [Google Scholar] [CrossRef]
- Nam, J.M.; Stoeva, S.I.; Mirkin, C.A. Bio-bar-code-based DNA detection with PCR-like sensitivity. J. Am. Chem. Soc. 2004, 126, 5932–5933. [Google Scholar] [CrossRef]
- Stoeva, S.I.; Lee, J.S.; Thaxton, C.S.; Mirkin, C.A. Multiplexed DNA detection with biobarcoded nanoparticle probes. Angew. Chem. Int. Ed. 2006, 45, 3303–3306. [Google Scholar] [CrossRef]
- Zhang, K.Y.; Lv, S.Z.; Lin, Z.Z.; Li, M.J.; Tang, D.P. Bio-bar-code-based photoelectrochemical immunoassay for sensitive detection of prostate-specific antigen using rolling circle amplification and enzymatic biocatalytic precipitation. Biosens. Bioelectron. 2018, 101, 159–166. [Google Scholar] [CrossRef]
- Delihas, N. Discovery and characterization of the first non-coding RNA that regulates gene expression, micF RNA: A historical perspective. World J. Biol. Chem. 2015, 6, 272–280. [Google Scholar] [CrossRef]
- Setten, R.L.; Rossi, J.J.; Han, S.P. The current state and future directions of RNAi-based therapeutics. Nat. Rev. Drug Discov. 2019, 18, 421–446. [Google Scholar] [CrossRef]
- Giljohann, D.A.; Seferos, D.S.; Prigodich, A.E.; Patel, P.C.; Mirkin, C.A. Gene regulation with polyvalent siRNA-nanoparticle conjugates. J. Am. Chem. Soc. 2009, 131, 2072–2073. [Google Scholar] [CrossRef]
- Patel, P.C.; Hao, L.; Yeung, W.S.; Mirkin, C.A. Duplex end breathing determines serum stability and intracellular potency of siRNA-Au NPs. Mol. Pharm. 2011, 8, 1285–1291. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.; Giljohann, D.A.; Chen, D.L.; Massich, M.D.; Wang, X.Q.; Iordanov, H.; Mirkin, C.A.; Paller, A.S. Topical delivery of siRNA-based spherical nucleic acid nanoparticle conjugates for gene regulation. Proc. Natl. Acad. Sci. USA 2012, 109, 11975–11980. [Google Scholar] [CrossRef] [PubMed]
- Jensen, S.A.; Day, E.S.; Ko, C.H.; Hurley, L.A.; Luciano, J.P.; Kouri, F.M.; Merkel, T.J.; Luthi, A.J.; Patel, P.C.; Cutler, J.I.; et al. Spherical nucleic acid nanoparticle conjugates as an RNAi-based therapy for glioblastoma. Sci. Transl. Med. 2013, 5, 209ra152. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Takemoto, H.; Yi, Y.; Zheng, M.; Maeda, Y.; Chaya, H.; Hayashi, K.; Mi, P.; Pittella, F.; Christie, R.J.; et al. Precise engineering of siRNA delivery vehicles to tumors using polyion complexes and gold nanoparticles. ACS Nano 2014, 8, 8979–8991. [Google Scholar] [CrossRef] [PubMed]
- Barnaby, S.N.; Lee, A.; Mirkin, C.A. Probing the inherent stability of siRNA immobilized on nanoparticle constructs. Proc. Natl. Acad. Sci. USA 2014, 111, 9739–9744. [Google Scholar] [CrossRef] [PubMed]
- Kranz, L.M.; Diken, M.; Haas, H.; Kreiter, S.; Loquai, C.; Reuter, K.C.; Meng, M.; Fritz, D.; Vascotto, F.; Hefesha, H.; et al. Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature 2016, 534, 396. [Google Scholar] [CrossRef] [PubMed]
- Koyfman, A.Y.; Braun, G.; Magonov, S.; Chworos, A.; Reich, N.O.; Jaeger, L. Controlled spacing of cationic gold nanoparticles by nanocrown RNA. J. Am. Chem. Soc. 2005, 127, 11886–11887. [Google Scholar] [CrossRef]
- Elbakry, A.; Zaky, A.; Liebl, R.; Rachel, R.; Goepferich, A.; Breunig, M. Layer-by-layer assembled gold nanoparticles for siRNA delivery. Nano Lett. 2009, 9, 2059–2064. [Google Scholar] [CrossRef]
- Kim, S.T.; Chompoosor, A.; Yeh, Y.C.; Agasti, S.S.; Solfiell, D.J.; Rotello, V.M. Dendronized gold nanoparticles for siRNA delivery. Small 2012, 8, 3253–3256. [Google Scholar] [CrossRef]
- Shaat, H.; Mostafa, A.; Moustafa, M.; Gamal-Eldeen, A.; Emam, A.; El-Hussieny, E.; Elhefnawi, M. Modified gold nanoparticles for intracellular delivery of anti-liver cancer siRNA. Int. J. Pharm. 2016, 504, 125–133. [Google Scholar] [CrossRef]
- Conde, J.; Rosa, J.; de la Fuente, J.M.; Baptist, P.V. Gold-nanobeacons for simultaneous gene specific silencing and intracellular tracking of the silencing events. Biomaterials 2013, 34, 2516–2523. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.F.; O’Driscoll, C.M.; Holmes, J.D.; Rahme, K. Bioconjugated gold nanoparticles enhance cellular uptake: A proof of concept study for siRNA delivery in prostate cancer cells. Int. J. Pharm. 2016, 509, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Hou, W.X.; Wei, P.; Kong, L.D.; Guo, R.; Wang, S.G.; Shi, X.Y. Partially PEGylated dendrimer-entrapped gold nanoparticles: A promising nanoplatform for highly efficient DNA and siRNA delivery. J. Mater. Chem. B 2016, 4, 2933–2943. [Google Scholar] [CrossRef]
- Majer, O.; Liu, B.; Barton, G.M. Nucleic acid-sensing TLRs: Trafficking and regulation. Curr. Opin. Immunol. 2017, 44, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Radovic-Moreno, A.F.; Chernyak, N.; Mader, C.C.; Nallagatla, S.; Kang, R.S.; Hao, L.L.; Walker, D.A.; Halo, T.L.; Merkel, T.J.; Rische, C.H.; et al. Immunomodulatory spherical nucleic acids. Proc. Natl. Acad. Sci. USA 2015, 112, 3892–3897. [Google Scholar] [CrossRef] [PubMed]
- Barnaby, S.N.; Perelman, G.A.; Kohlstedt, K.L.; Chinen, A.B.; Schatz, G.C.; Mirkin, C.A. Design Considerations for RNA Spherical Nucleic Acids (SNAs). Bioconjug. Chem. 2016, 27, 2124–2131. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, N.; Dasaradhi, P.V.N.; Mohmmed, A.; Malhotra, P.; Bhatnagar, R.K.; Mukherjee, S.K. RNA interference: Biology, mechanism, and applications. Microbiol. Mol. Biol. R 2003, 67, 657–685. [Google Scholar] [CrossRef]
- Hao, L.; Patel, P.C.; Alhasan, A.H.; Giljohann, D.A.; Mirkin, C.A. Nucleic acid-gold nanoparticle conjugates as mimics of microRNA. Small 2011, 7, 3158–3162. [Google Scholar] [CrossRef]
- Lee, J.S.; Green, J.J.; Love, K.T.; Sunshine, J.; Langer, R.; Anderson, D.G. Gold, poly(beta-amino ester) nanoparticles for small interfering RNA delivery. Nano Lett. 2009, 9, 2402–2406. [Google Scholar] [CrossRef]
- Allhenn, D.; Boushehri, M.A.; Lamprecht, A. Drug delivery strategies for the treatment of malignant gliomas. Int. J. Pharm. 2012, 436, 299–310. [Google Scholar] [CrossRef]
- Zhuo, Z.; Yu, Y.; Wang, M.; Li, J.; Zhang, Z.; Liu, J.; Wu, X.; Lu, A.; Zhang, G.; Zhang, B. Recent Advances in SELEX Technology and Aptamer Applications in Biomedicine. Int. J. Mol. Sci. 2017, 18, 2142. [Google Scholar] [CrossRef] [PubMed]
- Lupold, S.E.; Hicke, B.J.; Lin, Y.; Coffey, D.S. Identification and characterization of nuclease-stabilized RNA molecules that bind human prostate cancer cells via the prostate-specific membrane antigen. Cancer Res. 2002, 62, 4029–4033. [Google Scholar] [PubMed]
- Javier, D.J.; Nitin, N.; Levy, M.; Ellington, A.; Richards-Kortum, R. Aptamer-targeted gold nanoparticles as molecular-specific contrast agents for reflectance imaging. Bioconjug. Chem. 2008, 19, 1309–1312. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Jeong, Y.Y.; Jon, S. A Drug-Loaded Aptamer-Gold Nanoparticle Bioconjugate for Combined CT Imaging and Therapy of Prostate Cancer. ACS Nano 2010, 4, 3689–3696. [Google Scholar] [CrossRef] [PubMed]
- Ferapontova, E.E.; Olsen, E.M.; Gothelf, K.V. An RNA aptamer-based electrochemical biosensor for detection of theophylline in serum. J. Am. Chem. Soc. 2008, 130, 4256–4258. [Google Scholar] [CrossRef] [PubMed]
- Garst, A.D.; Edwards, A.L.; Batey, R.T. Riboswitches: Structures and Mechanisms. CSH Perspect. Biol. 2011, 3. [Google Scholar] [CrossRef] [PubMed]
- Seetharaman, S.; Zivarts, M.; Sudarsan, N.; Breaker, R.R. Immobilized RNA switches for the analysis of complex chemical and biological mixtures. Nat. Biotechnol. 2001, 19, 336–341. [Google Scholar] [CrossRef]
- Zhang, K.; Yang, X.J.; Zhao, W.; Xu, M.C.; Xu, J.J.; Chen, H.Y. Regulation and imaging of gene expression via an RNA interference antagonistic biomimetic probe. Chem. Sci. 2017, 8, 4973–4977. [Google Scholar] [CrossRef]
- Jiang, H.Y.; Ling, K.; Tao, X.J.; Zhang, Q.Q. Theophylline detection in serum using a self-assembling RNA aptamer-based gold nanoparticle sensor. Biosens. Bioelectron. 2015, 70, 299–303. [Google Scholar] [CrossRef]
- Yao, G.B.; Pei, H.; Li, J.; Zhao, Y.; Zhu, D.; Zhang, Y.N.; Lin, Y.F.; Huang, Q.; Fan, C.H. Clicking DNA to gold nanoparticles: Poly-adenine-mediated formation of monovalent DNA-gold nanoparticle conjugates with nearly quantitative yield. NPG Asia Mater. 2015, 7. [Google Scholar] [CrossRef]
- Ling, K.; Jiang, H.; Zhang, L.; Li, Y.; Yang, L.; Qiu, C.; Li, F.R. A self-assembling RNA aptamer-based nanoparticle sensor for fluorometric detection of Neomycin B in milk. Anal. Bioanal. Chem. 2016, 408, 3593–3600. [Google Scholar] [CrossRef] [PubMed]
- Moll, W.D.; Guo, P. Grouping of ferritin and gold nanoparticles conjugated to pRNA of the phage phi29 DNA-Packaging motor. J. Nanosci. Nanotechnol. 2007, 7, 3257–3267. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.D.; Qiu, J.R.; Shi, X.Y. Multifunctional PEI-entrapped gold nanoparticles enable efficient delivery of therapeutic siRNA into glioblastoma cells. J. Control. Release 2017, 259, E83–E84. [Google Scholar] [CrossRef]
- Gugliotti, L.A.; Feldheim, D.L.; Eaton, B.E. RNA-mediated control of metal nanoparticle shape. J. Am. Chem. Soc. 2005, 127, 17814–17818. [Google Scholar] [CrossRef] [PubMed]
- Jaeger, L.; Westhof, E.; Leontis, N.B. TectoRNA: Modular assembly units for the construction of RNA nano-objects. Nucleic Acids Res. 2001, 29, 455–463. [Google Scholar] [CrossRef] [PubMed]
- Grabow, W.W.; Zakrevsky, P.; Afonin, K.A.; Chworos, A.; Shapiro, B.A.; Jaeger, L. Self-assembling RNA nanorings based on RNAI/II inverse kissing complexes. Nano Lett. 2011, 11, 878–887. [Google Scholar] [CrossRef] [PubMed]
- Jedrzejczyk, D.; Gendaszewska-Darmach, E.; Pawlowska, R.; Chworos, A. Designing synthetic RNA for delivery by nanoparticles. J. Phys.Condens. Matter 2017, 29. [Google Scholar] [CrossRef]
- Chithrani, B.D.; Ghazani, A.A.; Chan, W.C.W. Determining the size and shape dependence of gold nanoparticle uptake into mammalian cells. Nano Lett. 2006, 6, 662–668. [Google Scholar] [CrossRef]
- Chithrani, B.D.; Chan, W.C.W. Elucidating the mechanism of cellular uptake and removal of protein-coated gold nanoparticles of different sizes and shapes. Nano Lett. 2007, 7, 1542–1550. [Google Scholar] [CrossRef]
- Patel, P.C.; Giljohann, D.A.; Daniel, W.L.; Zheng, D.; Prigodich, A.E.; Mirkin, C.A. Scavenger Receptors Mediate Cellular Uptake of Polyvalent Oligonucleotide-Functionalized Gold Nanoparticles. Bioconjug. Chem. 2010, 21, 2250–2256. [Google Scholar] [CrossRef]
- Wang, Z.D.; Zhang, J.Q.; Ekman, J.M.; Kenis, P.J.A.; Lu, Y. DNA-Mediated Control of Metal Nanoparticle Shape: One-Pot Synthesis and Cellular Uptake of Highly Stable and Functional Gold Nanoflowers. Nano Lett. 2010, 10, 1886–1891. [Google Scholar] [CrossRef] [PubMed]
- Choi, C.H.J.; Hao, L.L.; Narayan, S.P.; Auyeung, E.; Mirkin, C.A. Mechanism for the endocytosis of spherical nucleic acid nanoparticle conjugates. Proc. Natl. Acad. Sci. USA 2013, 110, 7625–7630. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Zhang, X.B.; Lv, Y.; Gong, L.; Wang, R.; Zhu, X.; Yang, R.; Tan, W. Functional DNA-containing nanomaterials: Cellular applications in biosensing, imaging, and targeted therapy. Acc. Chem. Res. 2014, 47, 1891–1901. [Google Scholar] [CrossRef] [PubMed]
- Kodiha, M.; Wang, Y.M.; Hutter, E.; Maysinger, D.; Stochaj, U. Off to the Organelles—Killing Cancer Cells with Targeted Gold Nanoparticles. Theranostics 2015, 5, 357–370. [Google Scholar] [CrossRef] [PubMed]
- Nemati, H.; Ghahramani, M.H.; Faridi-Majidi, R.; Izadi, B.; Bahrami, G.; Madani, S.H.; Tavoosidana, G. Using siRNA-based spherical nucleic acid nanoparticle conjugates for gene regulation in psoriasis. J. Control. Release 2017, 268, 259–268. [Google Scholar] [CrossRef] [PubMed]
- Tkachenko, A.G.; Xie, H.; Coleman, D.; Glomm, W.; Ryan, J.; Anderson, M.F.; Franzen, S.; Feldheim, D.L. Multifunctional gold nanoparticle-peptide complexes for nuclear targeting. J. Am. Chem. Soc. 2003, 125, 4700–4701. [Google Scholar] [CrossRef] [PubMed]
- Giljohann, D.A.; Seferos, D.S.; Patel, P.C.; Millstone, J.E.; Rosi, N.L.; Mirkin, C.A. Oligonucleotide loading determines cellular uptake of DNA-modified gold nanoparticles. Nano Lett. 2007, 7, 3818–3821. [Google Scholar] [CrossRef]
- Dai, Q.; Walkey, C.; Chan, W.C.W. Polyethylene Glycol Backfilling Mitigates the Negative Impact of the Protein Corona on Nanoparticle Cell Targeting. Angew. Chem. Int. Ed. 2014, 53, 5093–5096. [Google Scholar] [CrossRef]
- Panzarini, E.; Mariano, S.; Carata, E.; Mura, F.; Rossi, M.; Dini, L. Intracellular Transport of Silver and Gold Nanoparticles and Biological Responses: An Update. Int. J. Mol. Sci. 2018, 19, 1305. [Google Scholar] [CrossRef]
- Ohta, S.; Glancy, D.; Chan, W.C.W. DNA-controlled dynamic colloidal nanoparticle systems for mediating cellular interaction. Science 2016, 351, 841–845. [Google Scholar] [CrossRef]
- Mokhtarzadeh, A.; Vahidnezhad, H.; Youssefian, L.; Mosafer, J.; Baradaran, B.; Uitto, J. Applications of Spherical Nucleic Acid Nanoparticles as Delivery Systems. Trends Mol. Med. 2019. [Google Scholar] [CrossRef] [PubMed]
- Li, H.Y.; Chen, Z.; Ho, L.W.; Chan, P.S.; Li, Q.; Leung, S.C.; Zhang, B.; Lai, K.L.; Kwon, G.S.; Choi, C.H.J.; et al. Oligonucleotide-conjugated nanoparticles for targeted drug delivery via scavenger receptors class A: An in vitro assessment for proof-of-concept. Int. J. Pharm. 2017, 532, 647–655. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Chen, B.B.; He, M.; Li, X.T.; Chen, P.Y.; Hu, B. Study on uptake of gold nanoparticles by single cells using droplet microfluidic chip-inductively coupled plasma mass spectrometry. Talanta 2019, 200, 398–407. [Google Scholar] [CrossRef] [PubMed]
- Chithrani, D.B. Intracellular uptake, transport, and processing of gold nanostructures. Mol. Membr. Biol. 2010, 27, 299–311. [Google Scholar] [CrossRef] [PubMed]
- Chithrani, D.B.; Dunne, M.; Stewart, J.; Allen, C.; Jaffray, D.A. Cellular uptake and transport of gold nanoparticles incorporated in a liposomal carrier. Nanomed. Nanotechnol. 2010, 6, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Song, W.C.; Kim, K.R.; Park, M.; Lee, K.E.; Ahn, D.R. Backbone-modified oligonucleotides for tuning the cellular uptake behaviour of spherical nucleic acids. Biomater. Sci. 2017, 5, 412–416. [Google Scholar] [CrossRef]
- Dowdy, S.F. Overcoming cellular barriers for RNA therapeutics. Nat. Biotechnol 2017, 35, 222–229. [Google Scholar] [CrossRef]
- Bourquin, J.; Septiadi, D.; Vanhecke, D.; Balog, S.; Steinmetz, L.; Spuch-Calvar, M.; Taladriz-Blanco, P.; Petri-Fink, A.; Rothen-Rutishauser, B. Reduction of Nanoparticle Load in Cells by Mitosis but Not Exocytosis. ACS Nano 2019, 13, 7759–7770. [Google Scholar] [CrossRef]
- Levy, R.; Shaheen, U.; Cesbron, Y.; See, V. Gold nanoparticles delivery in mammalian live cells: A critical review. Nano Rev. 2010, 1. [Google Scholar] [CrossRef]
- Aoyama, Y.; Kanamori, T.; Nakai, T.; Sasaki, T.; Horiuchi, S.; Sando, S.; Niidome, T. Artificial viruses and their application to gene delivery. size-controlled gene coating with glycocluster nanoparticles. J. Am. Chem. Soc. 2003, 125, 3455–3457. [Google Scholar] [CrossRef]
- Jiang, W.; Kim, B.Y.S.; Rutka, J.T.; Chan, W.C.W. Nanoparticle-mediated cellular response is size-dependent. Nat. Nanotechnol. 2008, 3, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Osaki, F.; Kanamori, T.; Sando, S.; Sera, T.; Aoyama, Y. A quantum dot conjugated sugar ball and its cellular uptake. On the size effects of endocytosis in the subviral region. J. Am. Chem. Soc. 2004, 126, 6520–6521. [Google Scholar] [CrossRef] [PubMed]
- Cho, W.S.; Cho, M.; Jeong, J.; Choi, M.; Han, B.S.; Shin, H.S.; Hong, J.; Chung, B.H.; Jeong, J.; Cho, M.H. Size-dependent tissue kinetics of PEG-coated gold nanoparticles. Toxicol. Appl. Pharm. 2010, 245, 116–123. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.C.; Hajfathalian, M.; Maidment, P.S.N.; Hsu, J.C.; Naha, P.C.; Si-Mohamed, S.; Breuilly, M.; Kim, J.; Chhour, P.; Douek, P.; et al. Effect of Gold Nanoparticle Size on Their Properties as Contrast Agents for Computed Tomography. Sci. Rep. 2019, 9. [Google Scholar] [CrossRef] [PubMed]
- Yue, Z.G.; Wei, W.; Lv, P.P.; Yue, H.; Wang, L.Y.; Su, Z.G.; Ma, G.H. Surface Charge Affects Cellular Uptake and Intracellular Trafficking of Chitosan-Based Nanoparticles. Biomacromolecules 2011, 12, 2440–2446. [Google Scholar] [CrossRef]
- de la Fuente, J.M.; Berry, C.C. Tat peptide as an efficient molecule to translocate gold nanoparticles into the cell nucleus. Bioconjug. Chem 2005, 16, 1176–1180. [Google Scholar] [CrossRef]
- Gu, Y.J.; Cheng, J.P.; Lin, C.C.; Lam, Y.W.; Cheng, S.H.; Wong, W.T. Nuclear penetration of surface functionalized gold nanoparticles. Toxicol. Appl. Pharm. 2009, 237, 196–204. [Google Scholar] [CrossRef]
- Cesbron, Y.; Shaheen, U.; Free, P.; Levy, R. TAT and HA2 facilitate cellular uptake of gold nanoparticles but do not lead to cytosolic localisation. PLoS ONE 2015, 10, e0121683. [Google Scholar] [CrossRef]
- Feldherr, C.M.; Lanford, R.E.; Akin, D. Signal-mediated nuclear transport in simian virus 40-transformed cells is regulated by large tumor antigen. Proc. Natl. Acad. Sci. USA 1992, 89, 11002–11005. [Google Scholar] [CrossRef]
- Pujals, S.; Bastus, N.G.; Pereiro, E.; Lopez-Iglesias, C.; Punte, V.F.; Kogan, M.J.; Giralt, E. Shuttling Gold Nanoparticles into Tumoral Cells with an Amphipathic Proline-Rich Peptide. Chembiochem 2009, 10, 1025–1031. [Google Scholar] [CrossRef]
- Cho, W.S.; Cho, M.J.; Jeong, J.; Choi, M.; Cho, H.Y.; Han, B.S.; Kim, S.H.; Kim, H.O.; Lim, Y.T.; Chung, B.H.; et al. Acute toxicity and pharmacokinetics of 13 nm-sized PEG-coated gold nanoparticles. Toxicol. Appl. Pharm. 2009, 236, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Nativo, P.; Prior, I.A.; Brust, M. Uptake and intracellular fate of surface-modified gold nanoparticles. ACS Nano 2008, 2, 1639–1644. [Google Scholar] [CrossRef] [PubMed]
- Fraga, S.; Faria, H.; Soares, M.E.; Duarte, J.A.; Soares, L.; Pereira, E.; Costa-Pereira, C.; Teixeira, J.P.; de Lourdes Bastos, M.; Carmo, H. Influence of the surface coating on the cytotoxicity, genotoxicity and uptake of gold nanoparticles in human HepG2 cells. J. Appl. Toxicol. 2013, 33, 1111–1119. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Harrison, N.; Richards-Kortum, R.; Sokolov, K. Plasmonic nanosensors for imaging intracellular biomarkers in live cells. Nano Lett. 2007, 7, 1338–1343. [Google Scholar] [CrossRef]
- Cruz, E.; Kayser, V. Synthesis and Enhanced Cellular Uptake In Vitro of Anti-HER2 Multifunctional Gold Nanoparticles. Cancers 2019, 11, 870. [Google Scholar] [CrossRef]
- Malugin, A.; Ghandehari, H. Cellular uptake and toxicity of gold nanoparticles in prostate cancer cells: A comparative study of rods and spheres. J. Appl. Toxicol. 2010, 30, 212–217. [Google Scholar] [CrossRef]
- Yang, P.H.; Sun, X.S.; Chiu, J.F.; Sun, H.Z.; He, Q.Y. Transferrin-mediated gold nanoparticle cellular uptake. Bioconjug. Chem. 2005, 16, 494–496. [Google Scholar] [CrossRef]
- Soman, N.; Marsh, J.; Lanza, G.; Wickline, S. New mechanisms for non-porative ultrasound stimulation of cargo delivery to cell cytosol with targeted perfluorocarbon nanoparticles. Nanotechnology 2008, 19. [Google Scholar] [CrossRef][Green Version]
| AuNP Size (nm) | Surface Group | Cell Line | Toxicity | Subcellular Localization | Reference |
|---|---|---|---|---|---|
| 1, 4 | Phospholipid | HeLa | Not reported | Lysosomal, perinuclear/nuclear | [198] |
| 2, 8 | Tat peptide | HTERT-BJ1 | Low cytotoxicity below 10 µM | Cytoplasmic around the mitochondria and nuclear | [209] |
| 3, 7 | PEG | HeLa | Non toxic | Nuclear | [210] |
| 5, 10, 15 | CALNN, TAT and/or HA2 viral peptides | HeLa | Not reported | Cytoplasmic vesicles, lysosomal, endosomal, membranes | [211] |
| 10 | Oligonucleotides | HaCaT, A549, BALB/c 3T3, C166 | Not reported | Cytoplasmic, endosomal | [185] |
| 11–32 | Nucleoplasmin | BALB/c 3T3 A31, MOP-8, SV-T2 | Not reported | Nuclear and cytoplasmic | [212] |
| 12 | Sweet arrow peptide (SAP) | HeLa | Not reported | Endosomal | [213] |
| 13 | PEG | In vivo studies | Induction of acute inflammation and apoptosis | Cytoplasmic vesicles and lysosomal | [214] |
| 16 | PEG, CALNN, NLS, CPPs | HeLa | Not reported | Cytoplasmic, nuclear, lysosomal | [215] |
| 20 | citrate (Cit) compared with 11-mercaptoundecanoic acid (11-MUA) | HepG2 | Non-toxic, DNA damage in Cit-AuNPS | Cytoplasmic, endosomal | [216] |
| 20 | BSA with NLS, receptor-mediated endocytosis peptides (RME) | HepG2 | 5% death | Nuclear | [189] |
| 20 | Biotinylated Tat-HA2, PEG-SH, anti-actin antibodies | BALB/c 3T3 | Not reported | Cytoskeleton (cytoplasmic) | [217] |
| 20–50 | Citrate, PEG, CPP, Trastuzumab | DLD-1, SKOV-3, MDA-MB-231, SKBR-3, MCF-7 | Cytotoxicity dependent on the surface group. | Intracellular | [218] |
| 30–90 | PEG | PC-3 | Non toxic | Cytoplasmic and nuclear | [219] |
| 14–100 | Transferrin | STO, HeLa, SNB19, NPC | Non toxic | Endosomal | [181,182,220] |
| 100 | DPPE | C-32 | Non toxic | Endosomal | [221] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Graczyk, A.; Pawlowska, R.; Jedrzejczyk, D.; Chworos, A. Gold Nanoparticles in Conjunction with Nucleic Acids as a Modern Molecular System for Cellular Delivery. Molecules 2020, 25, 204. https://doi.org/10.3390/molecules25010204
Graczyk A, Pawlowska R, Jedrzejczyk D, Chworos A. Gold Nanoparticles in Conjunction with Nucleic Acids as a Modern Molecular System for Cellular Delivery. Molecules. 2020; 25(1):204. https://doi.org/10.3390/molecules25010204
Chicago/Turabian StyleGraczyk, Anna, Roza Pawlowska, Dominika Jedrzejczyk, and Arkadiusz Chworos. 2020. "Gold Nanoparticles in Conjunction with Nucleic Acids as a Modern Molecular System for Cellular Delivery" Molecules 25, no. 1: 204. https://doi.org/10.3390/molecules25010204
APA StyleGraczyk, A., Pawlowska, R., Jedrzejczyk, D., & Chworos, A. (2020). Gold Nanoparticles in Conjunction with Nucleic Acids as a Modern Molecular System for Cellular Delivery. Molecules, 25(1), 204. https://doi.org/10.3390/molecules25010204







