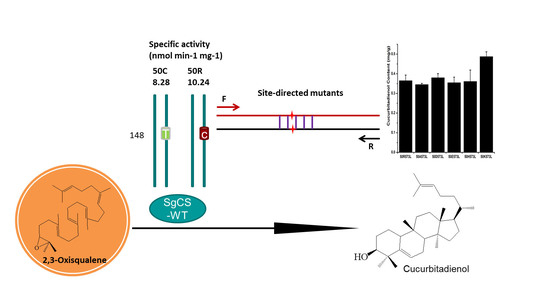
Identification of a Novel Specific Cucurbitadienol Synthase Allele in Siraitia grosvenorii Correlates with High Catalytic Efficiency
Abstract
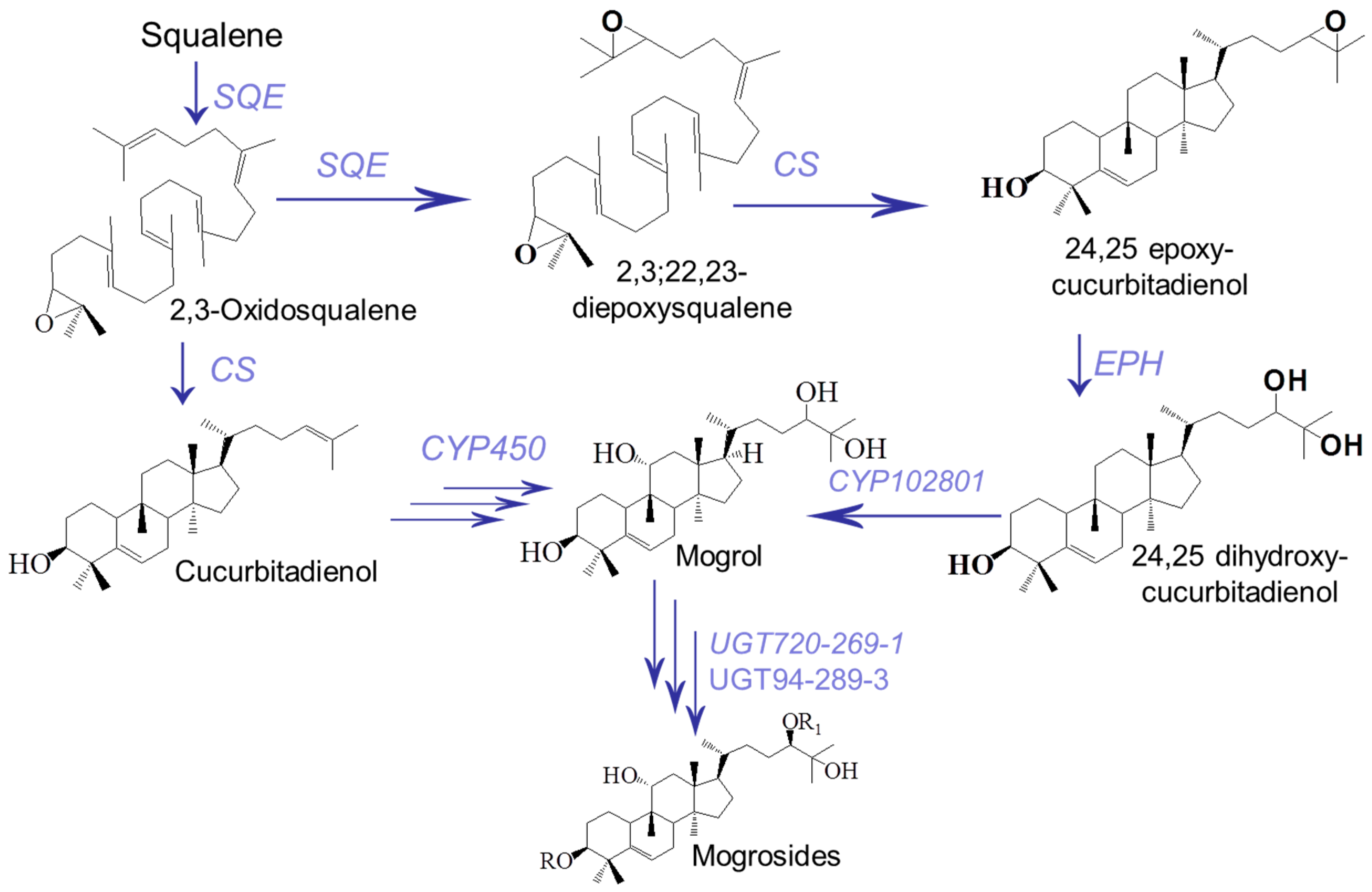
1. Introduction
2. Results
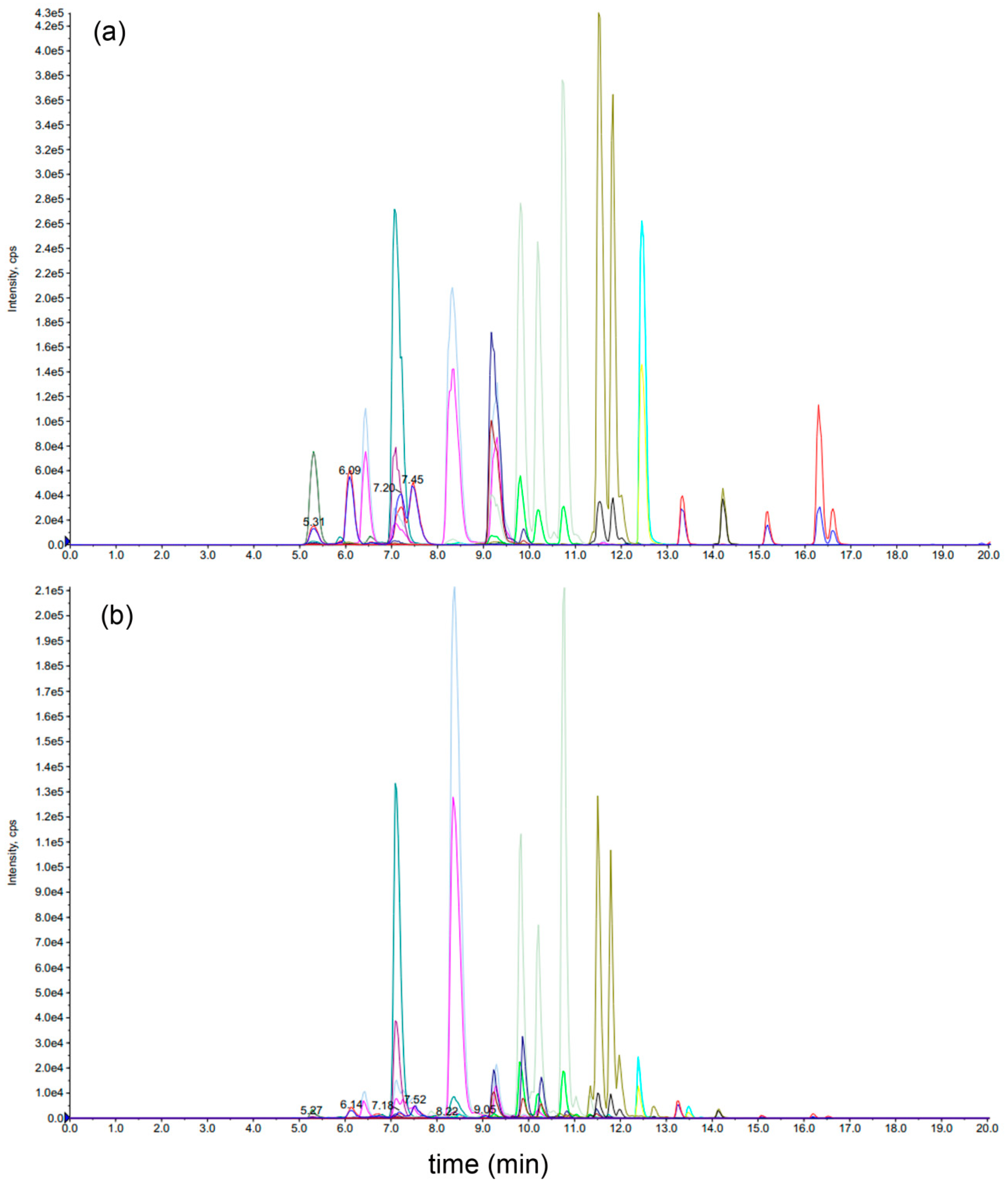
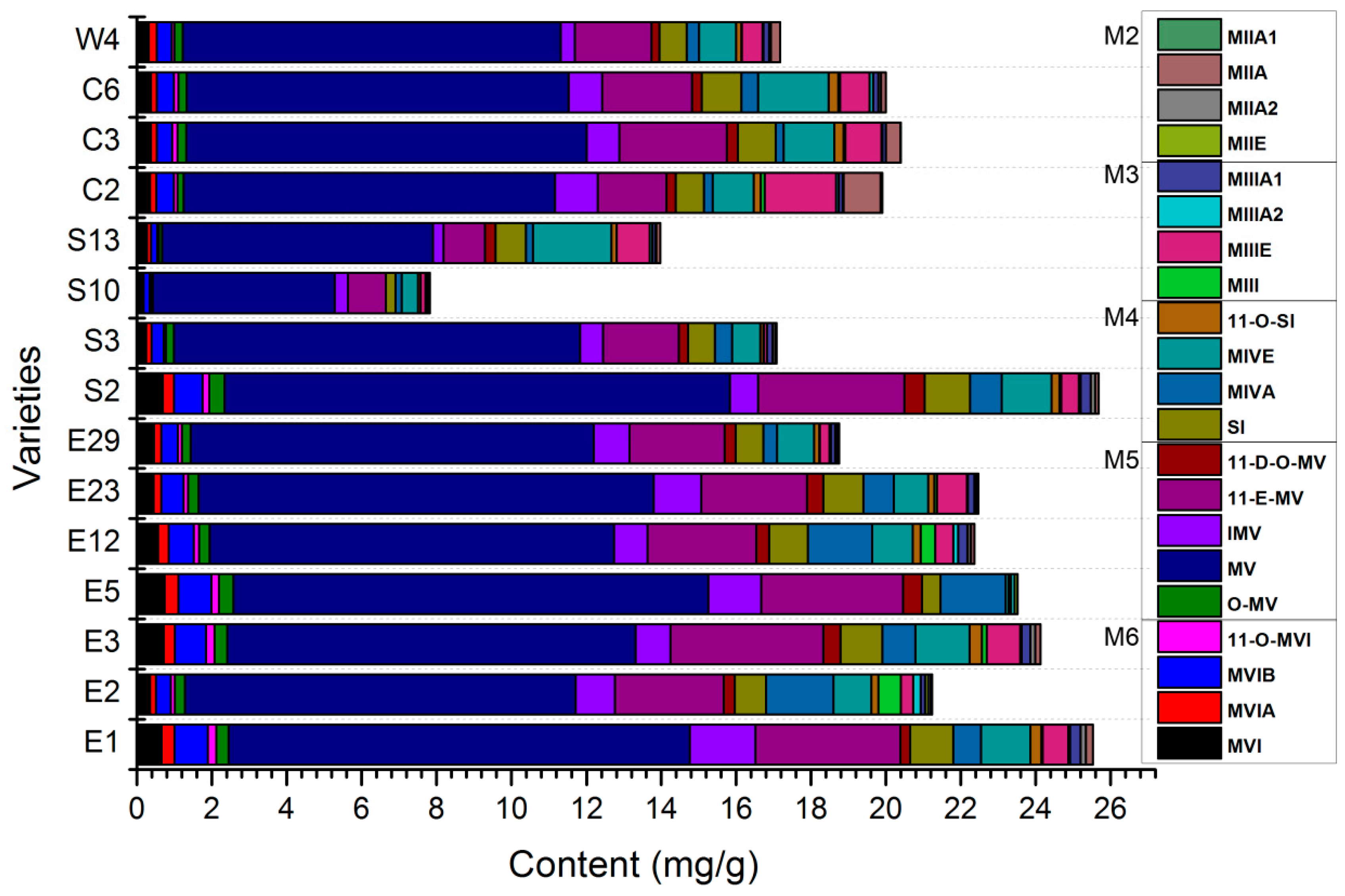
2.1. Determination of the Levels of 21 Mogrosides in 15 S. grosvenorii Varieties
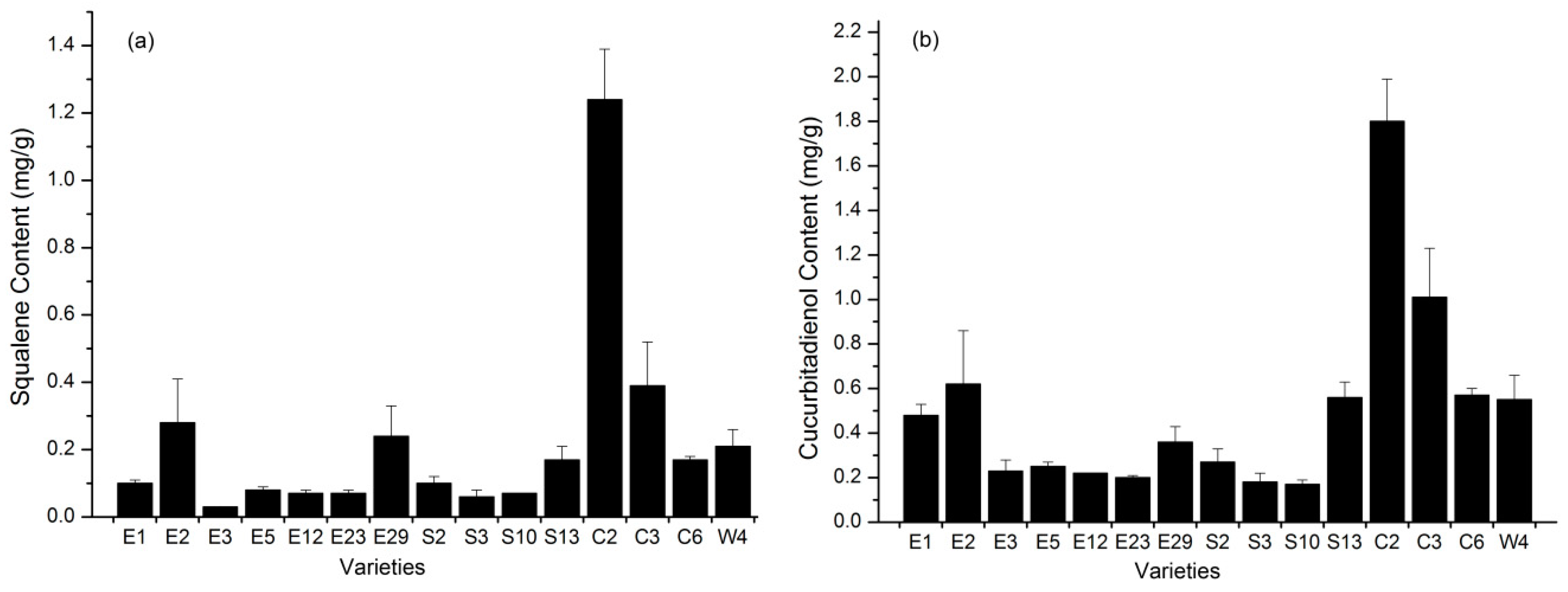
2.2. The Determination of Squalene and Cucurbitadienol Levels in the Fruit of S. grosvenorii
2.3. SNP Identification in ORF Region of the SgCS Gene
2.4. Activity Comparison
2.5. Improving the Activity of SgCS by Site-Directed Mutagenesis
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Sample Collection and Preparation
4.3. LC–MS/MS Analysis of 21 Mogrosides
4.4. SNP Analysis
4.5. Cloning 4 Different Copies of SgCS Genes in Yeast
4.6. SgCS Mutagenesis Experiments
4.7. Yeast Transformation and Cell Cultivation
4.8. In Vitro Activity
4.9. GC-MS Analysis of Yeast and Plant Extracts
4.10. Data Processing and Statistics
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| 11-D-O-MV | 11-deoxymogroside V |
| 11-E-MV | 11-epi-mogroside V |
| O-MV | 11-oxo-mogroside V |
| 11-O-SI | 11-O-siamenoside I |
| 11-O-MVI | 11-oxo-mogroside VI |
| CS | cucurbitadienol synthase |
| CYP450 | cytochrome P450s |
| EPH | epoxide hydrolases |
| IMV | isomogroside V |
| MIIA | mogroside IIA |
| MIIA1 | mogroside IIA1 |
| MIIA2 | mogroside IIA2 |
| MIIE | mogroside IIE |
| MIII | mogroside III |
| MIIIA1 | mogroside IIIA1 |
| MIIIA2 | mogroside IIIA2 |
| MIIIE | mogroside IIIE |
| MIVE | mogroside IVE |
| MIVA | mogroside IVA |
| MV | mogroside V |
| MVI | mogroside VI |
| MVIA | mogroside VI A |
| MVIB | mogroside VIB |
| SI | siamenoside I |
| SNP | single nucleotide polymorphism |
| SQE | squalene epoxidases |
| UGT | UDP-glucosyltransferase |
Appendix A


| Commom Name | CAS No. | -R1 | -R2 | -R3 |
|---|---|---|---|---|
| mogroside IIA | 1613527-65-3 | -H |  | 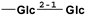 |
| mogroside IIA1 | 88901-44-4 | -H | 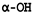 |  |
| mogroside IIA2 | 88901-45-5 |  | 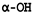 | -H |
| mogroside IIe | 88901-38-6 |  a a | 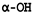 |  |
| mogroside III | 130567-83-8 |  | 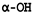 |  |
| mogroside IIIe | 88901-37-5 |  | 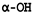 | 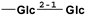 |
| mogroside IIIA1 | 88901-42-2 | -H | 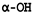 | 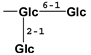 |
| mogroside IIIA2 | 88901-43-3 |  | 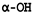 |  |
| 11-O-siamenoside I | 1793003-50-5 |  |  |  |
| mogroside IVa | 88901-41-1 |  |  |  |
| siamenoside I | 126105-12-2 |  | 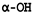 |  |
| mogroside IVE | 89590-95-4 |  | 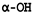 | 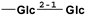 |
| 11-deoxymogroside V | 1707161-17-8 |  | -H |  |
| 11-epi-mogroside V | 2146088-12-0 |  |  |  |
| mogroside V | 88901-36-4 |  | 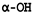 |  |
| 11-oxo-mogroside V | 126105-11-1 |  |  |  |
| isomogroside V | 1126032-65-2 | 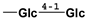 | 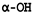 |  |
| 11-oxo-mogroside VI | 1421942-59-7 |  |  |  |
| mogroside VI | 89590-98-7 |  | 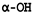 |  |
| mogroside VI A | 2146088-13-1 | 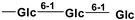 | 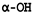 |  |
| mogroside VI B | 2149606-17-5 |  | 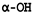 |  |
| MVI | MVIA | MVIB | 11-O-MVI | 11-O-MV | MV | IMV | 11-E-MV | 11-D-O-MV | SI | MIVA | MIVE | 11-O-SI | MIII | MIIIE | MIIIA2 | MIIIA1 | MIIE | MIIA2 | MIIA | MIIA1 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E1 | 0.66 ± 0.19 | 0.34 ± 0.10 | 0.89 ± 0.26 | 0.23 ± 0.07 | 0.33 ± 0.07 | 12.31 ± 2.83 | 1.75 ± 0.42 | 3.87 ± 1.01 | 0.26 ± 0.08 | 1.16 ± 0.26 | 0.74 ± 0.16 | 1.31 ± 0.30 | 0.30 ± 0.07 | 0.04 ± 0.01 | 0.68 ± 0.12 | 0.04 ± 0.01 | 0.29 ± 0.07 | ND | 0.14 ± 0.04 | 0.19 ± 0.05 | ND |
| E2 | 0.35 ± 0.02 | 0.15 ± 0.02 | 0.40 ± 0.02 | 0.11 ± 0.01 | 0.27 ± 0.00 | 10.43 ± 0.35 | 1.05 ± 0.07 | 2.91 ± 0.09 | 0.29 ± 0.02 | 0.83 ± 0.05 | 1.80 ± 0.05 | 1.02 ± 0.03 | 0.18 ± 0.01 | 0.61 ± 0.03 | 0.33 ± 0.03 | 0.19 ± 0.02 | 0.12 ± 0.00 | 0.09 ± 0.01 | 0.06 ± 0.01 | 0.04 ± 0.00 | ND |
| E3 | 0.71 ± 0.08 | 0.30 ± 0.02 | 0.84 ± 0.03 | 0.22 ± 0.01 | 0.34 ± 0.01 | 10.90 ± 0.48 | 0.93 ± 0.04 | 4.09 ± 0.13 | 0.45 ± 0.03 | 1.12 ± 0.05 | 0.88 ± 0.04 | 1.45 ± 0.10 | 0.33 ± 0.02 | 0.13 ± 0.01 | 0.89 ± 0.05 | 0.04 ± 0.01 | 0.23 ± 0.01 | 0.01 ± 0.00 | 0.12 ± 0.02 | 0.14 ± 0.01 | ND |
| E5 | 0.74 ± 0.06 | 0.36 ± 0.03 | 0.89 ± 0.07 | 0.19 ± 0.01 | 0.39 ± 0.03 | 12.68 ± 0.35 | 1.41 ± 0.04 | 3.80 ± 0.28 | 0.50 ± 0.05 | 0.50 ± 0.04 | 1.74 ± 0.12 | 0.08 ± 0.01 | 0.04 ± 0.00 | ND | 0.02 ± 0.00 | 0.09 ± 0.01 | ND | 0.08 ± 0.01 | ND | ND | ND |
| E12 | 0.56 ± 0.04 | 0.29 ± 0.01 | 0.67 ± 0.04 | 0.14 ± 0.00 | 0.28 ± 0.03 | 10.80 ± 0.43 | 0.89 ± 0.05 | 2.91 ± 0.20 | 0.34 ± 0.03 | 1.03 ± 0.09 | 1.72 ± 0.14 | 1.08 ± 0.06 | 0.21 ± 0.01 | 0.39 ± 0.03 | 0.49 ± 0.05 | 0.12 ± 0.01 | 0.25 ± 0.03 | 0.02 ± 0.00 | 0.08 ± 0.01 | 0.09 ± 0.00 | ND |
| E23 | 0.44 ± 0.02 | 0.21 ± 0.02 | 0.59 ± 0.02 | 0.12 ± 0.00 | 0.28 ± 0.01 | 12.15 ± 0.06 | 1.28 ± 0.05 | 2.82 ± 0.07 | 0.44 ± 0.01 | 1.07 ± 0.03 | 0.81 ± 0.03 | 0.91 ± 0.01 | 0.17 ± 0.01 | 0.07 ± 0.00 | 0.80 ± 0.02 | 0.03 ± 0.00 | 0.17 ± 0.01 | ND | 0.06 ± 0.01 | 0.05 ± 0.00 | ND |
| E29 | 0.46 ± 0.04 | 0.18 ± 0.00 | 0.45 ± 0.05 | 0.11 ± 0.01 | 0.23 ± 0.02 | 10.76 ± 0.36 | 0.96 ± 0.08 | 2.54 ± 0.14 | 0.29 ± 0.02 | 0.74 ± 0.05 | 0.37 ± 0.03 | 0.99 ± 0.03 | 0.14 ± 0.00 | 0.01 ± 0.00 | 0.26 ± 0.01 | 0.03 ± 0.00 | 0.12 ± 0.01 | ND | 0.06 ± 0.01 | 0.05 ± 0.01 | ND |
| S2 | 0.69 ± 0.07 | 0.30 ± 0.02 | 0.76 ± 0.05 | 0.18 ± 0.01 | 0.41 ± 0.02 | 13.49 ± 0.63 | 0.75 ± 0.01 | 3.91 ± 0.18 | 0.54 ± 0.05 | 1.21 ± 0.09 | 0.85 ± 0.06 | 1.33 ± 0.10 | 0.21 ± 0.01 | 0.05 ± 0.00 | 0.47 ± 0.04 | 0.05 ± 0.01 | 0.27 ± 0.02 | ND | 0.12 ± 0.01 | 0.09 ± 0.01 | ND |
| S3 | 0.24 ± 0.01 | 0.14 ± 0.01 | 0.34 ± 0.02 | 0.05 ± 0.00 | 0.22 ± 0.02 | 10.84 ± 0.45 | 0.61 ± 0.02 | 2.02 ± 0.13 | 0.25 ± 0.01 | 0.72 ± 0.04 | 0.46 ± 0.02 | 0.75 ± 0.04 | 0.09 ± 0.01 | 0.01 ± 0.00 | 0.08 ± 0.00 | ND | 0.15 ± 0.01 | ND | 0.07 ± 0.01 | 0.04 ± 0.00 | ND |
| S10 | 0.13 ± 0.00 | 0.04 ± 0.01 | 0.17 ± 0.01 | 0.03 ± 0.00 | 0.05 ± 0.00 | 4.86 ± 0.05 | 0.35 ± 0.02 | 1.01 ± 0.01 | 0.01 ± 0.00 | 0.26 ± 0.00 | 0.16 ± 0.00 | 0.44 ± 0.02 | 0.07 ± 0.00 | ND | 0.13 ± 0.00 | ND | 0.05 ± 0.00 | ND | 0.02 ± 0.01 | 0.04 ± 0.00 | ND |
| S13 | 0.27 ± 0.01 | 0.10 ± 0.01 | 0.17 ± 0.01 | 0.05 ± 0.00 | 0.08 ± 0.01 | 7.24 ± 0.20 | 0.28 ± 0.01 | 1.10 ± 0.02 | 0.28 ± 0.00 | 0.82 ± 0.02 | 0.18 ± 0.00 | 2.10 ± 0.01 | 0.13 ± 0.00 | 0.01 ± 0.00 | 0.88 ± 0.02 | 0.07 ± 0.00 | 0.08 ± 0.00 | ND | 0.02 ± 0.00 | 0.11 ± 0.00 | ND |
| C2 | 0.35 ± 0.03 | 0.16 ± 0.02 | 0.47 ± 0.04 | 0.10 ± 0.01 | 0.17 ± 0.02 | 9.91 ± 0.61 | 1.14 ± 0.10 | 1.83 ± 0.17 | 0.25 ± 0.03 | 0.76 ± 0.07 | 0.23 ± 0.03 | 1.10 ± 0.10 | 0.18 ± 0.01 | 0.11 ± 0.01 | 1.90 ± 0.15 | 0.08 ± 0.01 | 0.10 ± 0.01 | ND | 0.02 ± 0.00 | 0.99 ± 0.09 | 0.05 ± 0.01 |
| C3 | 0.37 ± 0.03 | 0.16 ± 0.02 | 0.41 ± 0.05 | 0.14 ± 0.01 | 0.23 ± 0.05 | 10.70 ± 0.46 | 0.87 ± 0.26 | 2.87 ± 0.60 | 0.29 ± 0.01 | 1.01 ± 0.14 | 0.22 ± 0.04 | 1.35 ± 0.11 | 0.25 ± 0.03 | 0.04 ± 0.05 | 0.97 ± 0.81 | 0.02 ± 0.00 | 0.10 ± 0.02 | ND | ND | 0.38 ± 0.48 | 0.02 ± 0.05 |
| C6 | 0.37 ± 0.02 | 0.16 ± 0.01 | 0.46 ± 0.02 | 0.11 ± 0.01 | 0.23 ± 0.01 | 10.20 ± 0.34 | 0.89 ± 0.04 | 2.40 ± 0.09 | 0.26 ± 0.02 | 1.06 ± 0.04 | 0.44 ± 0.02 | 1.88 ± 0.06 | 0.27 ± 0.01 | 0.04 ± 0.00 | 0.79 ± 0.03 | 0.10 ± 0.01 | 0.14 ± 0.01 | ND | 0.07 ± 0.01 | 0.13 ± 0.01 | ND |
| W4 | 0.31 ± 0.03 | 0.22 ± 0.02 | 0.40 ± 0.03 | 0.07 ± 0.01 | 0.22 ± 0.02 | 10.10 ± 0.56 | 0.37 ± 0.04 | 2.05 ± 0.13 | 0.21 ± 0.02 | 0.73 ± 0.04 | 0.33 ± 0.03 | 0.98 ± 0.04 | 0.14 ± 0.01 | 0.02 ± 0.00 | 0.55 ± 0.02 | 0.03 ± 0.01 | 0.15 ± 0.01 | ND | 0.04 ± 0 | 0.25 ± 0.01 | ND |
| Average Content | 0.44 | 0.21 | 0.53 | 0.12 | 0.25 | 10.49 | 0.90 | 2.67 | 0.31 | 0.87 | 0.73 | 1.12 | 0.18 | 0.10 | 0.62 | 0.06 | 0.15 | 0.01 | 0.06 | 0.17 | 0.00 |
| Coefficient of Variation (%) | 43.37 | 46.02 | 45.88 | 50.25 | 40.14 | 21.12 | 45.94 | 37.29 | 41.87 | 30.98 | 78.03 | 44.31 | 46.9 | 166.85 | 82.1 | 80.17 | 58.66 | -- | 88.59 | 157.39 | -- |
| E1 | E2 | E3 | E5 | E12 | E23 | E29 | S2 | S3 | S10 | S13 | C2 | C3 | C6 | W4 | Average Content | CV (%) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Squalene | 0.10 ± 0.01 | 0.28 ± 0.13 | 0.03 ± 0.00 | 0.08 ± 0.01 | 0.07 ± 0.01 | 0.07 ± 0.01 | 0.24 ± 0.09 | 0.10 ± 0.02 | 0.06 ± 0.02 | 0.07 ± 0.00 | 0.17 ± 0.04 | 1.24 ± 0.15 | 0.39 ± 0.13 | 0.17 ± 0.01 | 0.21 ± 0.05 | 0.22 | 135.03 |
| Cucurbitadienol | 0.48 ± 0.05 | 0.62 ± 0.24 | 0.23 ± 0.05 | 0.25 ± 0.02 | 0.22 ± 0.00 | 0.20 ± 0.01 | 0.36 ± 0.07 | 0.27 ± 0.06 | 0.18 ± 0.04 | 0.17 ± 0.02 | 0.56 ± 0.07 | 1.80 ± 0.19 | 1.01 ± 0.22 | 0.57 ± 0.03 | 0.55 ± 0.11 | 0.50 | 86.03 |
| SgCS-F | ATGTGGAGGTTAAAGGTC |
| SgCS-R | TTATTCAGTCAAAACCCG |
| pCEV- Seq-F | CGATGACCTCCCATTGATA |
| pCEV- Seq-R | CGTTCTTAATACTAACATAACT |
| 50A-F | ATTGTTGCAAGTTCATAAAGCTGCTAAAGCTTTTCATGATGATAGA |
| 50A-R | TCTATCATCATGAAAAGCTTTAGCAGCTTTATGAACTTGCAACAAT |
| 50C-F | ATTGTTGCAAGTTCATAAAGCTTGTAAAGCTTTTCATGATGATAGA |
| 50C-R | TCTATCATCATGAAAAGCTTTACAAGCTTTATGAACTTGCAACAAT |
| 50D-F | ATTGTTGCAAGTTCATAAAGCTGATAAAGCTTTTCATGATGATAGA |
| 50D-R | TCTATCATCATGAAAAGCTTTATCAGCTTTATGAACTTGCAACAAT |
| 50E-F | ATTGTTGCAAGTTCATAAAGCTGAAAAAGCTTTTCATGATGATAGA |
| 50E-R | TCTATCATCATGAAAAGCTTTTTCAGCTTTATGAACTTGCAACAAT |
| 50H-F | ATTGTTGCAAGTTCATAAAGCTCATAAAGCTTTTCATGATGATAGA |
| 50H-R | TCTATCATCATGAAAAGCTTTATGAGCTTTATGAACTTGCAACAAT |
| 50K-F | ATTGTTGCAAGTTCATAAAGCTAAAAAAGCTTTTCATGATGATAGA |
| 50K-R | TCTATCATCATGAAAAGCTTTTTTAGCTTTATGAACTTGCAACAAT |
References
- Chaturvedula, V.S.P.; Prakash, I. Cucurbitane Glycosides from Siraitia grosvenorii. J. Carbohydr. Chem. 2011, 30, 16–26. [Google Scholar] [CrossRef]
- Zhang, J.; Dai, L.; Yang, J.; Liu, C.; Men, Y.; Zeng, Y.; Cai, Y.; Zhu, Y.; Sun, Y. Oxidation of Cucurbitadienol Catalyzed by CYP87D18 in the Biosynthesis of Mogrosides from Siraitia grosvenorii. Plant Cell Physiol. 2016, 57, 1000–1007. [Google Scholar] [CrossRef]
- Li, C.; Lin, L.M.; Sui, F.; Wang, Z.M.; Huo, H.R.; Jiang, T.L. Chemistry and pharmacology of Siraitia grosvenorii: A review. Chin. J. Nat. Med. 2014, 12, 89–102. [Google Scholar] [CrossRef]
- Pawar, R.S.; Krynitsky, A.J.; Rader, J.I. Sweeteners from plants–with emphasis on Stevia rebaudiana (Bertoni) and Siraitia grosvenorii (Swingle). Anal. Bioanal. Chem. 2013, 405, 4397–4407. [Google Scholar] [CrossRef]
- Xia, Y.; Riverohuguet, M.E.; Hughes, B.H.; Marshall, W.D. Isolation of the sweet components from Siraitia grosvenorii. Food Chem. 2008, 107, 1022–1028. [Google Scholar] [CrossRef]
- Qing, Z.X.; Zhao, H.; Tang, Q.; Mo, C.M.; Huang, P.; Cheng, P.; Yang, P.; Yang, X.Y.; Liu, X.B.; Zheng, Y.J. Systematic identification of flavonols, flavonol glycosides, triterpene and siraitic acid glycosides from Siraitia grosvenorii using high-performance liquid chromatography/quadrupole-time-of-flight mass spectrometry combined with a screening strategy. J. Pharm. Biomed. Anal. 2017, 138, 240–248. [Google Scholar] [CrossRef]
- Fu, W.; Ma, X.; Tang, Q.; Mo, C. Karyotype analysis and genetic variation of a mutant in Siraitia grosvenorii. Mol. Biol. Rep. 2012, 39, 1247. [Google Scholar] [CrossRef]
- Bai, L.H.; Ma, X.J.; Mo, C.M.; Shi, L.; Feng, S.X.; Jiang, X.J. Study on quantitative assessment of Siraitia grosvenorii germplasms by general index. China J. Chin. Mater. Med. 2007, 32, 2482–2484. [Google Scholar]
- Tu, D.; Ma, X.; Zhao, H.; Mo, C.; Tang, Q.; Wang, L.; Huang, J.; Pan, L. Cloning and expression of SgCYP450-4 from Siraitia grosvenorii. Acta Pharm. Sin. B 2016, 6, 614–622. [Google Scholar] [CrossRef]
- Ma, X.J.; Mo, C.M.; Bai, L.H.; Feng, S.X. A New Siraitia grosvenorii Cultivar ‘Yongqing 1’. Acta Hortic. Sin. 2008, 35, 1855. [Google Scholar]
- Liu, W.J.; Mo, C.M.; Ma, X.J.; Zhang, H.Y.; Sun, B.X.; Jiang, X.J. Breeding of superior Siraitia grosvenorii cultivars producing glycosides-V. Guihaia 2010, 30, 881–883. [Google Scholar]
- Mo, C.M.; Ma, X.J.; Liu, W.J.; Bai, L.H.; Tang, Q.; Feng, S.X. Genetic effects of the main characteristics of Siraitia grosvenorii. Guihaia 2009, 29, 899–904. [Google Scholar]
- Pan, L.; Mo, C.; Ma, X.; Feng, S.; Tang, Q.; Bai, L.; Wei, R.; Xing, A.; Branch, L. The Construction of the Aseptic Strain of the Tissue Culture of Seedless Siraitia grosvenorii. Chin. Agric. Sci. Bull. 2013, 29, 150–155. [Google Scholar]
- Zhao, H.; Guo, J.; Tang, Q.; Guo, L.; Huang, L.; Ma, X. Cloning and expression analysis of squalene epoxidase genes from Siraitia grosvenorii. China J. Chin. Mater. Med. 2018, 43, 3255–3262. [Google Scholar]
- Dai, L.; Liu, C.; Zhu, Y.; Zhang, J.; Men, Y.; Zeng, Y.; Sun, Y. Functional Characterization of Cucurbitadienol Synthase and Triterpene Glycosyltransferase Involved in Biosynthesis of Mogrosides from Siraitia grosvenorii. Plant Cell Physiol. 2015, 56, 1172–1182. [Google Scholar] [CrossRef]
- Itkin, M.; Davidovich-Rikanati, R.; Cohen, S.; Portnoy, V.; Doron-Faigenboim, A.; Oren, E.; Freilich, S.; Tzuri, G.; Baranes, N.; Shen, S.; et al. The biosynthetic pathway of the nonsugar, high-intensity sweetener mogroside V from Siraitia grosvenorii. Proc. Natl. Acad. Sci. USA 2016, 113, E7619–E7628. [Google Scholar] [CrossRef]
- Zhao, H.; Tang, Q.; Mo, C.; Bai, L.; Tu, D.; Ma, X. Cloning and characterization of squalene synthase and cycloartenol synthase from Siraitia grosvenorii. Acta Pharm. Sin. B 2017, 7, 215–222. [Google Scholar] [CrossRef]
- Su, H.; Liu, Y.; Xiao, Y.; Tan, Y.; Gu, Y.; Liang, B.; Huang, H.; Wu, Y. Molecular and biochemical characterization of squalene synthase from Siraitia grosvenorii. Biotechnol. Lett. 2017, 39, 1–10. [Google Scholar] [CrossRef]
- Nicole, S.; Barcaccia, G.; Erickson, D.L.; Kress, J.W.; Lucchin, M. The coding region of the UFGT gene is a source of diagnostic SNP markers that allow single-locus DNA genotyping for the assessment of cultivar identity and ancestry in grapevine (Vitis vinifera L.). BMC Res. Notes 2013, 6, 502. [Google Scholar] [CrossRef]
- Primmer, C.R.; Borge, T.; Lindell, J.; Saetre, G.-P. Single-nucleotide polymorphism characterization in species with limited available sequence information: High nucleotide diversity revealed in the avian genome. Mol. Ecol. 2010, 11, 603–612. [Google Scholar] [CrossRef]
- Song, W.; Qiao, X.; Chen, K.; Wang, Y.; Ji, S.; Feng, J.; Li, K.; Lin, Y.; Ye, M. Biosynthesis-Based Quantitative Analysis of 151 Secondary Metabolites of Licorice to Differentiate Medicinal Glycyrrhiza Species and Their Hybrids. Anal. Chem. 2017, 89, 3146–3153. [Google Scholar] [CrossRef]
- Begovich, A.B.; Carlton, V.E.H.; Honigberg, L.A.; Schrodi, S.J.; Chokkalingam, A.P.; Alexander, H.C.; Ardlie, K.G.; Huang, Q.; Smith, A.M.; Spoerke, J.M. A Missense Single-Nucleotide Polymorphism in a Gene Encoding a Protein Tyrosine Phosphatase (PTPN22) Is Associated with Rheumatoid Arthritis. Am. J. Hum. Genet. 2004, 75, 330–337. [Google Scholar] [CrossRef]
- Hakim, I.R.; Kammoun, N.G.; Makhloufi, E.; Rebaï, A. Discovery and potential of SNP markers in characterization of Tunisian olive germplasm. Diversity 2009, 2, 2679–2685. [Google Scholar] [CrossRef]
- Hayashi, K.; Hashimoto, N.; Daigen, M.; Ashikawa, I. Development of PCR-based SNP markers for rice blast resistance genes at the Piz locus. Theor. Appl. Genet. 2004, 108, 1212–1220. [Google Scholar] [CrossRef]
- Choi, I.Y.; Hyten, D.L.; Matukumalli, L.K.; Song, Q.; Chaky, J.M.; Quigley, C.V.; Chase, K.; Lark, K.G.; Reiter, R.S.; Yoon, M.S. A soybean transcript map: Gene distribution, haplotype and single-nucleotide polymorphism analysis. Genet 2007, 176, 685. [Google Scholar] [CrossRef]
- Xia, M.; Han, X.; He, H.; Yu, R.; Zhen, G.; Jia, X.; Cheng, B.; Deng, X.W. Improved de novo genome assembly and analysis of the Chinese cucurbit Siraitia grosvenorii, also known as monk fruit or luo-han-guo. Gigascience 2018, 7, 1–9. [Google Scholar] [CrossRef]
- Luo, Z.; Shi, H.; Zhang, K.; Qin, X.; Guo, Y.; Ma, X. Liquid chromatography with tandem mass spectrometry method for the simultaneous determination of multiple sweet mogrosides in the fruits of Siraitia grosvenorii and its marketed sweeteners. J. Sep. Sci. 2016, 39, 4124–4135. [Google Scholar] [CrossRef]
- Zhang, H.; Yang, H.; Zhang, M.; Wang, Y.; Wang, J.; Yau, L.; Jiang, Z.; Hu, P. Identification of flavonol and triterpene glycosides in Luo-Han-Guo extract using ultra-high performance liquid chromatography/quadrupole time-of-flight mass spectrometry. J. Food Compos. Anal. 2012, 25, 142–148. [Google Scholar] [CrossRef]
- Lu, F.; Li, D.; Fu, C.; Liu, J.; Huang, Y.; Chen, Y.; Wen, Y.; Nohara, T. Studies on chemical fingerprints of Siraitia grosvenorii fruits (Luo Han Guo) by HPLC. J. Nat. Med. 2012, 66, 70–76. [Google Scholar] [CrossRef]
- Qiao, J.; Liu, J.; Liao, J.; Luo, Z.; Ma, X.; Ma, G. Identification of Key Amino Acid Residues Determining Product Specificity of 2,3-Oxidosqualene Cyclase in Siraitia grosvenorii. Catalysts 2018, 8, 577. [Google Scholar] [CrossRef]
- Abe, I.; Rohmer, M.; Prestwich, G.D. Enzymatic Cyclization of Squalene and Oxidosqualene to Sterols and Triterpenes. Chem. Rev. 1993, 93, 2189–2206. [Google Scholar] [CrossRef]
- Suzuki, M.; Xiang, T.; Ohyama, K.; Seki, H.; Saito, K.; Muranaka, T.; Hayashi, H.; Katsube, Y.; Kushiro, T.; Shibuya, M. Lanosterol synthase in dicotyledonous plants. Plant Cell Physiol. 2006, 47, 565–571. [Google Scholar] [CrossRef]
- Qiao, J.; Luo, Z.; Li, Y.; Ren, G.; Liu, C.; Ma, X. Effect of Abscisic Acid on Accumulation of Five Active Components in Root of Glycyrrhiza uralensis. Molecules 2017, 22, 1982. [Google Scholar] [CrossRef]
- Lodeiro, S.; Schulz-Gasch, T.; Matsuda, S.P. Enzyme redesign: Two mutations cooperate to convert cycloartenol synthase into an accurate lanosterol synthase. J. Am. Chem. Soc. 2005, 127, 14132–14133. [Google Scholar] [CrossRef]
- Hoshino, T. β-Amyrin biosynthesis: Catalytic mechanism and substrate recognition. Org. Biomol. Chem. 2017, 15, 1–55. [Google Scholar] [CrossRef]
- Qiao, J.; Luo, Z.; Cui, S.; Zhao, H.; Tang, Q.; Mo, C.; Ma, X.; Ding, Z. Modification of isoprene synthesis to enable production of curcurbitadienol synthesis in Saccharomyces cerevisiae. J. Ind. Microbiol. Biotechnol. 2018. [CrossRef]
- Shibuya, M.; Adachi, S.; Ebizuka, Y. Cucurbitadienol synthase, the first committed enzyme for cucurbitacin biosynthesis, is a distinct enzyme from cycloartenol synthase for phytosterol biosynthesis. Tetrahedron 2004, 60, 6995–7003. [Google Scholar] [CrossRef]
- Davidovich-Rikanati, R.; Shalev, L.; Baranes, N.; Meir, A.; Itkin, M.; Cohen, S.; Zimbler, K.; Portnoy, V.; Ebizuka, Y.; Shibuya, M. Recombinant yeast as a functional tool for understanding bitterness and cucurbitacin biosynthesis in watermelon (Citrullus spp.). Yeast 2015, 32, 103–114. [Google Scholar]
- Shang, Y.; Ma, Y.; Zhou, Y.; Zhang, H.; Duan, L.; Chen, H.; Zeng, J.; Zhou, Q.; Wang, S.; Gu, W. Biosynthesis, regulation, and domestication of bitterness in cucumber. Science 2014, 346, 1084–1088. [Google Scholar] [CrossRef]
- Kusano, M.; Abe, I.; Sankawa, U.; Ebizuka, Y. Purification and some properties of squalene-2,3-epoxide: Lanosterol cyclase from rat liver. Chem. Pharm. Bull. 1991, 39, 239. [Google Scholar] [CrossRef]
- Abe, I.; Ebizuka, Y.; Seo, S.; Sankawa, U. Purification of squalene-2,3-epoxide cyclases from cell suspension cultures of Rabdosia japonica Hara. FEBS Lett. 1989, 249, 100–104. [Google Scholar] [CrossRef]
- Zerikly, M.; Challis, G.L. Strategies for the discovery of new natural products by genome mining. ChemBioChem 2009, 10, 625–633. [Google Scholar] [CrossRef]
- Boutanaev, A.M.; Moses, T.; Zi, J.; Nelson, D.R.; Mugford, S.T.; Peters, R.J.; Osbourn, A. Investigation of terpene diversification across multiple sequenced plant genomes. Proc. Natl. Acad. Sci. USA 2015, 112, E81–E88. [Google Scholar] [CrossRef] [PubMed]
- Hutmacher, R.B.; Ulloa, M.; Wright, S.D.; Campbell, B.T.; Percy, R.; Wallace, T.; Myers, G.; Bourland, F.; Weaver, D.; Peng, C. Elite Upland Cotton Germplasm-Pool Assessment of Fusarium Wilt Resistance in California. Agron. J. 2013, 105, 1635. [Google Scholar] [CrossRef]
- Ma, X.J.; Mo, C.M. Prospects of molecular breeding in medical plants. China J. Chin. Mater. Med. 2017, 42, 2021–2031. [Google Scholar]
- Qi, T.; Ma, X.; Mo, C.M.; Wilson, I.; Song, C.; Zhao, H.; Yang, Y.F.; Fu, W.; Qiu, D.Y. An efficient approach to finding Siraitia grosvenorii triterpene biosynthetic genes by RNA-seq and digital gene expression analysis. BMC Genom. 2011, 12, 343. [Google Scholar]
- Vickers, C.E.; Bydder, S.F.; Zhou, Y.; Nielsen, L.K. Dual gene expression cassette vectors with antibiotic selection markers for engineering in Saccharomyces cerevisiae. Microb. Cell Fact. 2013, 12, 96. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds morgrosides and squalene are available from the authors. |

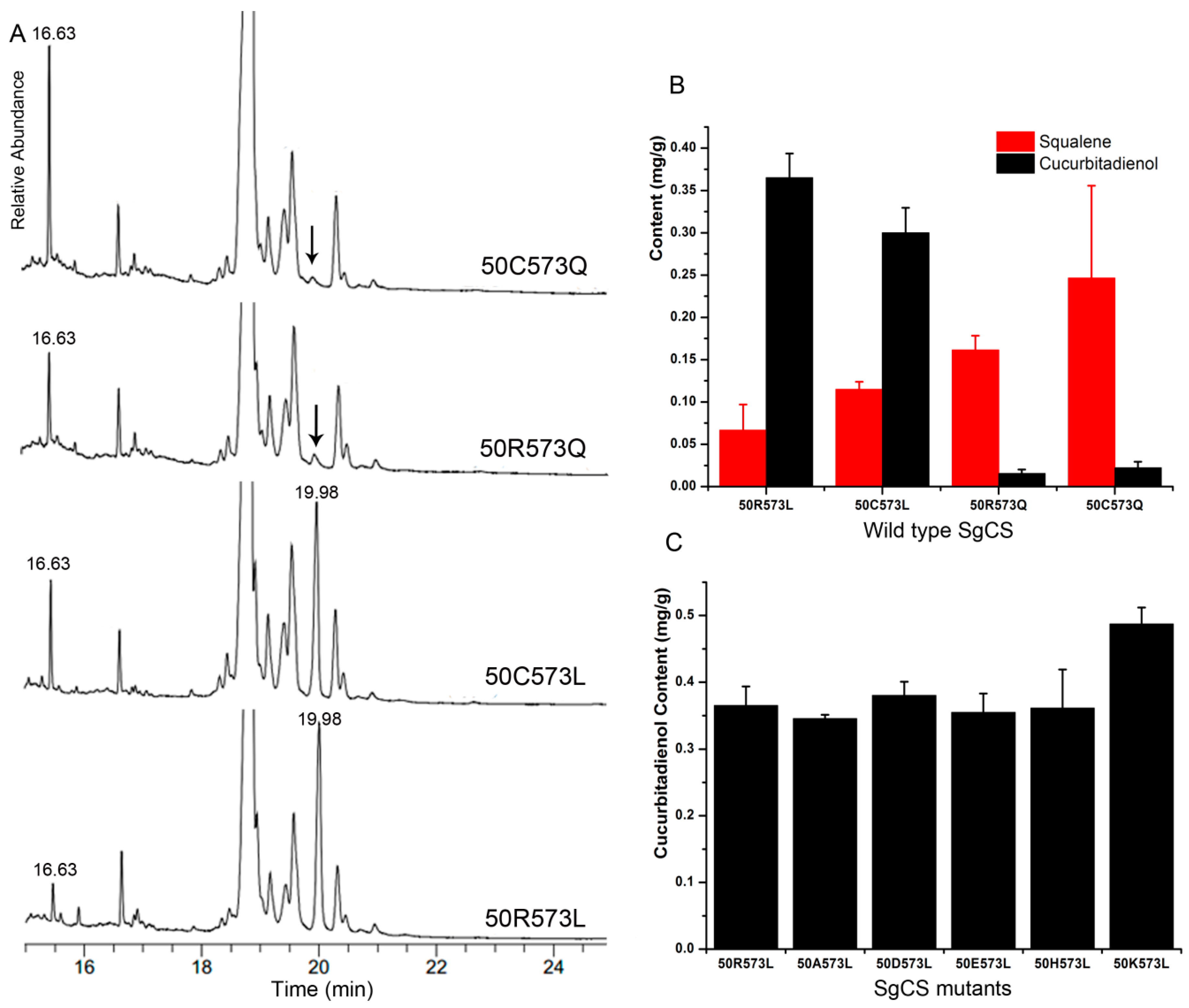
| Km (µM) | Kcat (min−1) | Kcat/Km (µM−1 min−1) | Specific Activity (nmol min−1 mg−1) | Relative Activity (%) | |
|---|---|---|---|---|---|
| 50R573L | 0.29 | 0.88 | 2.98 | 10.24 | 100 |
| 50C573L | 0.32 | 0.77 | 2.41 | 8.28 | 80.8 |
| 50R573Q | ND (a) | ND | ND | ND | - |
| 50C573Q | ND | ND | ND | ND | - |
| Analytes | Molecular Formula | tR (min) | m/z Precursor | m/z Productions | DP (V) | CE (eV) | |
|---|---|---|---|---|---|---|---|
| M2 | MIIA | C42H72O14 | 16.4 | 799.5 | 637.6 (a)/475.5 | −170 | −65 |
| MIIA1 | C42H72O14 | 16.7 | 799.5 | 637.6 (a)/475.5 | −170 | −65 | |
| MIIA2 | C42H72O14 | 15.27 | 799.5 | 637.6 (a)/475.5 | −170 | −65 | |
| MIIE | C42H72O14 | 13.4 | 799.5 | 637.6 (a)/475.5 | −170 | −65 | |
| M3 | MIII | C48H82O19 | 11.56 | 961.6 | 799.4 (a)/637.3 | −170 | −70 |
| MIIIE | C48H82O19 | 11.85 | 961.6 | 799.4 (a)/637.3 | −170 | −70 | |
| MIIIA1 | C48H82O19 | 14.31 | 961.6 | 799.4 (a)/637.3 | −170 | −70 | |
| MIIIA2 | C48H82O19 | 12.07 | 961.6 | 799.4 (a)/637.3 | −170 | −70 | |
| M4 | 11-O-SI | C54H90O24 | 9.32 | 1121.6 | 959.6 (a)/797.4 | −220 | −75 |
| SI | C54H92O24 | 9.82 | 1123.6 | 961.6 (a)/799.2 | −220 | −75 | |
| MIVA | C54H92O24 | 10.20 | 1123.6 | 961.6 (a)/799.2 | −220 | −75 | |
| MIVE | C54H92O24 | 10.77 | 1123.6 | 961.6 (a)/799.2 | −220 | −75 | |
| M5 | 11-D-O-MV | C60H102O28 | 12.48 | 1269.8 | 1107.7 (a)/945.6 | −230 | −90 |
| 11-E-MV | C60H100O29 | 7.12 | 1283.8 | 1121.6 (a)/959.5 | −220 | −85 | |
| MV | C60H102O29 | 8.44 | 1285.8 | 1123.7 (a)/961.7 | −220 | −90 | |
| O-MV | C60H102O29 | 6.52 | 1285.8 | 1123.7 (a)/961.7 | −220 | −90 | |
| IMV | C60H102O29 | 9.35 | 1285.8 | 1123.7 (a)/961.7 | −220 | −90 | |
| M6 | 11-O-MVI | C66H110O34 | 5.29 | 1445.8 | 1283.5 (a)/1121.7 | −240 | −100 |
| MVI | C66H112O34 | 6.15 | 1447.8 | 1285.6 (a)/1123.6 | −230 | −105 | |
| MVIA | C66H112O34 | 7.33 | 1447.8 | 1285.6 (a)/1123.6 | −230 | −105 | |
| MVIB | C66H112O34 | 7.13 | 1447.8 | 1285.6 (a)/1123.6 | −230 | −105 | |
| Source temperature (°C) | 500 | ||||||
| Ionization voltage (V) | −4500 | ||||||
| Ion source (GS1) setting (psi) | 50 | ||||||
| Ion source (GS2) setting (psi) | 40 | ||||||
| Curtain gas setting (psi) | 15 | ||||||
| CAD | high | ||||||
| Dwell time (ms) | 400 | ||||||
| EP (V) | −10 | ||||||
| CXP (V) | −15 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qiao, J.; Luo, Z.; Gu, Z.; Zhang, Y.; Zhang, X.; Ma, X. Identification of a Novel Specific Cucurbitadienol Synthase Allele in Siraitia grosvenorii Correlates with High Catalytic Efficiency. Molecules 2019, 24, 627. https://doi.org/10.3390/molecules24030627
Qiao J, Luo Z, Gu Z, Zhang Y, Zhang X, Ma X. Identification of a Novel Specific Cucurbitadienol Synthase Allele in Siraitia grosvenorii Correlates with High Catalytic Efficiency. Molecules. 2019; 24(3):627. https://doi.org/10.3390/molecules24030627
Chicago/Turabian StyleQiao, Jing, Zuliang Luo, Zhe Gu, Yanling Zhang, Xindan Zhang, and Xiaojun Ma. 2019. "Identification of a Novel Specific Cucurbitadienol Synthase Allele in Siraitia grosvenorii Correlates with High Catalytic Efficiency" Molecules 24, no. 3: 627. https://doi.org/10.3390/molecules24030627
APA StyleQiao, J., Luo, Z., Gu, Z., Zhang, Y., Zhang, X., & Ma, X. (2019). Identification of a Novel Specific Cucurbitadienol Synthase Allele in Siraitia grosvenorii Correlates with High Catalytic Efficiency. Molecules, 24(3), 627. https://doi.org/10.3390/molecules24030627