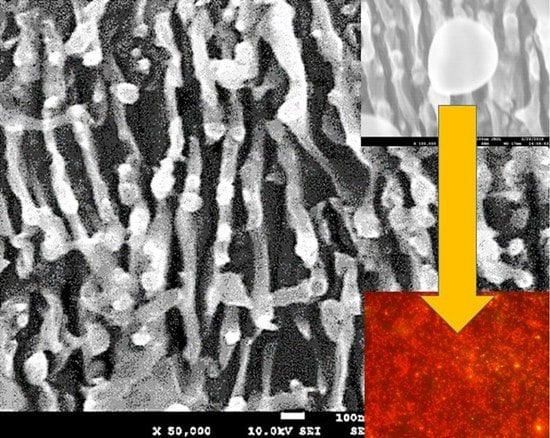
Surface-Enhanced IR-Absorption Microscopy of Staphylococcus aureus Bacteria on Bactericidal Nanostructured Si Surfaces
Abstract
:1. Introduction
2. Experimental Results
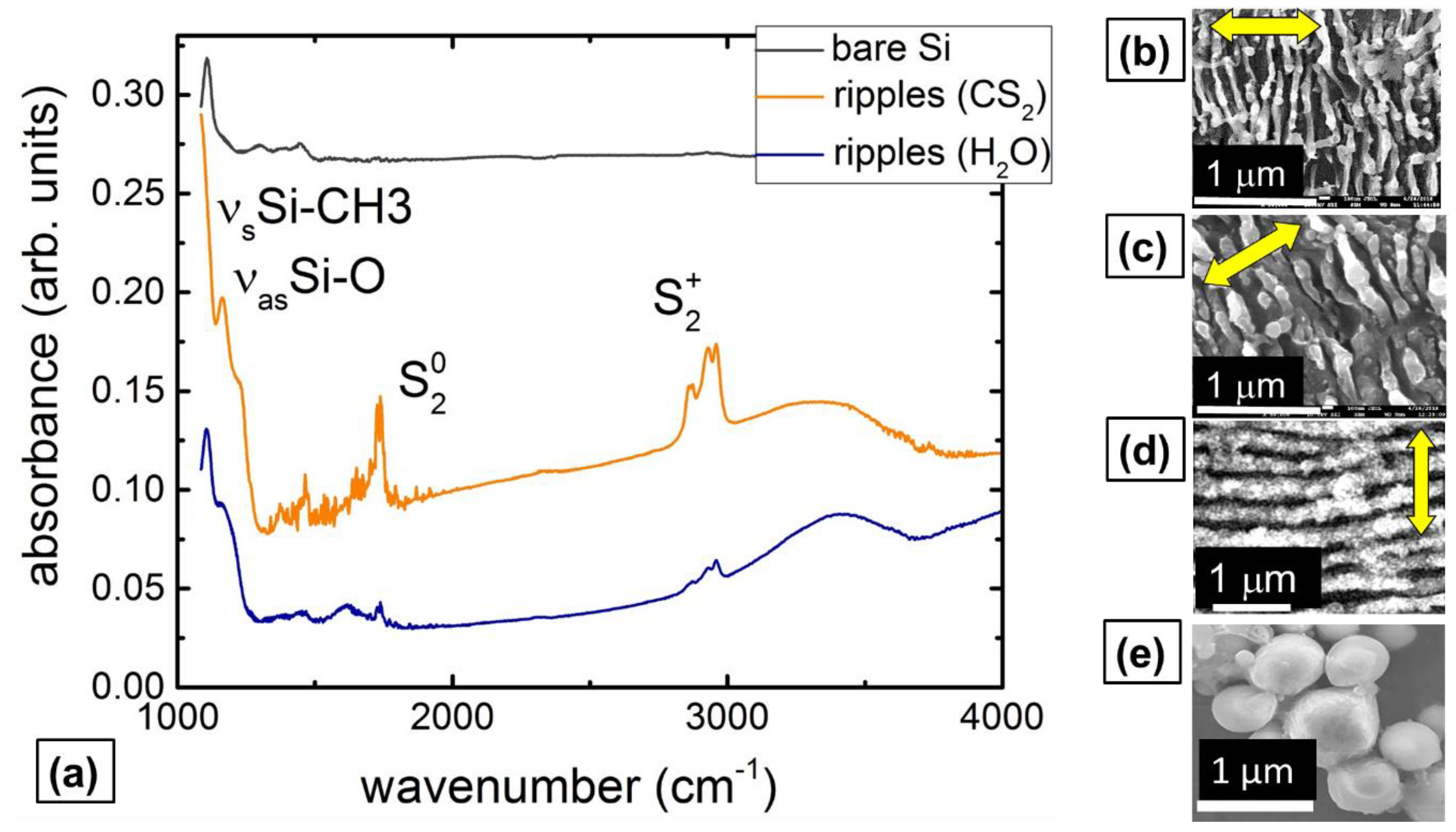
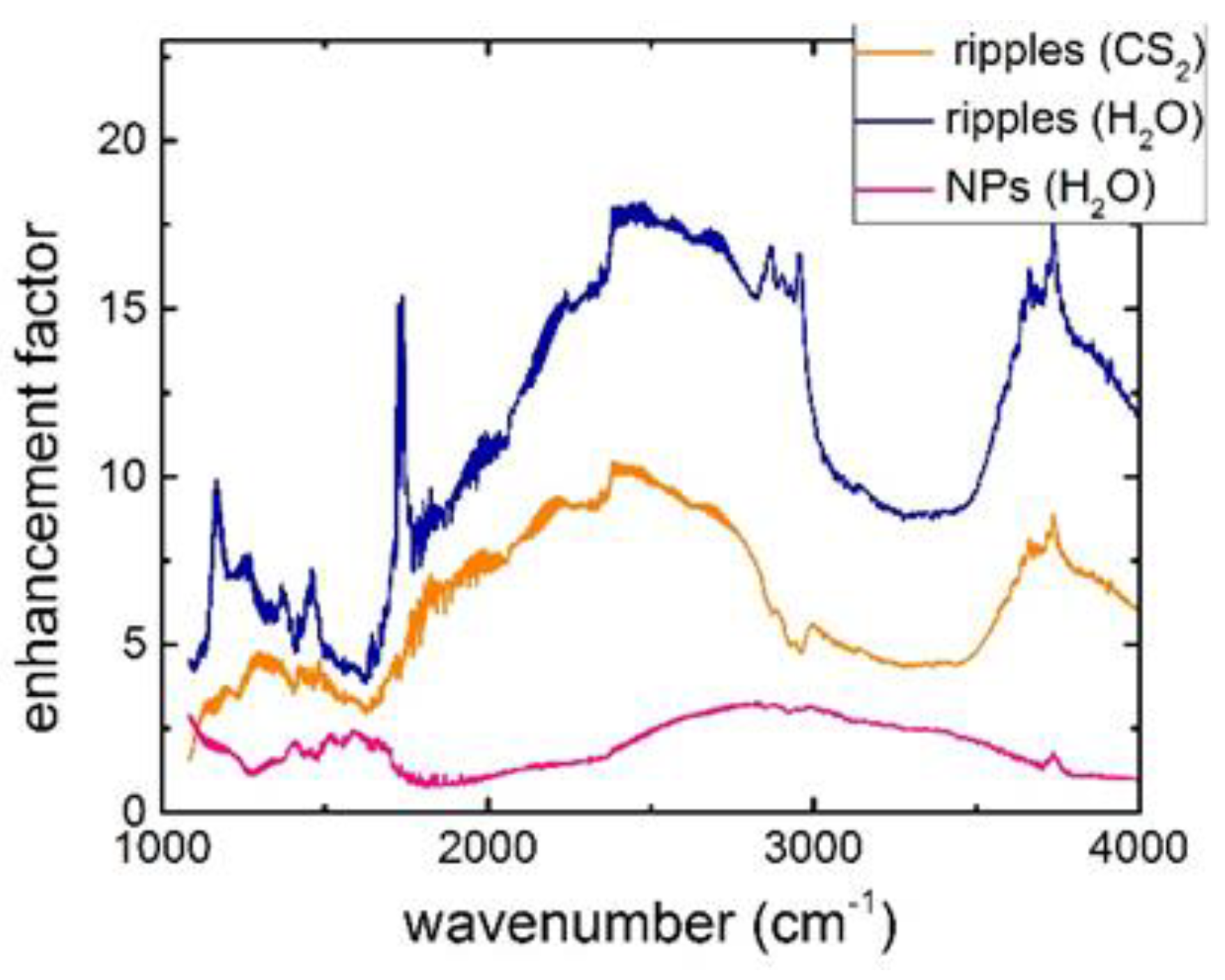
2.1. IR Characterization of Bactericidal Si Nanoripples and Nanoparticle Coatings
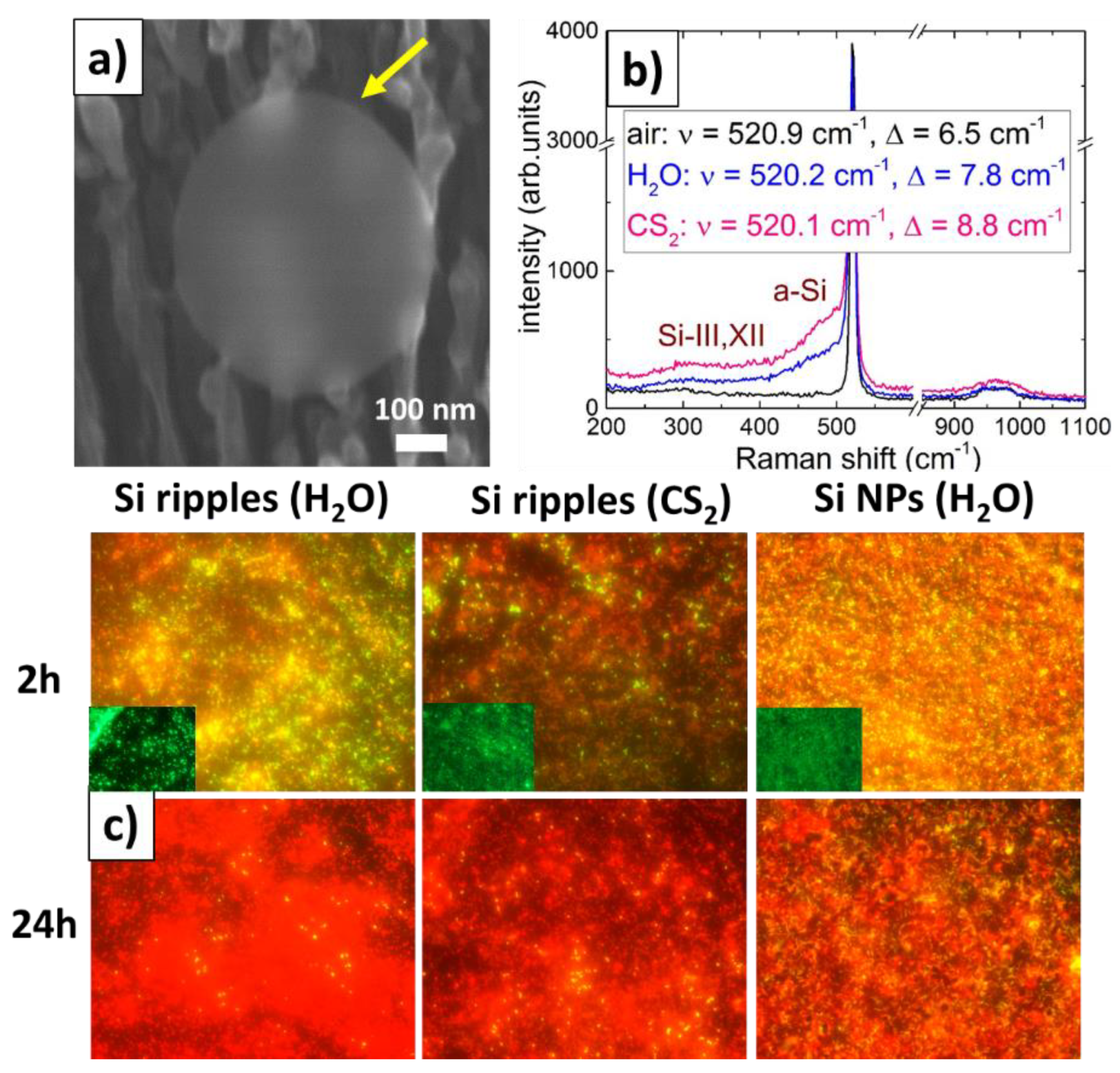
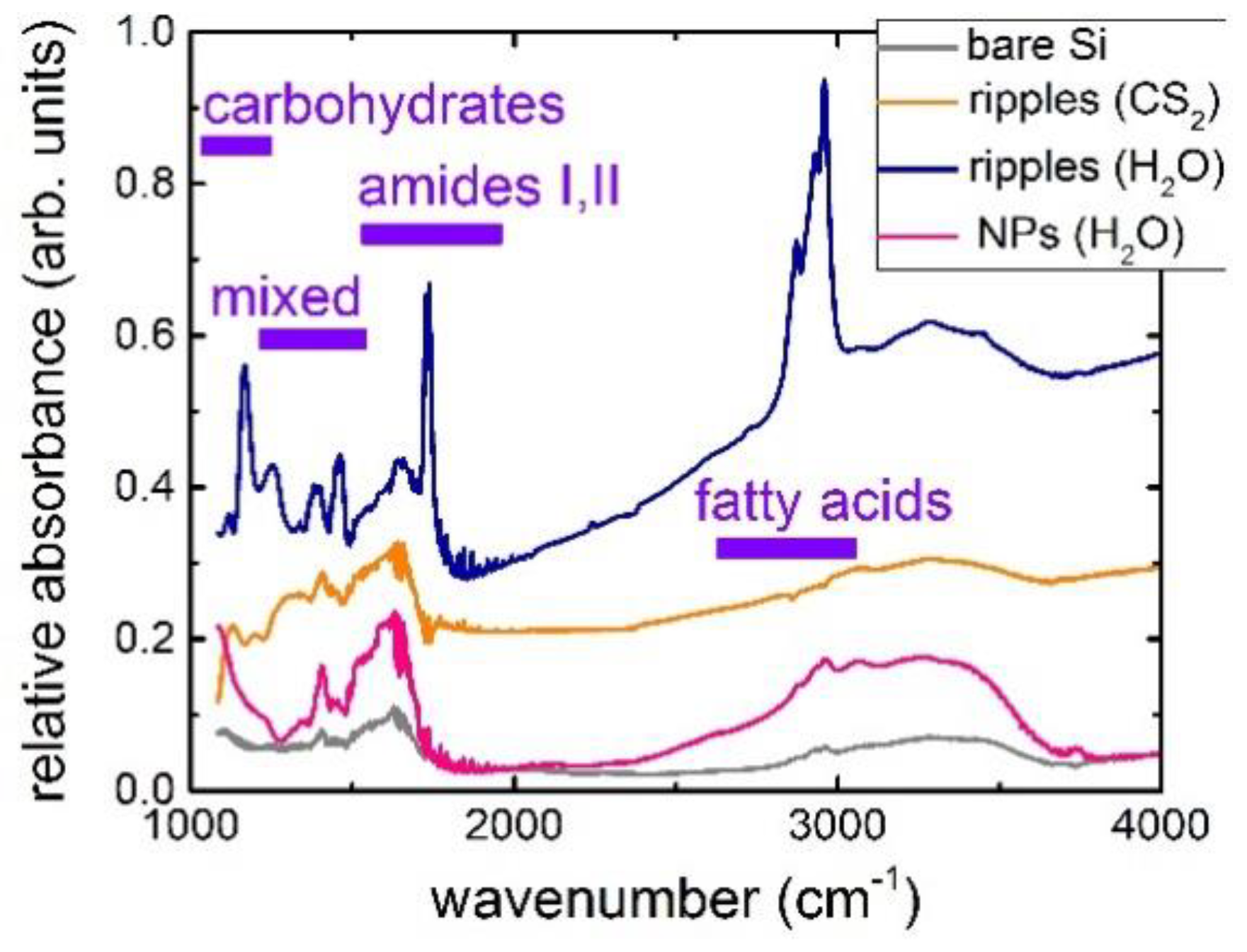
2.2. IR Characterization of Staphylococcus Aureus Bacterial Biofilms on Bactericidal Si Nanoripples and Nanoparticle Coatings
- in the window between 3000 and 2800 cm−1 (W1, the ‘fatty acid region I’), dominated by the -CH3, >CH, and =CH stretching vibrations of the functional groups usually present in the fatty acid components of the various membrane amphiphiles;
- in the window between 1800 and 1500 cm−1 (W2, the ‘amide region’), dominated by the amide I and amide II bands of proteins and peptides;
- in the window between 1500 and 1200 cm−1 (W3, the ‘mixed region’), a spectral region containing information from proteins, fatty acids and phosphate-carrying compounds, including the window between 1500 and 1400 cm−1 (W31, the ‘fatty acid region II’), dominated by the -CH3 and -CH2 bending vibrations of the same functional groups as expressed in W1;
- in the window between 1200 and 900 cm−1 (W4, the ‘polysaccharide region’), dominated by the fingerprint-like absorption bands of the carbohydrates present within the cell wall.
3. Concluding Remarks
4. Materials and Methods
Author Contributions
Funding
Conflicts of Interest
References
- Fu, P.P.; Xia, Q.; Hwang, H.M.; Ray, P.C.; Yu, H. Mechanisms of nanotoxicity: Generation of reactive oxygen species. J. Food Drug Anal. 2014, 22, 64–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, L.; Gu, L.; Howell, S.B.; Sailor, M.J. Porous Silicon Nanoparticle Photosensitizers for Singlet Oxygen and their Phototoxicity against Cancer Cells. ACS Nano 2011, 5, 3651–3659. [Google Scholar] [CrossRef] [PubMed]
- Timoshenko, V.Y.; Kudryavtsev, A.A.; Osminkina, L.A.; Vorontsov, A.S.; Ryabchikov, Y.V.; Belogorokhov, I.A.; Kovalev, D.; Kashkarov, P.K. Silicon nanocrystals as photosensitizers of active oxygen for biomedical applications. Jetp Lett. 2006, 83, 423–426. [Google Scholar] [CrossRef]
- Chernousova, S.; Epple, M. Silver as antibacterial agent: Ion, nanoparticle, and metal. Angew. Chem. Int. Ed. 2013, 52, 1636–1653. [Google Scholar] [CrossRef] [PubMed]
- Nastulyavichus, A.; Kudryashov, S.; Smirnov, N.; Saraeva, I.; Rudenko, A.; Tolordava, E.; Ionin, A.; Romanova, Y.; Zayarny, D. Antibacterial coatings of Se and Si nanoparticles. Appl. Surf. Sci. 2019, 469, 220–225. [Google Scholar] [CrossRef]
- Ivanova, E.P.; Hasan, J.; Webb, H.K.; Gervinskas, G.; Juodkazis, S.; Truong, V.K.; Wu, A.H.F.; Lamb, R.N.; Baulin, V.A.; Watson, G.S.; et al. Bactericidal activity of black silicon. Nat. Commun. 2013, 4, 2838. [Google Scholar] [CrossRef]
- Kudryashov, S.I.; Nguyen, L.V.; Kirilenko, D.A.; Brunkov, P.N.; Rudenko, A.A.; Busleev, N.I.; Saraeva, I.N.; Nastulyavichus, A.A.; Ionin, A.A.; Tolordava, E.R.; et al. Large-scale Laser Fabrication of Anti-Fouling Si Surface Nanosheet Arrays via Nanoplasmonic Ablative Self-organization in Liquid CS2 Tracked by Sulfur Dopant. ACS Appl. Nano Mater. 2018, 1, 2461–2468. [Google Scholar] [CrossRef]
- Linklater, D.P.; Juodkazis, S.; Ivanova, E.P. Nanofabrication of Mechano-Bactericidal Surfaces. Nanoscale 2017, 9, 16564–16585. [Google Scholar] [CrossRef]
- Smirnov, N.A.; Kudryashov, S.I.; Nastulyavichus, A.A.; Rudenko, A.A.; Saraeva, I.N.; Tolordava, E.R.; Gonchukov, S.A.; Romanova, Y.M.; Ionin, A.A.; Zayarny, D.A. Antibacterial properties of silicon nanoparticles. Laser Phys. Lett. 2018, 15, 105602. [Google Scholar] [CrossRef]
- Milla, M.J.; Barho, F.; González-Posada, F.; Cerutti, L.; Charlot, B.; Bomers, M.; Neubrech, F.; Tournie, E.; Taliercio, T. Surface-enhanced infrared absorption with Si-doped InAsSb/GaSb nano-antennas. Opt. Express 2017, 25, 26651–26661. [Google Scholar] [CrossRef]
- Caldarola, M.; Albella, P.; Cortés, E.; Rahmani, M.; Roschuk, T.; Grinblat, G.; Oulton, R.F.; Bragas, A.V.; Maier, S.A. Non-plasmonic nanoantennas for surface enhanced spectroscopies with ultra-low heat conversion. Nat. Commun. 2015, 6, 7915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, L.; Chen, M.; Zhao, X.; Zhang, Z.; Xia, J.; Xu, H.; Sun, M. Visualized method of chemical enhancement mechanism on SERS and TERS. J. Raman Spectrosc. 2014, 45, 533–540. [Google Scholar] [CrossRef]
- Mitsai, E.; Kuchmizhak, A.; Pustovalov, E.; Sergeev, A.; Mironenko, A.; Bratskaya, S.; Linklater, D.P.; Balčytis, A.; Ivanova, E.; Juodkazis, S. Chemically non-perturbing SERS detection of a catalytic reaction with black silicon. Nanoscale 2018, 10, 9780–9787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mosier-Boss, P.A. Review on SERS of Bacteria. Biosensors 2017, 7, 51. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Zhou, H.; Hu, Z.; Yu, G.; Yang, D.; Zhao, J. Label and label-free based surface-enhanced Raman scattering for pathogen bacteria detection: A review. Biosens. Bioelectron. 2017, 94, 131–140. [Google Scholar] [CrossRef]
- Zheng, X.S.; Jahn, I.J.; Weber, K.; Cialla-May, D.; Popp, J. Label-free SERS in biological and biomedical applications: Recent progress, current challenges and opportunities. Spectrochim. Acta Part. A Mol. Biomol. Spectrosc. 2018, 197, 56–77. [Google Scholar] [CrossRef]
- Lamprell, H.; Mazerolles, G.; Kodjo, A.; Chamba, J.F.; Noel, Y.; Beuvier, E. Discrimination of Staphylococcus aureus strains from different species of Staphylococcus using Fourier transform infrared (FTIR) spectroscopy. Int. J. Food Microbiol. 2006, 108, 125–129. [Google Scholar] [CrossRef]
- Danilov, P.A.; Gonchukov, S.A.; Ionin, A.A.; Khmelnitskii, R.A.; Kudryashov, S.I.; Nguyen, T.T.H.; Rudenko, A.A.; Saraeva, I.N.; Zayarny, D.A. Background-free, highly-sensitive surface-enhanced IR absorption of rhodamine 6G molecules deposited onto array of microholes in thin silver film. Laser Phys. Lett. 2016, 13, 055602. [Google Scholar] [CrossRef]
- Bonse, J.; Krüger, J.; Höhm, S.; Rosenfeld, A. Femtosecond laser-induced periodic surface structures. J. Laser Appl. 2012, 24, 042006. [Google Scholar] [CrossRef]
- Tsibidis, G.D.; Skoulas, E.; Stratakis, E. Ripple formation on nickel irradiated with radially polarized femtosecond beams. Opt. Lett. 2015, 40, 5172–5175. [Google Scholar] [CrossRef] [Green Version]
- Pai, P.G.; Chao, S.S.; Takagi, Y.; Lucovsky, G. Infrared spectroscopic study of SiO x films produced by plasma enhanced chemical vapor deposition. J. Vac. Sci. Technol. A Vac. Surf. Film. 1986, 4, 689–694. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.T.; Lee, J. Characterization of amorphous SiC: H films deposited from hexamethyldisilazane. Thin Solid Film. 1997, 303, 173–179. [Google Scholar] [CrossRef]
- Grimmeiss, H.G.; Janzen, E.; Skarstam, B. Deep sulfur-related centers in silicon. J. Appl. Phys. 1980, 51, 4212–4217. [Google Scholar] [CrossRef]
- Ionin, A.A.; Kudryashov, S.I.; Levchenko, A.O.; Nguyen, L.V.; Saraeva, I.N.; Rudenko, A.A.; Ageev, A.A.; Potorochin, D.V.; Veiko, V.P.; Borisov, E.V.; et al. Correlated topographic and structural modification on Si surface during multi-shot femtosecond laser exposures: Si nanopolymorphs as potential local structural nanomarkers. Appl. Surf. Sci. 2017, 416, 988–995. [Google Scholar] [CrossRef]
- Helm, D.; Labischinski, H.; Schallehn, G.; Naumann, D. Classification and identification of bacteria by Fourier-transform infrared spectroscopy. Microbiology 1991, 137, 69–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krishnamurthy, K.; Tewari, J.C.; Irudayaraj, J.; Demirci, A. Microscopic and spectroscopic evaluation of inactivation of Staphylococcus aureus by pulsed UV light and infrared heating. Food Bioprocess. Technol. 2010, 3, 93. [Google Scholar] [CrossRef]
- Faghihzadeh, F.; Anaya, N.M.; Schifman, L.A.; Oyanedel-Craver, V. Fourier transform infrared spectroscopy to assess molecular-level changes in microorganisms exposed to nanoparticles. Nanotechnol. Environ. Eng. 2016, 1, 1. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are available from the authors. |
| Structure/Chemical Composition (at. %) | Silicon Si | Oxygen O | Carbon C | Sulfur S |
|---|---|---|---|---|
| Si ripples (air) | 67 ± 1 | 15 ± 1 | 17 ± 1 | 0 |
| Si nanoripples (CS2) | 75 ± 1 | 10 ± 1 | 14 ± 1 | 0.9 ± 0.3 |
| Si nanoripples (H2O) | 92 ± 1 | 2 ± 1 | 6 ± 1 | 0 |
| Si nanoparticles (H2O) | 84 ± 1 | 16 ± 1 | 0 | 0 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kudryashov, S.I.; Nastulyavichus, A.A.; Tolordava, E.R.; Kirichenko, A.N.; Saraeva, I.N.; Rudenko, A.A.; Romanova, Y.M.; Panarin, A.Y.; Ionin, A.A.; Itina, T.E. Surface-Enhanced IR-Absorption Microscopy of Staphylococcus aureus Bacteria on Bactericidal Nanostructured Si Surfaces. Molecules 2019, 24, 4488. https://doi.org/10.3390/molecules24244488
Kudryashov SI, Nastulyavichus AA, Tolordava ER, Kirichenko AN, Saraeva IN, Rudenko AA, Romanova YM, Panarin AY, Ionin AA, Itina TE. Surface-Enhanced IR-Absorption Microscopy of Staphylococcus aureus Bacteria on Bactericidal Nanostructured Si Surfaces. Molecules. 2019; 24(24):4488. https://doi.org/10.3390/molecules24244488
Chicago/Turabian StyleKudryashov, Sergey I., Alena A. Nastulyavichus, Eteri R. Tolordava, Alexey N. Kirichenko, Irina N. Saraeva, Andrey A. Rudenko, Yulia M. Romanova, Andrey Yu. Panarin, Andrey A. Ionin, and Tatiana E. Itina. 2019. "Surface-Enhanced IR-Absorption Microscopy of Staphylococcus aureus Bacteria on Bactericidal Nanostructured Si Surfaces" Molecules 24, no. 24: 4488. https://doi.org/10.3390/molecules24244488