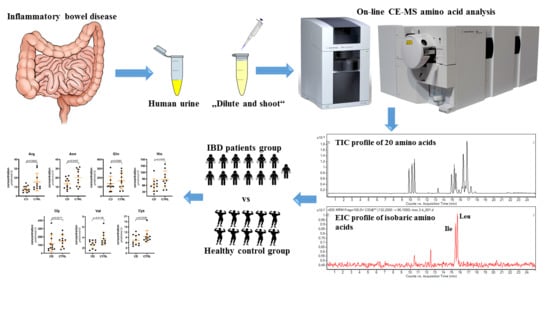
Profiling of Amino Acids in Urine Samples of Patients Suffering from Inflammatory Bowel Disease by Capillary Electrophoresis-Mass Spectrometry
Abstract
:1. Introduction
2. Results and Discussion
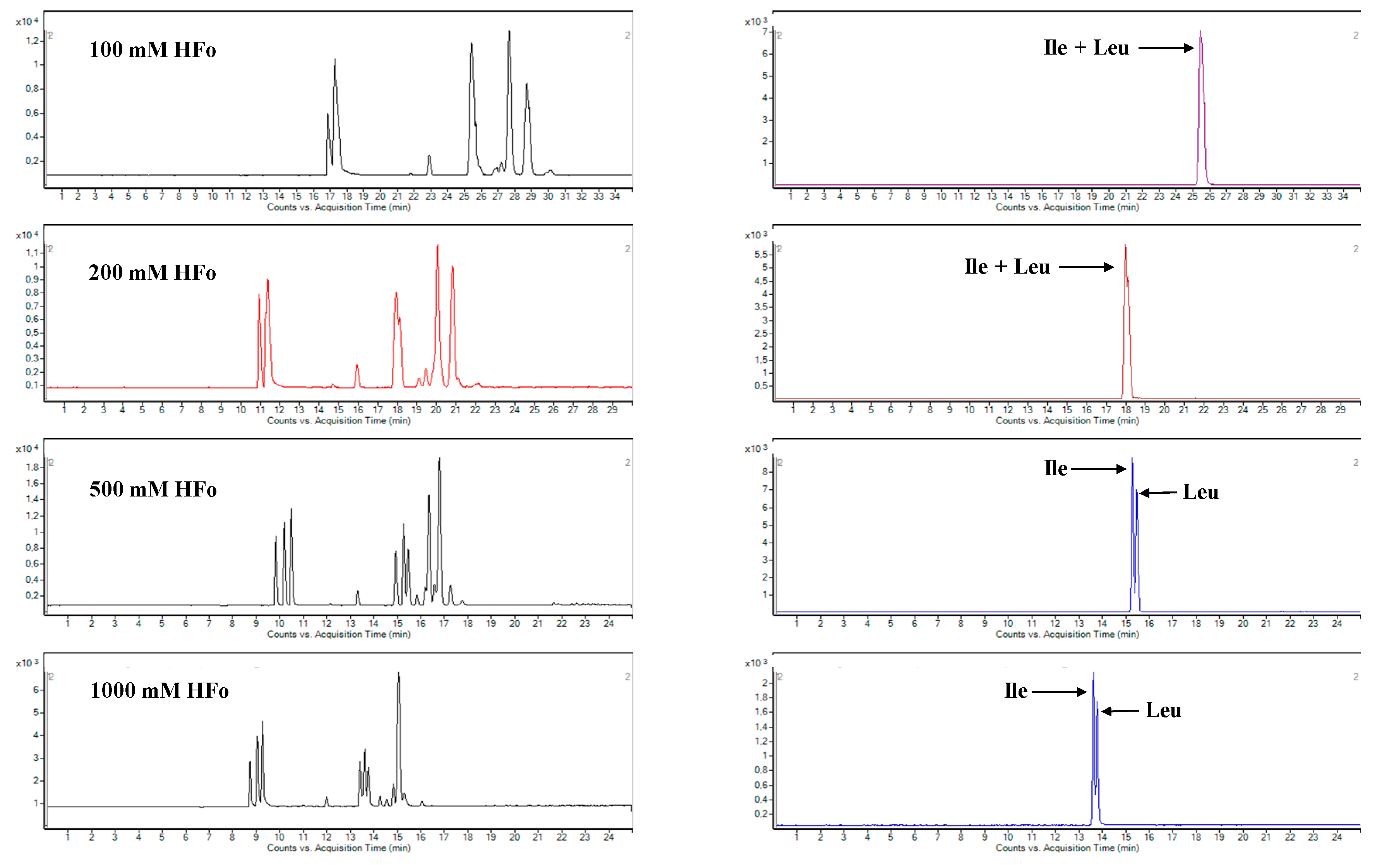
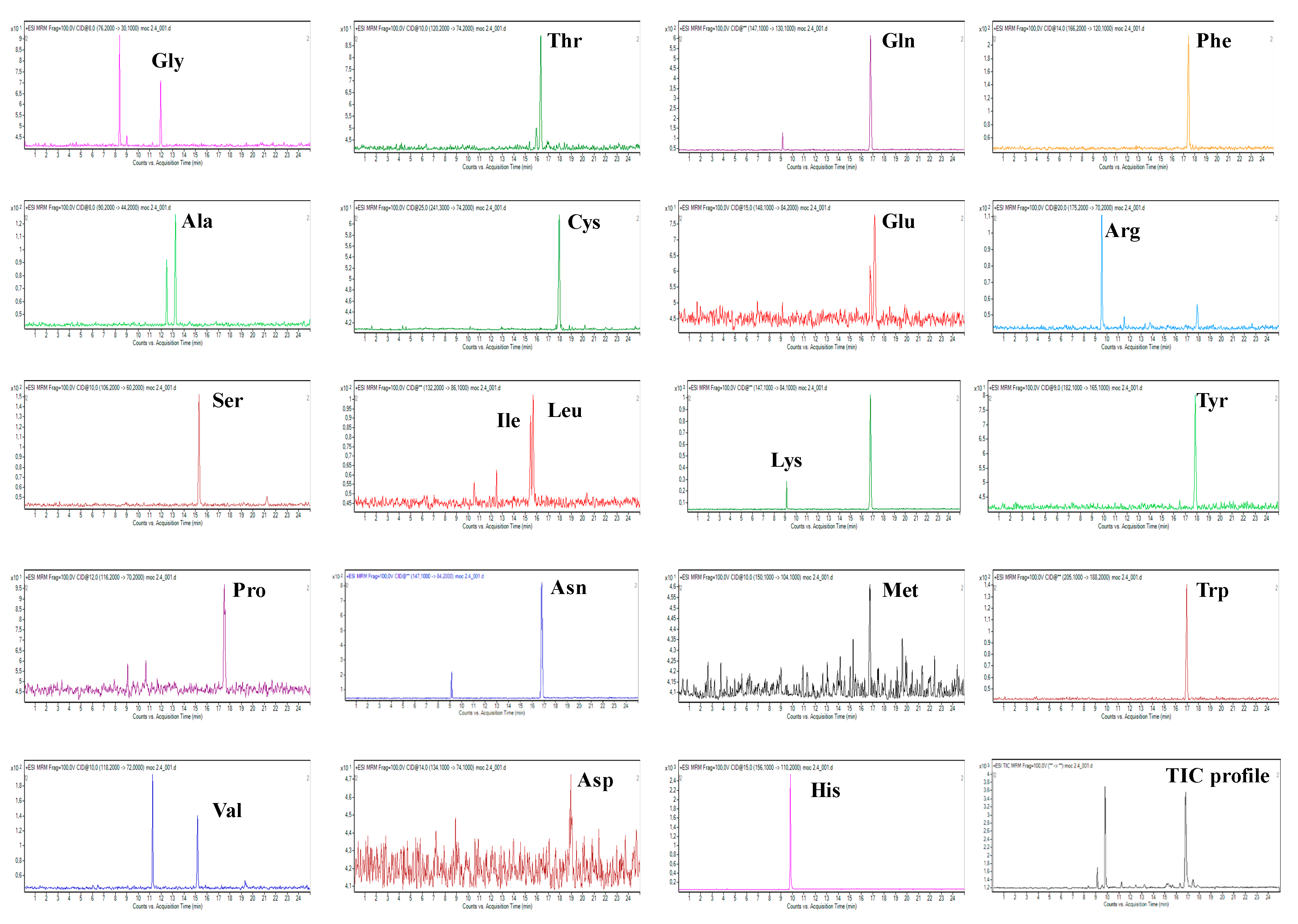
2.1. Development of CZE-ESI-MS/MS Method
2.2. Evaluation of Performance Parameters
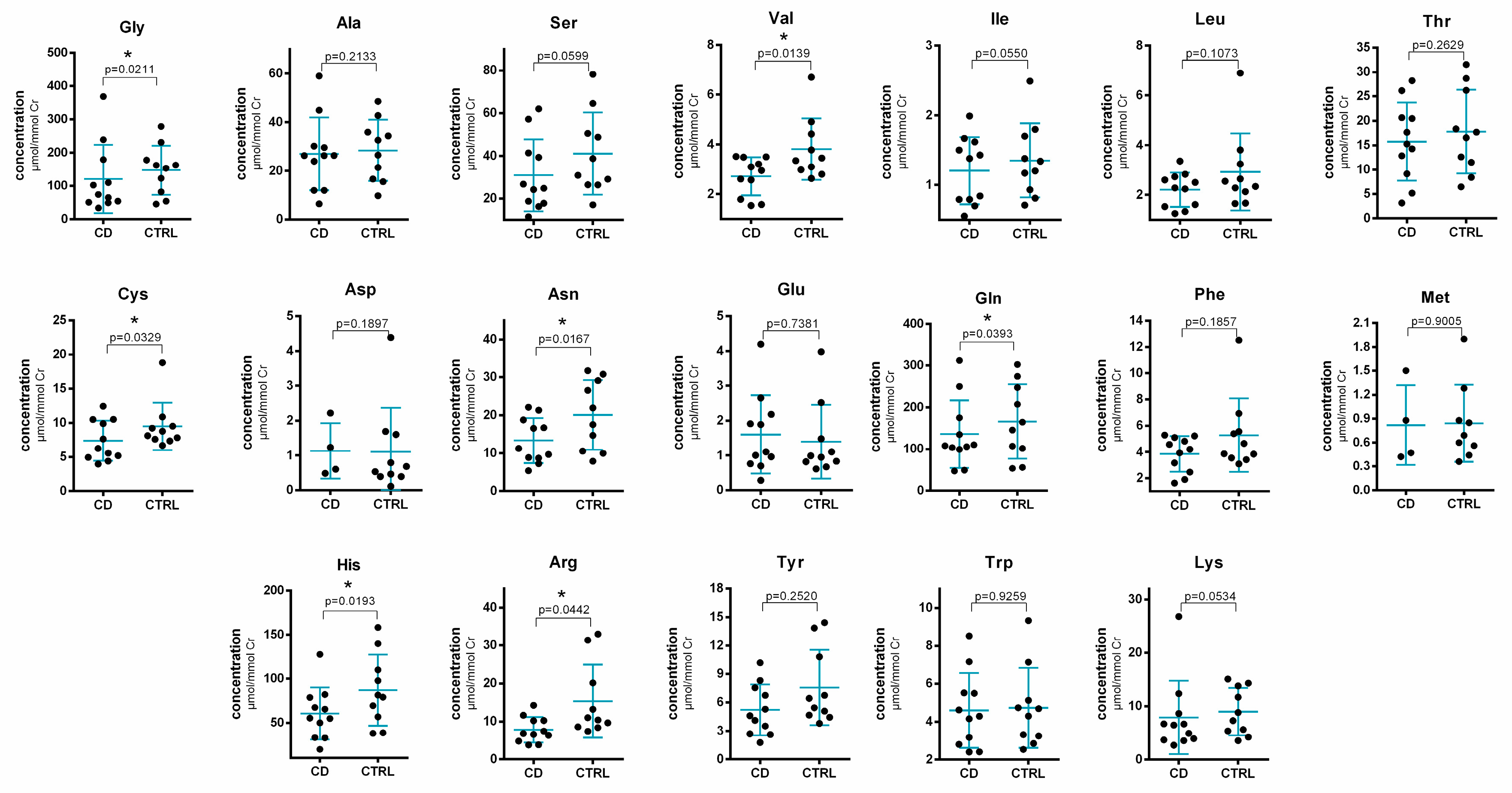
2.3. Method Application
3. Materials and Methods
3.1. CE-MS/MS Instrumentation and Separation Conditions
3.2. UHPLC-MS Instrumentation and Separation Conditions
3.3. Chemicals and Samples
3.4. Procedures for Sample and Standard Solution Preparation
3.4.1. Standard Solutions, Calibration Solutions and Quality Control (QC) Samples
3.4.2. Urine Sample Collection and Preparation
3.5. Creatinine
3.6. Statistical Data Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Mischak, H.; Coon, J.J.; Novak, J.; Weissinger, E.M.; Schanstra, J.P.; Dominiczak, A.F. Capillary electrophoresis—Mass spectrometry as a powerful tool in biomarker discovery and clinical diagnosis: An update of recent developments. Mass Spectrom. Rev. 2009, 28, 703–724. [Google Scholar] [CrossRef] [PubMed]
- Ramautar, R. Capillary electrophoresis-mass spectrometry for clinical metabolomics. Adv. Clin. Chem. 2016, 74, 1–34. [Google Scholar] [PubMed]
- López-López, Á.; López-Gonzálvez, Á.; Barker-Tejeda, T.C.; Barbas, C. A review of validated biomarkers obtained through metabolomics. Expert Rev. Mol. Diagn. 2018, 18, 557–575. [Google Scholar] [CrossRef] [PubMed]
- Markuszewski, M.J.; Waszczuk-Jankowska, M.; Stuck, W.; Kośliński, P. Application of electromigration techniques: Metabolomics-determination of potential biomarkers using electromigration techniques. In Electromigration Techniques, 1st ed.; Buszewski, B., Dziubakiewicz, E., Szumski, M., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 271–285. [Google Scholar]
- Wu, G. Amino acids: Metabolism, functions and nutrition. Amino Acids 2009, 37, 1–17. [Google Scholar] [CrossRef]
- Song, Y.; Xu, C.; Kuroki, H.; Lio, Y.; Tsunoda, M. Recent trends in analytical methods for the determination of amino acids in biological samples. J. Pharm. Biomed. Anal. 2018, 147, 35–49. [Google Scholar] [CrossRef]
- Mayboroda, O.A.; Neusüß, C.; Pelzing, M.; Zurek, G.; Derks, R.; Meulenbelt, I.; Kloppenburg, M.; Slagboom, E.P.; Deelder, A.M. Amino acid profiling in urine by capillary zone electrophoresis—Mass spectrometry. J. Chromatogr. A 2007, 1159, 149–153. [Google Scholar] [CrossRef]
- Ramautar, R.; Mayboroda, O.A.; Derks, R.J.E.; van Niewkoop, C.; van Dissel, J.T.; Somsen, G.W.; Deelder, A.M.; de Jong, G.J. Capillary electrophoresi-time of flight-mass spectrometry using noncovalently bilayer-coated capillaries for the analysis of amino acids in human urine. Electrophoresis 2008, 29, 2714–2722. [Google Scholar] [CrossRef]
- Chalcraft, K.R.; Britz-McKibbin, P. Newborn screening of inborn errors of metabolism by capillary electrophoresis-electrospray ionization-mass spectrometry: A second-tier method with improved specificity and sensitivity. Anal. Chem. 2009, 81, 307–314. [Google Scholar] [CrossRef]
- MacLennan, M.S.; Kok, M.G.M.; Soliman, L.; So, A.; Hurtado-Coll, A.; Chen, D.D.Y. Capillary electrophoresis-mass spectrometry for targeted and untargeted analysis of the sub-5 kDa urine metabolome of patients with prostate or bladder cancer: A feasibility study. J. Chromatogr. B 2018, 1074-1075, 79–85. [Google Scholar] [CrossRef]
- Cieslarova, Z.; Lopes, F.S.; do Lago, C.L.; Franca, M.C.; Simionato, A.V.C. Capillary electrophoresis tandem mass spectrometry determination of glutamic acid and homocysteine’s metabolites: Potential biomarkers of amyotrophic lateral sclerosis. Talanta 2017, 170, 63–68. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, X.; Hu, C.-A.A. Therapeutic potential of amino acids in inflammatory bowel disease. Nutrients 2017, 9, 920. [Google Scholar] [CrossRef]
- Vidal-Lletjós, S.; Beaumont, M.; Tomé, D.; Benamouzing, R.; Blacier, F.; Lan, A. Dietary protein and amino acid supplementation in inflammatory bowel disease course: What impact on the colonic mucosa? Nutrients 2017, 9, 310. [Google Scholar] [CrossRef]
- Papada, E.; Torović, L.; Amerikanou, C.; Gioxari, A.; Kalogeropoulos, N.; Kaliora, A.C. Metabolomic analysis of amino acids in inflammatory bowel disease (IBD): The role in disease management. Clin. Nutr. ESPEN 2018, 24, 183. [Google Scholar] [CrossRef]
- Nikolaus, S.; Schulte, B.; Al-Massad, N.; Thieme, T.; Schulte, D.M.; Bethge, J.; Rehman, A.; Tran, F.; Aden, K.; Häsler, R.; et al. Increased tryptophan metabolism is associated with activity of inflammatory bowel diseases. Gastroenterology 2017, 153, 1504–1516. [Google Scholar] [CrossRef]
- Rodrigues, K.T.; Tavares, M.F.M.; Van Schepdael, A. CE-MS for the analysis of amino acids. Methods Mol. Biol. 2018, 1730, 305–313. [Google Scholar]
- Soga, T.; Kakazu, Y.; Robert, M.; Tomita, M.; Nishioka, T. Qualitative and quantitative analysis of amino acids by capillary electrophoresis-electrospray ionization-tandem mass spectrometry. Electrophoresis 2004, 25, 1964–1972. [Google Scholar] [CrossRef]
- Soliman, L.C.; Hui, Y.; Hewavitharana, A.K.; Chen, D.D.Y. Monitoring potential prostate cancer biomarkers in urine by capillary electrophoresis-tandem mass spectrometry. J. Chromatogr. A 2012, 1267, 162–169. [Google Scholar] [CrossRef]
- Rodrigues, K.T.; Mekahli, D.; Tavares, M.F.M.; Van Schepdael, A. Development and validation of a CE-MS method for the targeted assessment of amino acids in urine. Electrophoresis 2016, 37, 1039–1047. [Google Scholar] [CrossRef]
- Cavazza, A.; Corradini, C.; Marini, M.; Roda, L.G.; Valenti, A. Capillary electrophoresis coupled with mass spectrometry for the evaluation of substance P enzymatic degradation by SaOS-2 human osteosarcoma. J. Chromatogr. B 2011, 879, 2501–2506. [Google Scholar] [CrossRef]
- Jeong, J.-S.; Kim, S.-K.; Park, S.-R. Capillary electrophoresis mass spectrometry with sheathless electrospray ionization for high sensitivity analysis of underivatized amino acids. Electrophoresis 2012, 33, 2112–2121. [Google Scholar] [CrossRef]
- Hirayama, A.; Tomita, M.; Soga, T. Sheathless capillary electrophoresis-mass spectrometry with a high-sensitivity porous sprayer for cationic metabolome analysis. Analyst 2012, 137, 5026–5033. [Google Scholar] [CrossRef]
- Martin, F.P.; Su, M.M.; Xie, G.X.; Guiraud, S.P.; Kussmann, M.; Godin, J.P.; Jia, W.; Nydegger, A. Urinary metabolic insights into host-gut microbial interactions in healthy and IBD children. World J. Gastroenterol. 2017, 23, 3643–3654. [Google Scholar] [CrossRef]
- Williams, H.R.T.; Cox, I.J.; Walker, D.G.; Nort, B.V.; Patel, V.M.; Marshall, S.E.; Jewell, D.P.; Ghosh, S.; Thomas, H.J.W.; Teare, J.P.; et al. Characterization of inflammatory bowel disease with urinary metabolic profiling. Am. J. Gastroenterol. 2009, 104, 1435–1444. [Google Scholar] [CrossRef]
- Stephens, N.S.; Siffledeen, J.; Su, X.; Murdoch, T.B.; Fedorak, R.N.; Slupsky, C.M. Urinary NMR metabolomic profiles disriminate inflammatory bowel disease from healthy. J. Crohns Colitis 2013, 7, e42–e48. [Google Scholar] [CrossRef]
- Schicho, R.; Shaykhutdinov, R.; Ngo, J.; Nazyrova, A.; Schneider, C.; Panaccione, R.; Kaplan, G.G.; Vogel, H.J.; Storr, M. Quantitative metabolomic profiling of serum, plasma and urine by 1H NMR spectroscopy discriminates between patients with inflammatory bowel disease and healthy individuals. J. Proteome Res. 2012, 11, 3344–3357. [Google Scholar] [CrossRef]
- Dawiskiba, T.; Deja, S.; Mulak, A.; Zabek, A.; Javień, E.; Pawelka, D.; Banasik, M.; Mastalerz-Migas, A.; Balcerzak, W.; Kaliszewski, K.; et al. Serum and urine metabolomic fingerprinting in diagnostics of inflammatory bowel diseases. World J. Gastroenterol. 2014, 20, 163–174. [Google Scholar] [CrossRef]
- Bjerrum, J.T.; Nielsen, O.H.; Hao, F.; Tang, H.; Nicholson, J.K.; Wang, Y.; Olsen, J. Metabonomics in ulcerative colitis: Diagnostics, biomarker identification and insight into the pathophysiology. J. Proteome Res. 2010, 9, 954–962. [Google Scholar] [CrossRef]
- Martin, F.P.; Ezri, J.; Cominetti, O.; Da Silva, L.; Kussmann, M.; Godin, J.P.; Nydegger, A. Urinary metabolic phenotyping reveals differences in the metabolic status of healthy and inflammatory bowel disease (IBD) children in relation to growth and disease activity. Int. J. Mol. Sci. 2016, 17, 1310. [Google Scholar] [CrossRef]
- Soga, T.; Heiger, D.N. Amino acid analysis by capillary electrophoresis electrospray ionization mass spectrometry. Anal. Chem. 2000, 72, 1236–1241. [Google Scholar] [CrossRef]
- U.S. Department of Health and Human Services Food and Drug Administration, Center for Drug Evaluation and Research (CDER) Center for Veterinary Medicine (CVM). Bioanalytical Method Validation Guidance for Industry; US FDA: Rockville, MD, USA, 2018.
- Kaspar, H.; Dettmer, K.; Chan, Q.; Daniels, S.; Nimkar, S.; Daviglus, M.L.; Stamler, J.; Elliott, P.; Oefner, P.J. Urinary amino acid analysis: A comparison of iTRAQ® - LC-MS/MS, GC-MS and amino acid analyzer. J. Chromatogr. B 2009, 877, 1838–1846. [Google Scholar] [CrossRef]
- Bouatra, S.; Aziat, F.; Mandal, R.; Guo, A.C.; Wilson, M.R.; Knox, C.; Bjorndahl, T.C.; Krishnamurthy, R.; Saleem, F.; Liu, P.; et al. The human urine metabolome. PLoS ONE 2013, 8, e73076. [Google Scholar] [CrossRef]
- Walker, R.; Whittlesea, C. Clinical Pharmacy and Therapeutics, 5th ed.; Churchill Livingstone Elsevier: London, UK, 2012. [Google Scholar]
- Alonso, A.; Julià, A.; Vinaixa, M.; Domènech, E.; Fernández-Nebro, A.; Cañete, J.D.; Ferrándiz, C.; Tornero, J.; Gisbert, J.P.; Nos, P.; et al. Urine metabolome profiling of immune-mediated inflammatory diseases. BMC Med. 2016, 14, 133. [Google Scholar] [CrossRef]
- Piestansky, J.; Galba, J.; Olesova, D.; Kovacech, B.; Kovac, A. Determination of immunogenic proteins in biopharmaceuticals by UHPLC-MS amino acid analysis. BMC Chem. 2019, 13, 64. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |
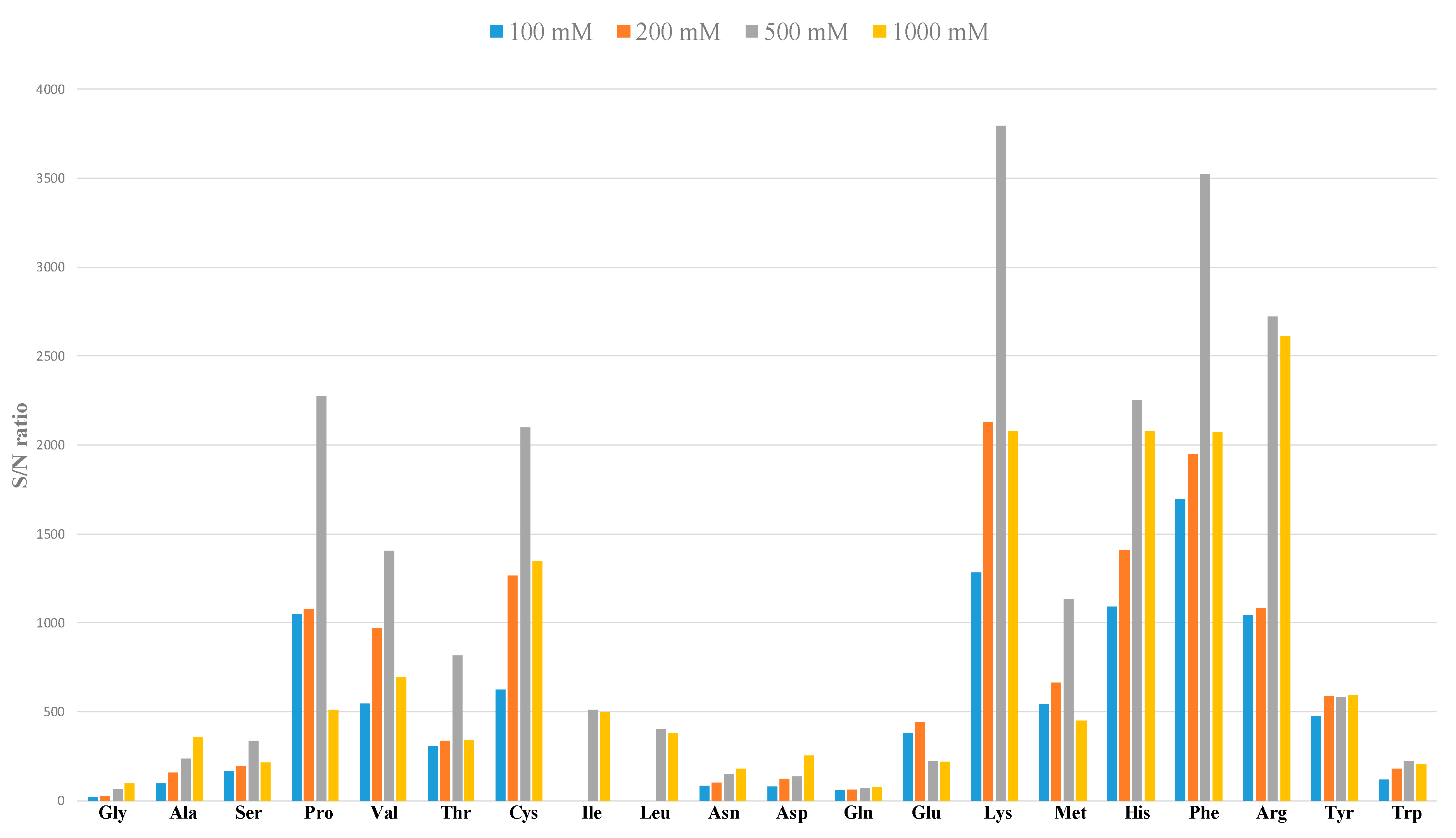
| Analyte | Parent Ion–Product Ion Transition (m/z) | Range (µM) | Calibration Line | Linearity (r2) | LOD (µM) | LLOQ (µM) | tm (min) | N |
|---|---|---|---|---|---|---|---|---|
| Glycine | 76.2→30.1 | 15–500 | y = 3.3694x + 1.5964 | 0.9965 | 11.03 | 18.38 | 10.63 | 111,289 |
| Alanine | 90.2→44.2 | 15–500 | y = 35.668x − 37.131 | 0.9992 | 1.33 | 2.22 | 11.53 | 101,866 |
| Serine | 106.2→60.2 | 3–500 | y = 33.646x + 142.79 | 0.9987 | 2.44 | 4.06 | 12.90 | 121,801 |
| Proline | 116.2→70.2 | 3–500 | y = 424.642x + 108.53 | 0.9974 | 0.63 | 1.05 | 14.31 | 260,363 |
| Valine | 118.2→72.0 | 3–500 | y = 168.106x − 133.02 | 0.9965 | 0.70 | 1.16 | 12.81 | 109,815 |
| Threonine | 120.2→74.2 | 15–500 | y = 34.036x + 1.223 | 0.9958 | 1.71 | 2.85 | 13.57 | 117,865 |
| Cysteine | 241.3→74.2 | 6.25–250 | y = 50.700x − 49.235 | 0.9986 | 0.47 | 0.79 | 14.56 | 98,824 |
| Isoleucine | 132.2→69.2 | 3–500 | y = 30.944x − 34.896 | 0.9992 | 1.33 | 2.22 | 13.00 | 123,659 |
| Leucine | 132.2→86.1 | 3–500 | y = 198.105x − 28.692 | 0.9963 | 1.57 | 2.61 | 13.11 | 106,688 |
| Asparagine | 133.1→74.0 | 3–500 | y = 12.194x + 23.981 | 0.9973 | 5.19 | 8.65 | 14.86 | 79,716 |
| Aspartic acid | 134.1→74.1 | 15–500 | y = 27.142x − 36.341 | 0.9930 | 7.14 | 11.91 | 15.19 | 190,107 |
| Glutamine | 147.1→84.1 | 15–500 | y = 15.483x + 94.291 | 0.9935 | 7.48 | 12.47 | 13.78 | 81,339 |
| Lysine | 147.1→84.2 | 3–500 | y = 216.476x + 34.134 | 0.9973 | 0.38 | 0.64 | 8.54 | 82,399 |
| Glutamic acid | 148.1→84.2 | 3–500 | y = 101.390x − 34.946 | 0.9983 | 1.91 | 3.19 | 14.07 | 126,768 |
| Methionine | 150.1→104.1 | 3–500 | y = 47.415x + 8.5213 | 0.9975 | 0.50 | 0.84 | 13.82 | 124,994 |
| Histidine | 156.1→110.2 | 15–500 | y = 306.972x + 34.669 | 0.9988 | 0.02 | 0.04 | 9.02 | 91,906 |
| Phenylalanine | 166.2→120.1 | 3–500 | y = 539.879x − 206.72 | 0.9993 | 0.38 | 0.64 | 14.24 | 119,378 |
| Arginine | 175.2→70.2 | 3–500 | y = 236.186x + 78.621 | 0.9969 | 0.34 | 0.57 | 8.82 | 85,396 |
| Tyrosine | 182.1→165.1 | 3–500 | y = 70.794x + 15.698 | 0.9992 | 1.08 | 1.8 | 14.47 | 113,759 |
| Tryptophan | 205.1→118.1 | 3–500 | y = 68.235x + 107.91 | 0.9984 | 1.92 | 3.21 | 14.01 | 91,491 |
| Analyte | Parent Ion–Product Ion Transition (m/z) | Range (µM) | Calibration Line | Linearity (r2) | LOD (µM) | LLOQ (µM) | tm (min) | N |
|---|---|---|---|---|---|---|---|---|
| Glycine | 76.2→30.1 | 15–500 | y = 2.6075x + 25.297 | 0.9976 | 5.45 | 9.08 | 11.96 | 95,241 |
| Alanine | 90.2→44.2 | 15–500 | y = 30.587x + 429.8 | 0.9997 | 1.62 | 2.71 | 13.24 | 97,838 |
| Serine | 106.2→60.2 | 3–500 | y = 26.391x + 247.5 | 0.9983 | 2.61 | 4.36 | 15.30 | 117,903 |
| Proline | 116.2→70.2 | 3–500 | y = 269.53x + 493.84 | 0.9984 | 0.40 | 0.66 | 17.51 | 99,395 |
| Valine | 118.2→72.0 | 3–500 | y = 126.42x + 165.05 | 0.9994 | 0.51 | 0.85 | 15.18 | 113,496 |
| Threonine | 120.2→74.2 | 15–500 | y = 29.596x + 266.15 | 0.9995 | 1.31 | 2.18 | 16.35 | 92,125 |
| Cysteine | 241.3→74.2 | 6.25–250 | y = 43.708x + 25.297 | 0.9993 | 0.31 | 0.52 | 17.96 | 99,185 |
| Isoleucine | 132.2→69.2 | 3–500 | y = 135.27x + 20.916 | 0.9991 | 0.23 | 0.38 | 15.45 | 156,381 |
| Leucine | 132.2→86.1 | 3–500 | y = 114.7x + 92.705 | 0.9979 | 0.93 | 1.54 | 15.68 | 116,776 |
| Asparagine | 133.1 →74.0 | 3–500 | y = 14.272x + 241.43 | 0.9968 | 1.24 | 2.07 | 16.33 | 80,394 |
| Aspartic acid | 134.1→74.1 | 15–500 | y = 23.42x + 291.32 | 0.9995 | 8.73 | 14.56 | 16.69 | 102,863 |
| Glutamine | 147.1→84.1 | 15–500 | y = 19.263x + 908.9 | 0.9897 | 1.37 | 2.28 | 16.81 | 93,292 |
| Lysine | 147.1→84.2 | 3–500 | y = 213.13x + 161.59 | 0.9944 | 0.45 | 0.75 | 9.14 | 97,211 |
| Glutamic acid | 148.1→84.2 | 3–500 | y = 109.51x + 210.19 | 0.9984 | 1.45 | 2.42 | 17.16 | 83,677 |
| Methionine | 150.1→104.1 | 3–500 | y = 40.467x + 29.661 | 0.9993 | 0.63 | 1.04 | 16.77 | 106,854 |
| Histidine | 156.1→110.2 | 15–500 | y = 293.33x + 700.5 | 0.9994 | 0.33 | 0.55 | 9.82 | 84,402 |
| Phenylalanine | 166.2→120.1 | 3–500 | y = 376.93x + 342.59 | 0.9991 | 0.32 | 0.53 | 17.45 | 101,095 |
| Arginine | 175.2→70.2 | 3–500 | y = 189.47x + 97.003 | 0.9992 | 0.22 | 0.37 | 9.56 | 86,366 |
| Tyrosine | 182.1→165.1 | 3–500 | y = 41.35x + 76.958 | 0.9980 | 1.39 | 2.32 | 17.86 | 95,190 |
| Tryptophan | 205.1→118.1 | 3–500 | y = 81.434x + 425.82 | 0.9993 | 0.84 | 1.40 | 16.96 | 100,132 |
| Analyte (QC Level) | Intra-Assay (n = 5) | Inter-Assay (n = 15) | Recovery (%) | ||||
|---|---|---|---|---|---|---|---|
| Mean Found Concentration (µM) | RSD (%) | RE (% Nom.) | Mean Found Concentration (µM) | RSD (%) | RE (% Nom.) | ||
| Glycine | |||||||
| QC low | 16.01 | 5.92 | 6.74 | 14.97 | 5.80 | −0.19 | 87.40 |
| QC medium | 147.29 | 5.47 | −1.81 | 140.76 | 6.65 | −6.16 | 92.31 |
| QC high | 380.42 | 6.01 | −4.90 | 386.94 | 1.62 | −3.26 | 101.99 |
| Alanine | |||||||
| QC low | 14.28 | 9.70 | −4.81 | 15.10 | 9.03 | 0.62 | 90.87 |
| QC medium | 147.36 | 3.57 | −1.76 | 145.97 | 3.50 | −2.69 | 90.49 |
| QC high | 407.07 | 2.32 | 1.77 | 411.73 | 3.99 | 2.93 | 94.50 |
| Serine | |||||||
| QC low | 14.20 | 2.60 | −5.32 | 14.62 | 4.61 | −2.53 | 94.55 |
| QC medium | 153.23 | 1.17 | 2.15 | 149.90 | 4.55 | −0.06 | 98.32 |
| QC high | 403.69 | 2.41 | 0.92 | 404.24 | 6.73 | 1.06 | 95.54 |
| Proline | |||||||
| QC low | 13.56 | 2.25 | −9.63 | 13.78 | 2.77 | −8.17 | 93.44 |
| QC medium | 151.94 | 3.54 | 1.29 | 151.50 | 6.22 | 1.00 | 92.82 |
| QC high | 399.40 | 4.92 | −0.15 | 402.24 | 5.88 | 0.56 | 94.03 |
| Valine | |||||||
| QC low | 14.32 | 1.26 | −4.53 | 14.53 | 2.30 | −3.15 | 95.54 |
| QC medium | 153.54 | 2.53 | 2.36 | 144.69 | 8.61 | −3.54 | 95.06 |
| QC high | 406.49 | 3.12 | 1.62 | 391.09 | 5.34 | −2.23 | 95.66 |
| Threonine | |||||||
| QC low | 15.57 | 2.06 | 3.76 | 15.48 | 2.55 | 3.17 | 89.61 |
| QC medium | 151.58 | 2.00 | 1.06 | 149.93 | 8.50 | −0.05 | 96.39 |
| QC high | 405.95 | 2.57 | 1.49 | 394.41 | 3.97 | −1.40 | 96.75 |
| Cysteine | |||||||
| QC low | 7.95 | 4.19 | 5.97 | 7.19 | 10.55 | −4.12 | 95.37 |
| QC medium | 79.33 | 2.83 | 5.78 | 77.27 | 5.86 | 3.02 | 101.25 |
| QC high | 206.96 | 0.91 | 3.48 | 204.89 | 0.93 | 2.44 | 99.99 |
| Isoleucine | |||||||
| QC low | 14.58 | 4.49 | −2.80 | 14.18 | 6.85 | −5.50 | 96.91 |
| QC medium | 148.98 | 3.87 | −0.68 | 144.84 | 3.35 | −3.44 | 92.57 |
| QC high | 402.74 | 4.38 | 0.69 | 392.06 | 5.31 | −1.99 | 93.42 |
| Leucine | |||||||
| QC low | 14.32 | 3.67 | −4.55 | 15.17 | 6.92 | 1.10 | 93.93 |
| QC medium | 153.09 | 3.54 | 2.06 | 140.17 | 11.32 | −6.55 | 94.89 |
| QC high | 428.14 | 2.88 | 7.03 | 409.89 | 6.84 | 2.47 | 97.88 |
| Asparagine | |||||||
| QC low | 13.87 | 3.62 | −7.53 | 14.37 | 11.38 | −4.23 | 97.34 |
| QC medium | 149.27 | 9.66 | −0.48 | 142.63 | 15.12 | −4.92 | 95.17 |
| QC high | 384.25 | 9.87 | −3.94 | 368.74 | 11.58 | −7.81 | 95.08 |
| Aspartic acid | |||||||
| QC low | 14.77 | 11.86 | −1.57 | 15.27 | 14.19 | 1.80 | 96.61 |
| QC medium | 135.74 | 9.66 | −9.51 | 143.26 | 12.89 | −4.49 | 92.55 |
| QC high | 388.46 | 4.31 | −2.89 | 394.17 | 8.20 | −1.46 | 97.08 |
| Glutamine | |||||||
| QC low | 15.74 | 5.73 | 4.91 | 15.78 | 6.27 | 5.18 | 91.39 |
| QC medium | 168.69 | 0.55 | 12.46 | 146.84 | 8.48 | −2.11 | 98.13 |
| QC high | 403.84 | 4.89 | 0.96 | 385.69 | 4.92 | −3.58 | 101.07 |
| Lysine | |||||||
| QC low | 13.15 | 3.18 | −12.37 | 14.36 | 13.13 | −4.27 | 85.07 |
| QC medium | 148.80 | 0.54 | −0.80 | 140.64 | 6.44 | −6.24 | 92.77 |
| QC high | 415.27 | 0.91 | 3.82 | 391.76 | 7.13 | −2.06 | 93.50 |
| Glutamic acid | |||||||
| QC low | 13.10 | 8.82 | −12.67 | 14.14 | 12.52 | −5.76 | 85.37 |
| QC medium | 136.29 | 4.59 | −9.14 | 132.94 | 4.45 | −11.37 | 94.66 |
| QC high | 396.10 | 5.53 | −0.98 | 383.54 | 10.43 | −4.12 | 97.82 |
| Methionine | |||||||
| QC low | 14.53 | 6.01 | −3.14 | 13.75 | 10.14 | −8.33 | 102.05 |
| QC medium | 163.29 | 3.63 | 8.86 | 148.17 | 8.65 | −1.22 | 100.27 |
| QC high | 407.77 | 3.10 | 1.94 | 381.27 | 8.36 | −4.68 | 98.70 |
| Histidine | |||||||
| QC low | 15.32 | 1.39 | 2.14 | 15.37 | 1.09 | 2.43 | 97.01 |
| QC medium | 145.08 | 2.97 | −3.28 | 143.03 | 2.87 | −4.65 | 97.51 |
| QC high | 395.78 | 4.89 | −1.06 | 386.85 | 2.15 | −3.29 | 96.18 |
| Phenylalanine | |||||||
| QC low | 14.19 | 1.71 | −5.42 | 14.74 | 6.88 | −1.73 | 96.97 |
| QC medium | 149.71 | 3.93 | −0.19 | 141.60 | 5.00 | −5.60 | 94.12 |
| QC high | 402.79 | 2.11 | 0.70 | 395.23 | 2.71 | −1.20 | 96.29 |
| Arginine | |||||||
| QC low | 12.82 | 2.45 | −14.53 | 14.22 | 14.06 | −5.22 | 86.30 |
| QC medium | 141.78 | 2.36 | −5.48 | 139.90 | 2.54 | −6.74 | 93.76 |
| QC high | 399.97 | 3.83 | −0.01 | 392.12 | 4.21 | −1.97 | 96.52 |
| Tyrosine | |||||||
| QC low | 14.46 | 2.20 | −3.63 | 12.86 | 10.63 | −14.26 | 89.19 |
| QC medium | 160.07 | 1.93 | 6.71 | 148.59 | 10.78 | −0.94 | 97.28 |
| QC high | 421.06 | 3.42 | 5.26 | 403.01 | 6.44 | 0.75 | 97.25 |
| Tryptophan | |||||||
| QC low | 14.92 | 2.93 | −0.54 | 14.22 | 6.84 | −5.22 | 97.04 |
| QC medium | 154.52 | 3.49 | 3.01 | 146.82 | 7.57 | −2.12 | 100.37 |
| QC high | 381.42 | 3.45 | −4.64 | 394.32 | 5.66 | −1.42 | 98.35 |
| Room Temperature Stability (24 h) | Freeze-Thaw Stability (3 cycles) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| QC Low | QC Medium | QC High | QC Low | QC Medium | QC High | |||||||
| Analyte | Found (μM) | Accuracy (%RE) | Found (μM) | Accuracy (%RE) | Found (μM) | Accuracy (%RE) | Found (μM) | Accuracy (%RE) | Found (μM) | Accuracy (%RE) | Found (μM) | Accuracy (%RE) |
| Gly | 14.64 | −2.40 | 145.32 | −3.12 | 374.30 | −6.43 | 14.79 | −1.41 | 138.42 | −7.72 | 370.53 | −7.37 |
| Ala | 14.12 | −5.86 | 154.73 | 3.16 | 386.75 | −3.31 | 14.60 | −2.66 | 145.56 | −2.96 | 357.90 | −10.53 |
| Ser | 14.92 | −0.55 | 157.30 | 4.87 | 376.52 | −5.87 | 15.59 | 3.90 | 144.91 | −3.40 | 363.74 | −9.07 |
| Pro | 14.86 | −0.95 | 152.39 | 1.59 | 388.29 | −2.93 | 15.29 | 1.92 | 150.52 | 0.35 | 386.73 | −3.32 |
| Val | 15.53 | 3.53 | 149.41 | −0.39 | 394.89 | −1.28 | 15.28 | 1.85 | 145.22 | −3.18 | 390.97 | −2.26 |
| Thr | 15.02 | 0.08 | 155.39 | 3.59 | 389.64 | −2.59 | 15.19 | 1.28 | 151.65 | 1.10 | 388.38 | −2.90 |
| Cys | 7.40 | −1.34 | 77.00 | 2.67 | 190.38 | −4.81 | 7.15 | −4.69 | 71.13 | −5.17 | 192.74 | −3.63 |
| Ile | 14.23 | −5.16 | 155.00 | 3.33 | 382.15 | −4.46 | 14.30 | −4.68 | 149.28 | −0.48 | 374.61 | −6.35 |
| Leu | 14.10 | −6.03 | 152.15 | 1.43 | 393.09 | −1.73 | 15.10 | 0.64 | 147.27 | −1.82 | 405.10 | 1.27 |
| Asn | 15.44 | 2.89 | 161.33 | 7.56 | 371.20 | −7.20 | 16.47 | 9.78 | 152.48 | 1.65 | 366.67 | −8.33 |
| Asp | 13.73 | −8.48 | 148.65 | −0.90 | 385.64 | −3.59 | 14.77 | −1.55 | 160.39 | 6.93 | 390.99 | −2.25 |
| Gln | 13.64 | −9.07 | 132.85 | −11.44 | 387.83 | −3.04 | 15.62 | 4.15 | 144.06 | −3.96 | 371.99 | −7.00 |
| Glu | 13.39 | −10.74 | 136.96 | −8.70 | 397.61 | −0.60 | 15.69 | 4.61 | 133.82 | −10.78 | 376.95 | −5.76 |
| Lys | 13.98 | −6.83 | 142.07 | −5.29 | 361.19 | −9.71 | 14.23 | −5.19 | 142.96 | −4.69 | 388.88 | −2.78 |
| Met | 13.87 | −7.54 | 148.54 | −0.97 | 395.56 | −1.11 | 14.02 | −6.55 | 140.39 | −6.41 | 364.36 | −8.91 |
| His | 15.18 | 1.16 | 154.24 | 2.83 | 390.41 | −2.40 | 14.80 | −1.35 | 139.13 | −7.24 | 362.44 | −9.39 |
| Phe | 14.96 | −0.27 | 154.91 | 3.27 | 386.71 | −3.32 | 15.98 | 6.49 | 142.55 | −4.97 | 361.24 | −9.69 |
| Arg | 14.63 | −2.50 | 154.02 | 2.68 | 383.41 | −4.15 | 14.81 | −1.32 | 142.20 | −5.20 | 372.78 | −6.81 |
| Tyr | 15.43 | 2.82 | 155.02 | 3.35 | 413.04 | 3.26 | 13.97 | −6.90 | 157.89 | 5.26 | 409.93 | 2.48 |
| Trp | 14.46 | −3.64 | 149.86 | −0.10 | 393.38 | −1.66 | 13.86 | −7.60 | 138.59 | −7.61 | 377.94 | −5.52 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piestansky, J.; Olesova, D.; Galba, J.; Marakova, K.; Parrak, V.; Secnik, P.; Secnik, P., Jr.; Kovacech, B.; Kovac, A.; Zelinkova, Z.; et al. Profiling of Amino Acids in Urine Samples of Patients Suffering from Inflammatory Bowel Disease by Capillary Electrophoresis-Mass Spectrometry. Molecules 2019, 24, 3345. https://doi.org/10.3390/molecules24183345
Piestansky J, Olesova D, Galba J, Marakova K, Parrak V, Secnik P, Secnik P Jr., Kovacech B, Kovac A, Zelinkova Z, et al. Profiling of Amino Acids in Urine Samples of Patients Suffering from Inflammatory Bowel Disease by Capillary Electrophoresis-Mass Spectrometry. Molecules. 2019; 24(18):3345. https://doi.org/10.3390/molecules24183345
Chicago/Turabian StylePiestansky, Juraj, Dominika Olesova, Jaroslav Galba, Katarina Marakova, Vojtech Parrak, Peter Secnik, Peter Secnik, Jr., Branislav Kovacech, Andrej Kovac, Zuzana Zelinkova, and et al. 2019. "Profiling of Amino Acids in Urine Samples of Patients Suffering from Inflammatory Bowel Disease by Capillary Electrophoresis-Mass Spectrometry" Molecules 24, no. 18: 3345. https://doi.org/10.3390/molecules24183345