Rapid and Nondestructive Measurement of Rice Seed Vitality of Different Years Using Near-Infrared Hyperspectral Imaging
Abstract
1. Introduction
2. Results and Discussion
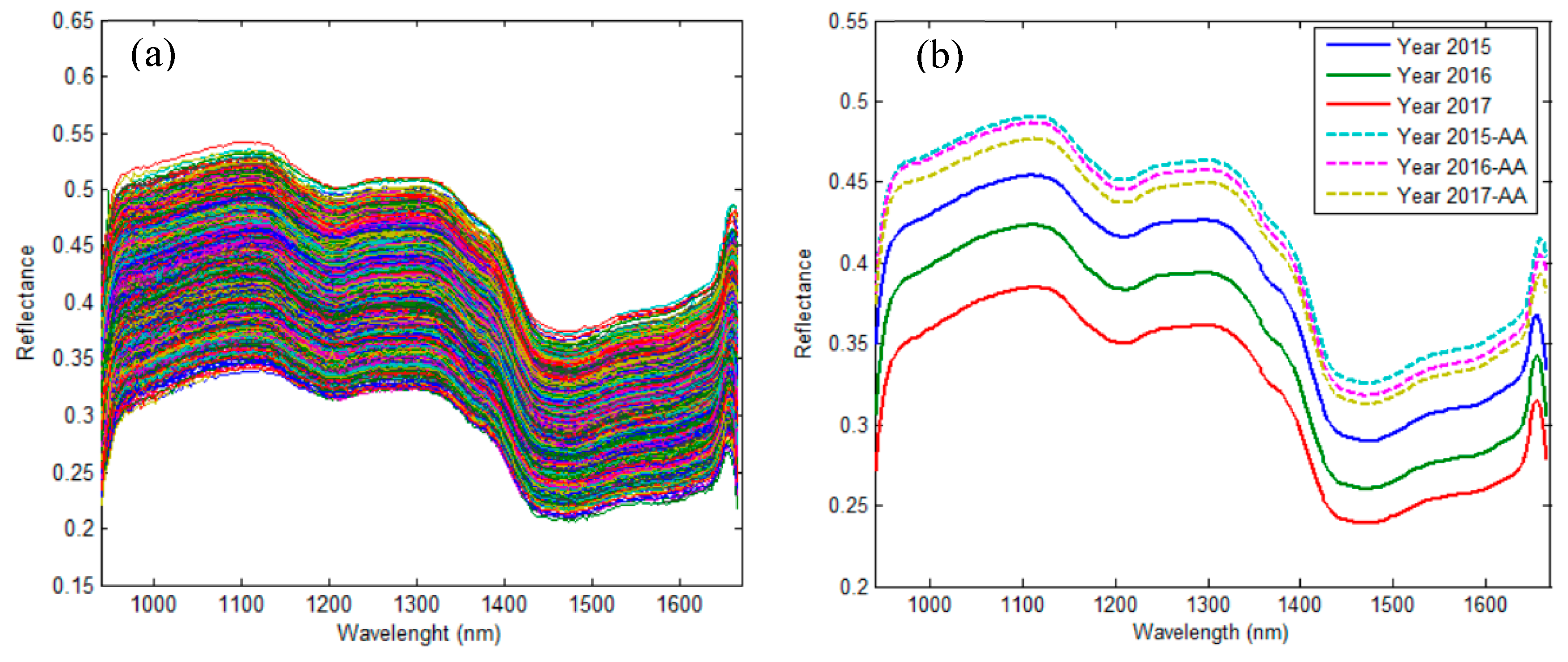
2.1. Spectral Interpretation
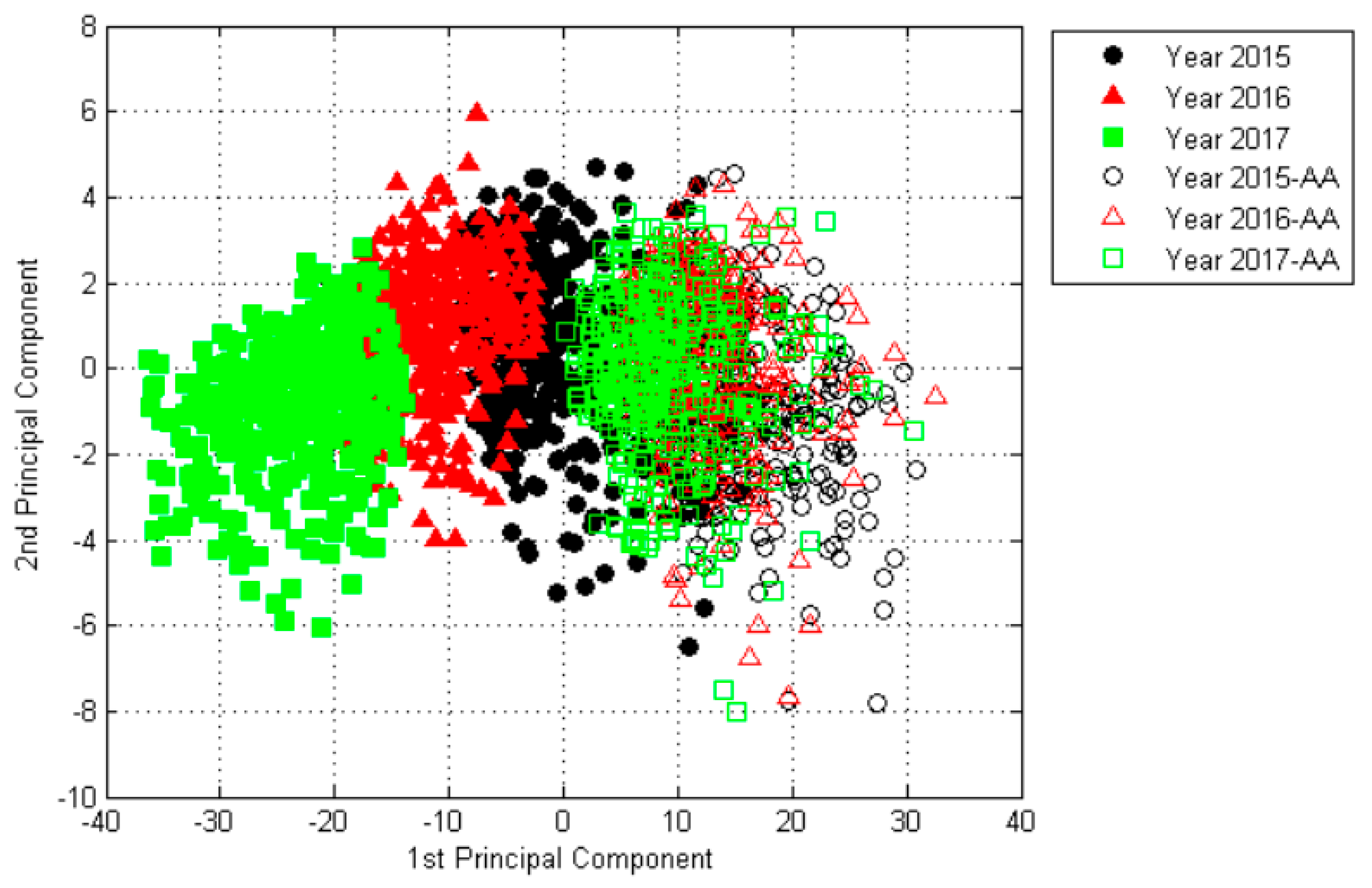
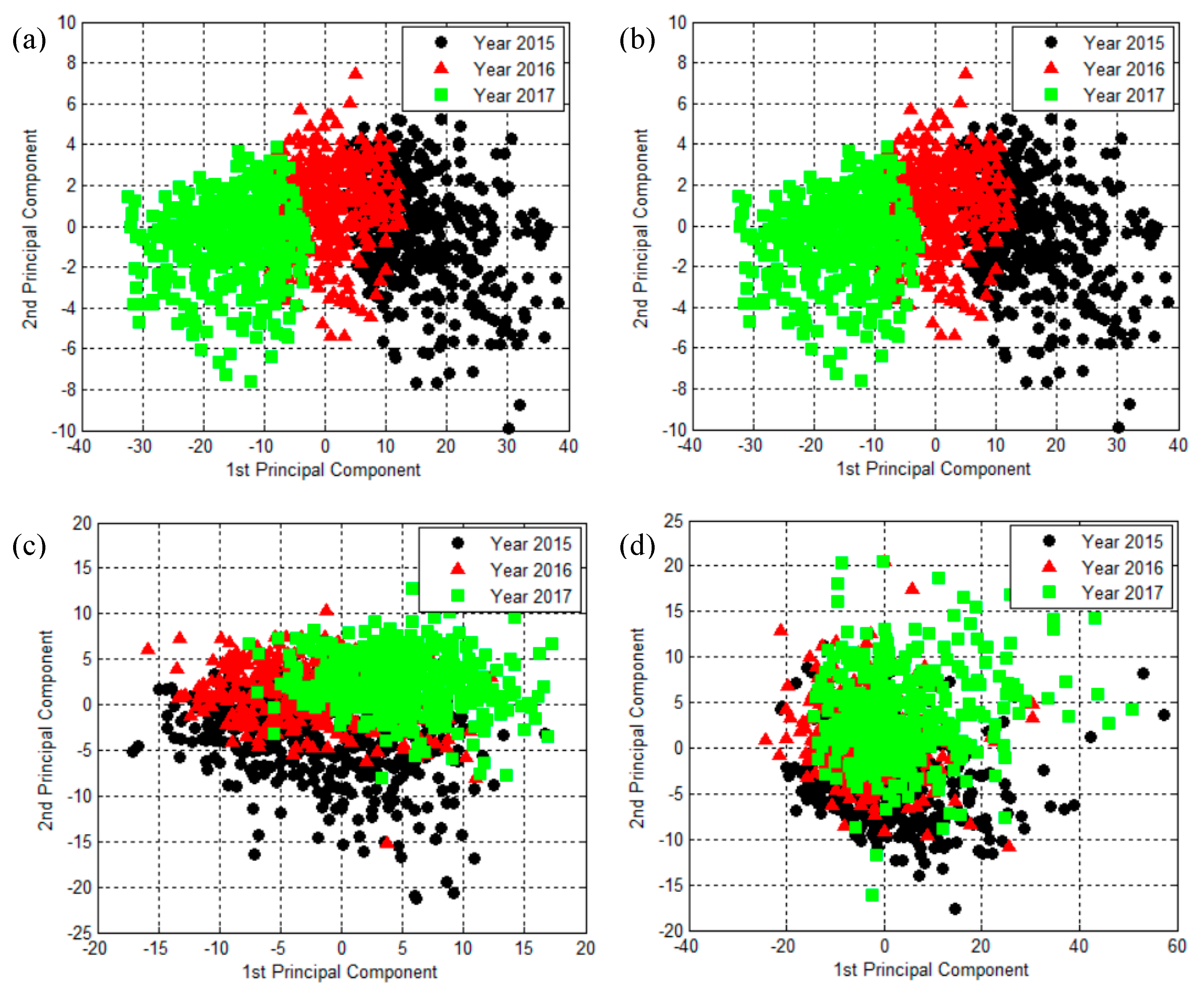
2.2. The Results of Principal Component Analysis
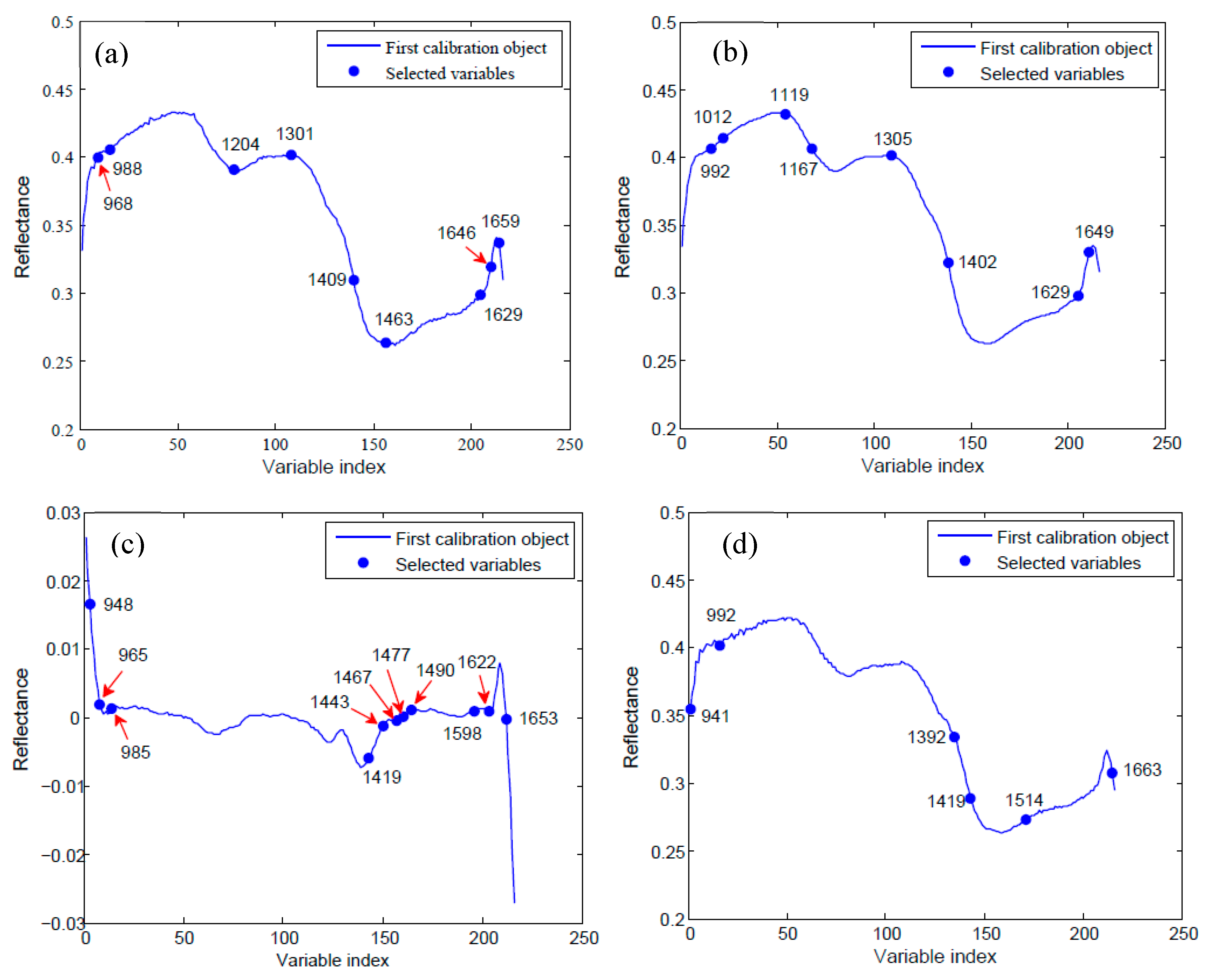
2.3. Optimal Wavelengths Selection
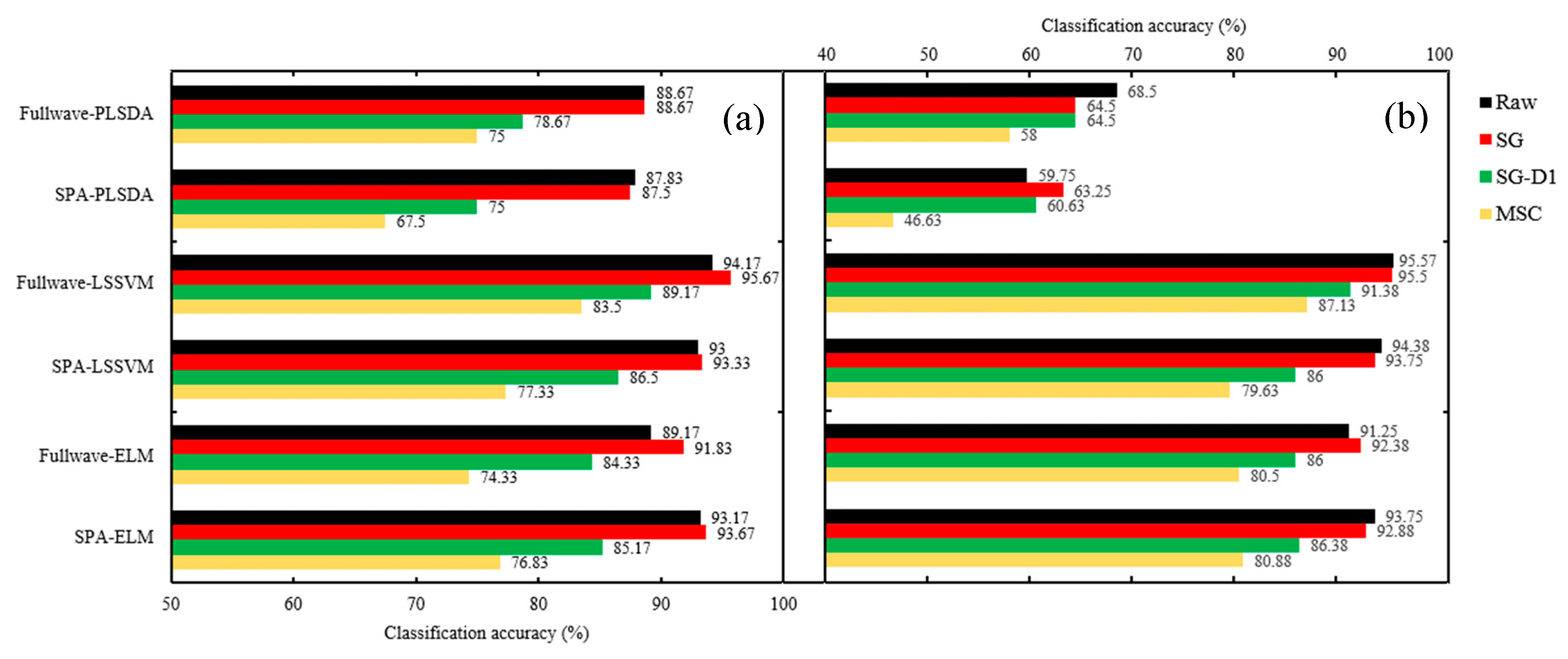
2.4. Classification Model Results
2.4.1. Assessment of Seed Vitality of Three Different Years
2.4.2. Identifying Non-Viable Seeds from Viable Seeds of Different Years.
3. Materials and Methods
3.1. Samples and Sample Preparation
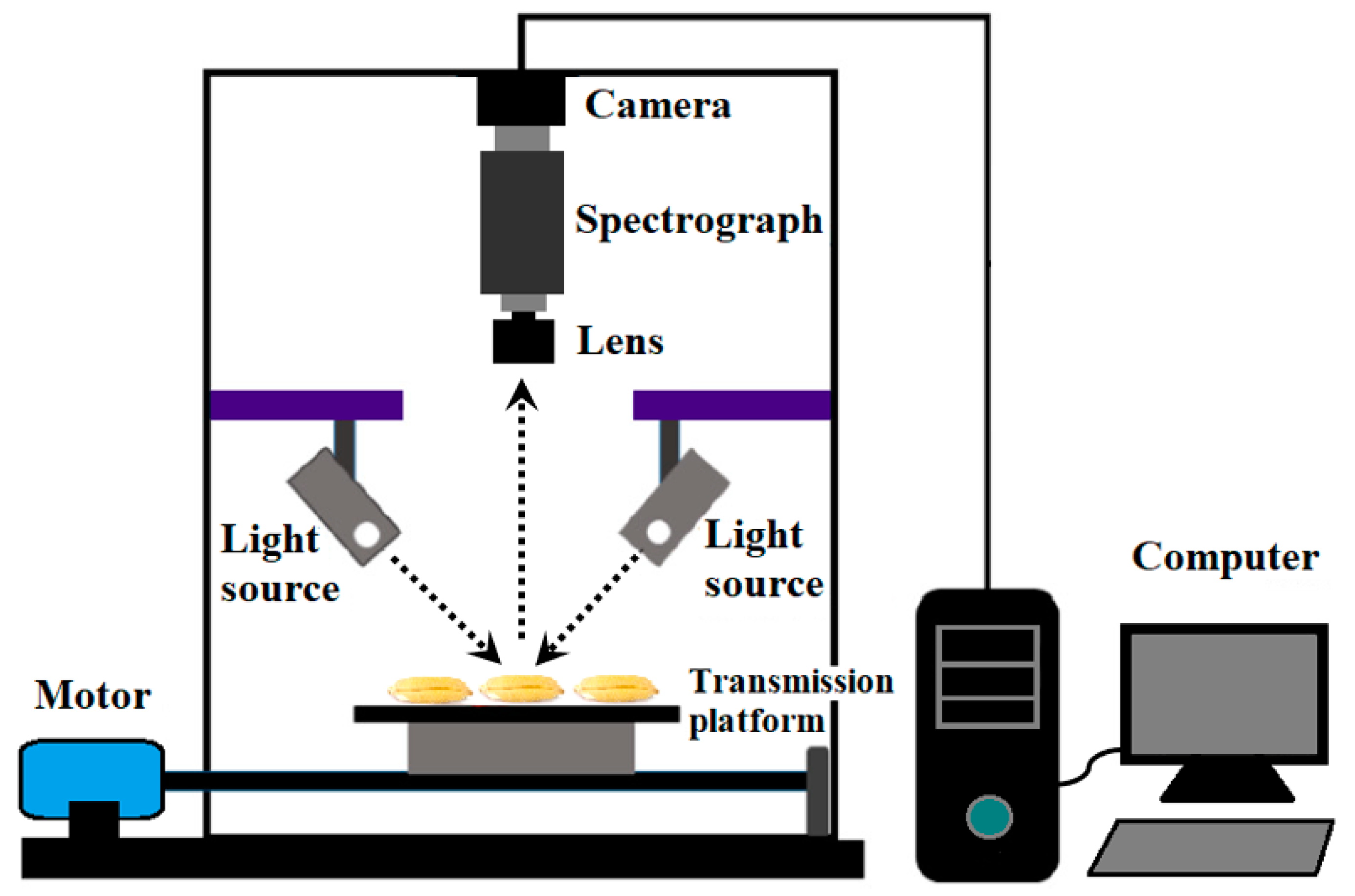
3.2. Hyperspectral Image Collection
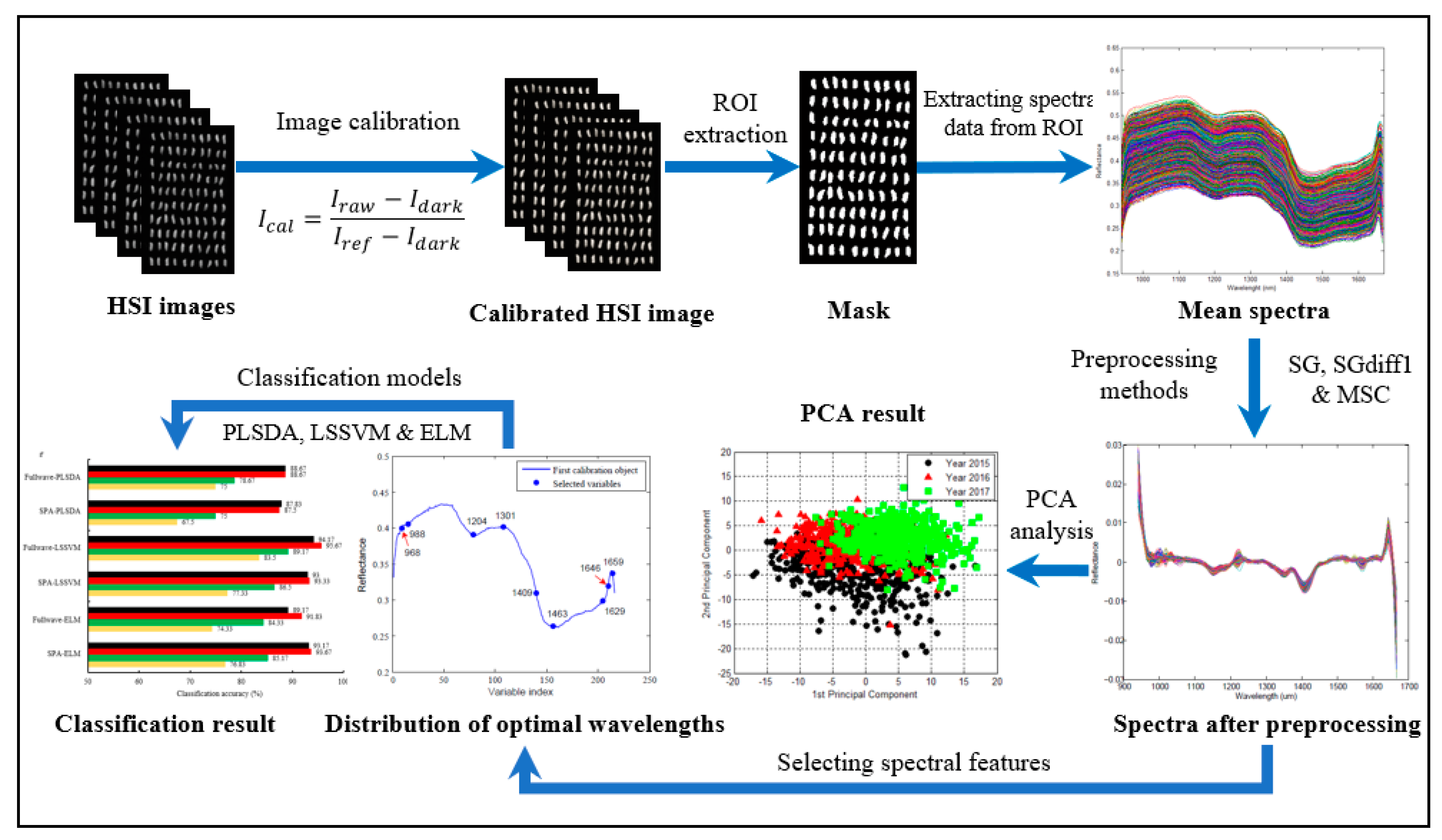
3.3. Data Extraction and Preprocessing
3.4. Spectral Feature Selection
3.5. Construction and Analysis of Classification Models
3.6. Germination Test
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Database of Crops from Food and Agriculture Organization of the United Nations. Available online: http://www.fao.org/faostat/en/#data/QC (accessed on 20 April 2019).
- World Population (2019 and Historical) from Worldometers. Available online: http://www.worldometers.info/world-population/#table-historical (accessed on 20 April 2019).
- Wakholi, C.; Kandpal, L.M.; Lee, H.; Bae, H.; Park, E.; Kim, M.S.; Mo, C.; Lee, W.; Cho, B. Rapid assessment of corn seed viability using short wave infrared line-scan hyperspectral imaging and chemometrics. Sens. Actuators B Chem. 2018, 255, 498–507. [Google Scholar] [CrossRef]
- Ramesh, K.; Rao, A.N.; Chauhan, B.S. Role of crop competition in managing weeds in rice, wheat, and maize in India: A review. Crop Prot. 2017, 95, 14–21. [Google Scholar] [CrossRef]
- Angélica, B.R.; Julio, M.F. Onion seed vigor in relation to plant growth and yield. Hortic. Bras. 2003, 21, 220–226. [Google Scholar]
- Finch-Savage, W.E.; Bassel, G.W. Seed vigour and crop establishment: Extending performance beyond adaptation. J. Exp. Bot. 2016, 67, 567–591. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, P.; Joshi, D.K.; Nagarajan, S.; Moharir, A.V. Characterization of germinating and non-viable soybean seeds by nuclear magnetic resonance (NMR) spectroscopy. Seed Sci. Res. 2004, 14, 355–362. [Google Scholar] [CrossRef]
- Al-Turki, T.A.; Baskin, C.C. Determination of seed viability of eight wild Saudi Arabian species by germination and X-ray tests. Saudi J. Biol. Sci. 2017, 24, 822–829. [Google Scholar] [CrossRef][Green Version]
- Al-Hammad, B.A.; Al-Ammari, B.S. Seed viability of five wild Saudi Arabian species by germination and X-ray tests. Saudi J. Biol. Sci. 2017, 24, 1424–1429. [Google Scholar] [CrossRef]
- Braga, R.A.; Dal Fabbro, I.M.; Borem, F.M.; Rabelo, G.; Arizaga, R.; Rabal, H.J.; Trivi, M. Assessment of Seed Viability by Laser Speckle Techniques. Biosyst. Eng. 2003, 86, 287–294. [Google Scholar] [CrossRef]
- Fatonah, K.; Suliansyah, I.; Rozen, N. Electrical conductivity for seed vigor test in sorghum (Sorghum bicolor). Cell Biol. Dev. 2017, 1, 6–12. [Google Scholar] [CrossRef]
- Feng, X.; Yu, C.; Chen, Y.; Peng, J.; Ye, L.; Shen, T.; Wen, H.; He, Y. Non-Destructive Determination of Shikimic Acid Concentration in Transgenic Maize Exhibiting Glyphosate Tolerance Using Chlorophyll Fluorescence and Hyperspectral Imaging. Front. Plant Sci. 2018, 9, 468. [Google Scholar] [CrossRef]
- Doherty, B.; Daveri, A.; Clementi, C.; Romani, A.; Bioletti, S.; Brunetti, B.; Sgamellotti, A.; Miliani, C. The Book of Kells: A non-invasive MOLAB investigation by complementary spectroscopic techniques. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2013, 115, 330–336. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, N.; Whitworth, M.B.; Fisk, I.D. Near-Infrared spectroscopy and hyperspectral imaging for non-destructive quality assessment of cereal grains. Appl. Spectrosc. Rev. 2018, 53, 667–687. [Google Scholar] [CrossRef]
- Park, B. Future Trends in Hyperspectral Imaging. NIR News 2016, 27, 35–38. [Google Scholar] [CrossRef]
- Ambrose, A.; Lohumi, S.; Lee, W.; Cho, B.K. Comparative nondestructive measurement of corn seed viability using Fourier transform near-infrared (FT-NIR) and Raman spectroscopy. Sens. Actuators B Chem. 2016, 224, 500–506. [Google Scholar] [CrossRef]
- Han, L.; Mao, P.; Wang, X.; Wang, Y. Study on vigour test of oat seeds with near infrared reflectance spectroscopy. J. Infrared Millim. Waves 2008, 27, 86–90. [Google Scholar] [CrossRef]
- Kandpal, L.M.; Lohumi, S.; Kim, M.S.; Kang, J.; Cho, B. Near-infrared hyperspectral imaging system coupled with multivariate methods to predict viability and vigor in muskmelon seeds. Sens. Actuators B Chem. 2016, 229, 534–544. [Google Scholar] [CrossRef]
- Al-Amery, M.; Geneve, R.L.; Sanches, M.F.; Armstrong, P.R.; Maghirang, E.B.; Lee, C.; Vieira, R.D.; Hildebrand, D.F. Near-infrared spectroscopy used to predict soybean seed germination and vigour. Seed Sci. Res. 2018, 28, 245–252. [Google Scholar] [CrossRef]
- Qi, X.W.; Li, W.K.; Li, W.; Li, H. Study on the Vigour Testing of Soybean Seed Based on Near Infrared Spectroscopy Technology. Appl. Mech. Mater. 2011, 58, 458–462. [Google Scholar] [CrossRef]
- Lohumi, S.; Mo, C.; Kang, J.; Hong, S.; Cho, B. Nondestructive Evaluation for the Viability of Watermelon (Citrullus lanatus) Seeds Using Fourier Transform Near Infrared Spectroscopy. J. Biosyst. Eng. 2013, 38, 312–317. [Google Scholar] [CrossRef]
- Sun, J.; Lu, X.; Mao, H.; Jin, X.; Wu, X. A method for rapid identification of rice origin by hyperspectral imaging technology. J. Food Process Eng. 2015, 40, e12297. [Google Scholar] [CrossRef]
- Liu, W.; Liu, C.; Ma, F.; Lu, X.; Yang, J.; Zheng, L. Online Variety Discrimination of Rice Seeds Using Multispectral Imaging and Chemometric Methods. J. Appl. Spectrosc. 2016, 82, 993–999. [Google Scholar] [CrossRef]
- Wang, L.; Liu, D.; Pu, H.; Sun, D.; Gao, W.; Xiong, Z. Use of Hyperspectral Imaging to Discriminate the Variety and Quality of Rice. Food Anal. Methods 2015, 8, 515–523. [Google Scholar] [CrossRef]
- Qiu, Z.; Chen, J.; Zhao, Y.; Zhu, S.; He, Y.; Zhang, C. Variety Identification of Single Rice Seed Using Hyperspectral Imaging Combined with Convolutional Neural Network. Appl. Sci. 2018, 8, 212. [Google Scholar] [CrossRef]
- Chen, S.; Huang, C.; Huang, C.; Yang, C.; Wu, T.; Tsai, Y.; Miao, P. Determination of Nitrogen Content in Rice Crop Using Multi-Spectral Imaging. In Proceedings of the 2003 ASAE Annual Meeting, Las Vegas, NV, USA, 27–30 July 2003; Available online: https://elibrary.asabe.org/azdez.asp?JID=5&AID=13741&CID=lnv2003&T=2 (accessed on 21 April 2019).
- Lin, L.H.; Lu, F.M.; Chang, Y.C. Development of a Near-Infrared Imaging System for Determination of Rice Moisture. Cereal Chem. 2006, 83, 498–504. [Google Scholar] [CrossRef]
- Liu, M.; Liu, X.; Wu, M.; Li, L.; Xiu, L. Integrating spectral indices with environmental parameters for estimating heavy metal concentrations in rice using a dynamic fuzzy neural-network model. Comput. Geosci. 2011, 37, 1642–1652. [Google Scholar] [CrossRef]
- Abdi, H.; Williams, L.J. Principal component analysis. WIREs Comput. Stat. 2010, 2, 433–459. [Google Scholar] [CrossRef]
- Tsenkova, R.; Munćan, J.; Pollner, B.; Kovacs, Z. Essentials of Aquaphotomics and Its Chemometrics Approaches. Front. Chem. 2018, 6, 363. [Google Scholar] [CrossRef]
- Aenugu, H.P.R.; Kumar, D.S.; Srisudharson, N.P.; Ghosh, S.; Banji, D. Near infra red spectroscopy—An overview. Int. J. ChemTech Res. 2011, 3, 825–836. [Google Scholar]
- Lee, J.; Welti, R.; Roth, M.; Schapaugh, W.T.; Li, J.; Trick, H.N. Enhanced seed viability and lipid compositional changes during natural ageing by suppressing phospholipase Dα in soybean seed. Plant Biotechnol. J. 2012, 10, 164–173. [Google Scholar] [CrossRef]
- Eldin, A.B. Near Infra Red Spectroscopy; INTECH Open Access Publisher: London, UK, 2011; Available online: https://www.intechopen.com/books/wide-spectra-of-quality-control/near-infra-red-spectroscopy (accessed on 19 April 2019).
- Uarrota, V.G.; Moresco, R.; Coelho, B.; Nunes, E.C.; Peruch, L.A.M.; Neubert, E.O.; Rocha, M.; Maraschin, M. Metabolomics combined with chemometric tools (PCA, HCA, PLS-DA and SVM) for screening cassava (Manihot esculenta Crantz) roots during postharvest physiological deterioration. Food Chem. 2014, 161, 67–78. [Google Scholar] [CrossRef]
- Ambrose, A.; Lee, W.H.; Cho, B.K. Effect of microwave heat treatment on inhibition of corn seed germination. J. Biosyst. Eng. 2015, 40, 224–231. [Google Scholar] [CrossRef]
- Maleki, M.R.; Mouazen, A.M.; Ramon, H.; Baerdemaeker, J. Multiplicative scatter correction during on-line measurement with near infrared spectroscopy. Biosyst. Eng. 2007, 96, 427–433. [Google Scholar] [CrossRef]
- Araújo, M.C.U.; Saldanha, T.C.B.; Galvão, R.K.H.; Yoneyama, T.; Chame, H.C.; Visani, V. The successive projections algorithm for variable selection in spectroscopic multicomponent analysis. Chemom. Intell. Lab. Syst. 2001, 57, 65–73. [Google Scholar] [CrossRef]
- Zhang, J.; Dai, L.; Cheng, F. Classification of Frozen Corn Seeds Using Hyperspectral VIS/NIR Reflectence Imaging. Molecules 2019, 24, 149. [Google Scholar] [CrossRef] [PubMed]
- Khosravani, H.S.; Nabipour, M. Application of LSSVM algorithm as a novel tool for prediction of density of bitumen and heavy n-alkane mixture. Pet. Sci. Technol. 2018, 36, 1137–1142. [Google Scholar] [CrossRef]
- Huang, G.B.; Zhu, Q.Y.; Siew, C.K. Extreme learning machine: Theory and applications. Neurocomputing 2006, 70, 489–501. [Google Scholar] [CrossRef]
- International Seed Testing Association. International Rules for Seed Testing 2018; International Seed Testing Association: Bassersdorf, Switzerland, 2018. [Google Scholar]
Sample Availability: Samples of the compounds are not available from the authors. |
| Years of Seed | Treatment | Germination Number | Non-Germination Number | Germination Rate (GR) | Vitality Index (VI) |
|---|---|---|---|---|---|
| 2015 | − | 113 | 27 | 80.71% | 154.15 |
| AA | 0 | 140 | 0 | 0 | |
| 2016 | − | 130 | 10 | 92.86% | 225.6 |
| AA | 0 | 140 | 0 | 0 | |
| 2017 | − | 133 | 7 | 95% | 261.26 |
| AA | 0 | 140 | 0 | 0 |
| IVY | INV | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PLS-DA | LS-SVM | ELM | PLS-DA | LS-SVM | ELM | ||||||||
| Full. | Sel. | Full. | Sel. | Full. | Sel. | Full. | Sel. | Full. | Sel. | Full. | Sel. | ||
| Raw | Cal. | 92.17 | 86.83 | 96.67 | 95.83 | 95.5 | 93.5 | 69.75 | 58.38 | 96 | 95.13 | 94.75 | 94.13 |
| Pre. | 88.67 | 87.83 | 94.17 | 93 | 89.17 | 93.17 | 68.5 | 59.75 | 95.57 | 94.38 | 91.25 | 93.75 | |
| SG | Cal. | 87.75 | 87 | 97.5 | 94.33 | 95.67 | 94.17 | 62.63 | 62.13 | 96.38 | 93.5 | 95.25 | 93.13 |
| Pre. | 88.67 | 87.5 | 95.67 | 93.33 | 91.83 | 93.67 | 64.5 | 63.25 | 95.5 | 93.75 | 92.38 | 92.88 | |
| SG-D1 | Cal. | 79.17 | 73.67 | 94.67 | 86.17 | 90.17 | 85.5 | 66.25 | 61.13 | 95.75 | 87.13 | 91 | 86.38 |
| Pre. | 78.67 | 75 | 89.17 | 86.5 | 84.33 | 85.17 | 64.5 | 60.63 | 91.38 | 86 | 86 | 86.38 | |
| MSC | Cal. | 78.83 | 64.67 | 87.33 | 78 | 82.83 | 79 | 61.25 | 48.75 | 94.25 | 77.88 | 86.25 | 80.63 |
| Pre. | 75 | 67.5 | 83.5 | 77.33 | 74.33 | 76.83 | 58 | 46.63 | 87.13 | 79.63 | 80.5 | 80.88 | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, X.; Feng, X.; Sun, D.; Liu, F.; Bao, Y.; He, Y. Rapid and Nondestructive Measurement of Rice Seed Vitality of Different Years Using Near-Infrared Hyperspectral Imaging. Molecules 2019, 24, 2227. https://doi.org/10.3390/molecules24122227
He X, Feng X, Sun D, Liu F, Bao Y, He Y. Rapid and Nondestructive Measurement of Rice Seed Vitality of Different Years Using Near-Infrared Hyperspectral Imaging. Molecules. 2019; 24(12):2227. https://doi.org/10.3390/molecules24122227
Chicago/Turabian StyleHe, Xiantao, Xuping Feng, Dawei Sun, Fei Liu, Yidan Bao, and Yong He. 2019. "Rapid and Nondestructive Measurement of Rice Seed Vitality of Different Years Using Near-Infrared Hyperspectral Imaging" Molecules 24, no. 12: 2227. https://doi.org/10.3390/molecules24122227
APA StyleHe, X., Feng, X., Sun, D., Liu, F., Bao, Y., & He, Y. (2019). Rapid and Nondestructive Measurement of Rice Seed Vitality of Different Years Using Near-Infrared Hyperspectral Imaging. Molecules, 24(12), 2227. https://doi.org/10.3390/molecules24122227









