A Machine Learning Approach for the Discovery of Ligand-Specific Functional Mechanisms of GPCRs
Abstract
1. Introduction
2. Results
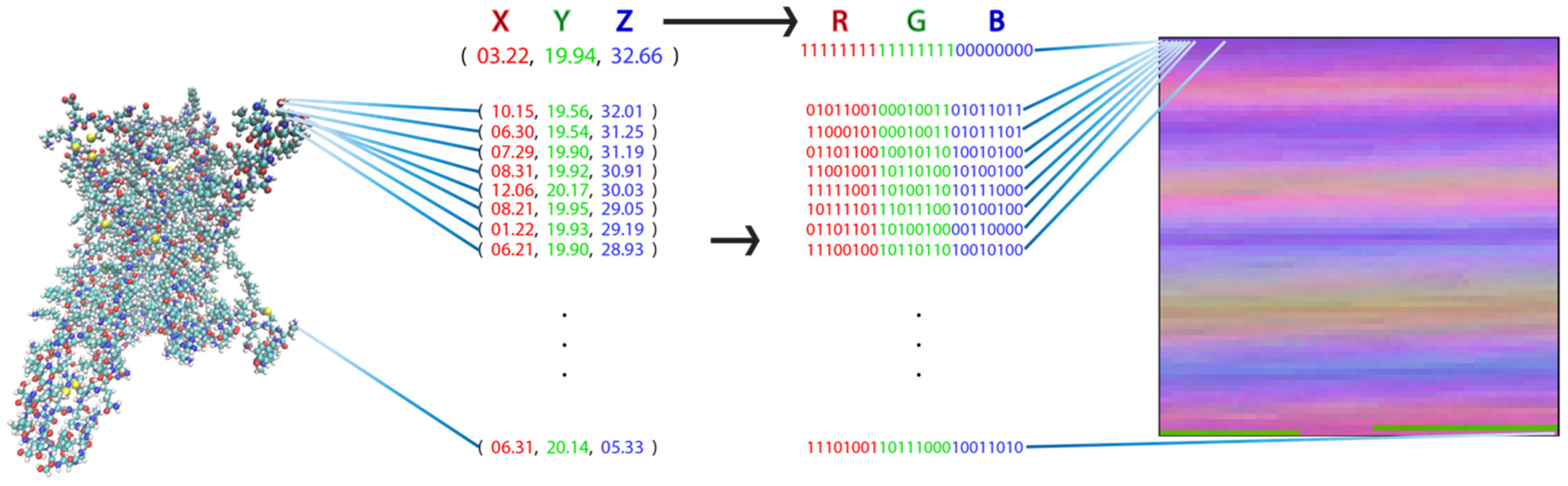
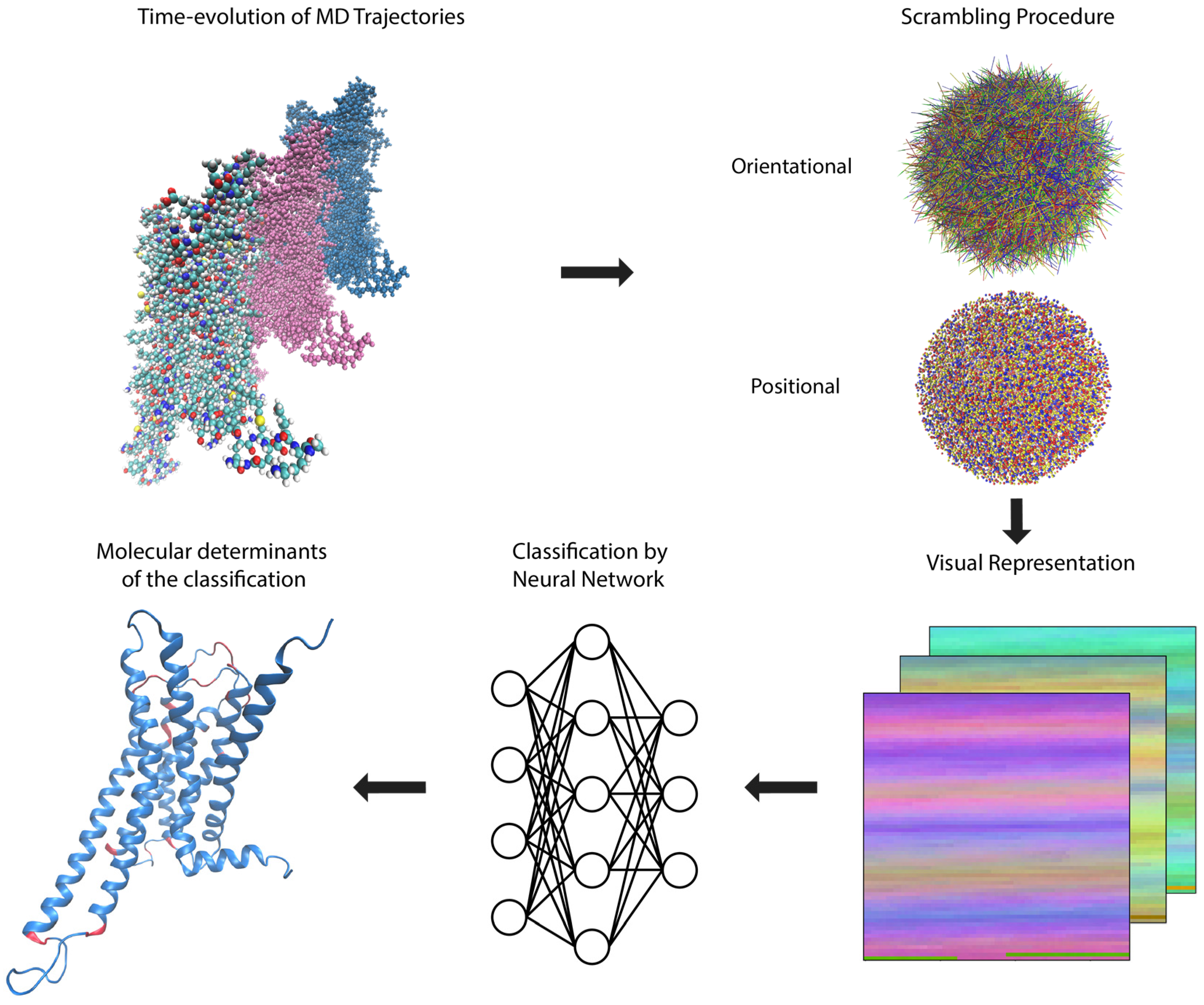
2.1. Transformation of MD Trajectories into Pixel Representations Readable by DNNs
2.2. Training and Application of DNNs Able to Recognize Ligand-Dependent Receptor Conformations
2.2.1. General Protocol
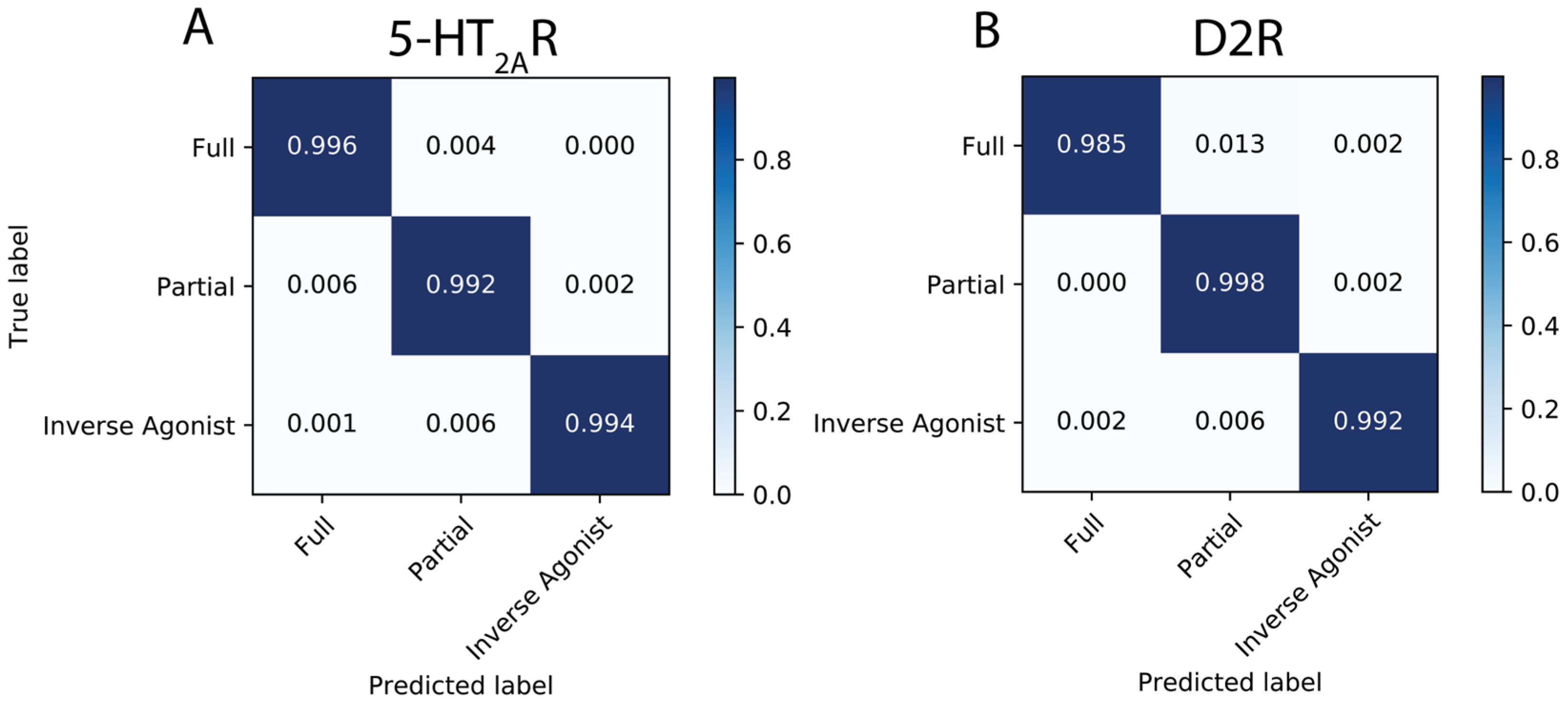
2.2.2. Pharmacological Classification of the MD Trajectories of 5-HT2AR-ligand Complexes
2.2.3. Pharmacological Classification of D2R-Ligand Trajectories
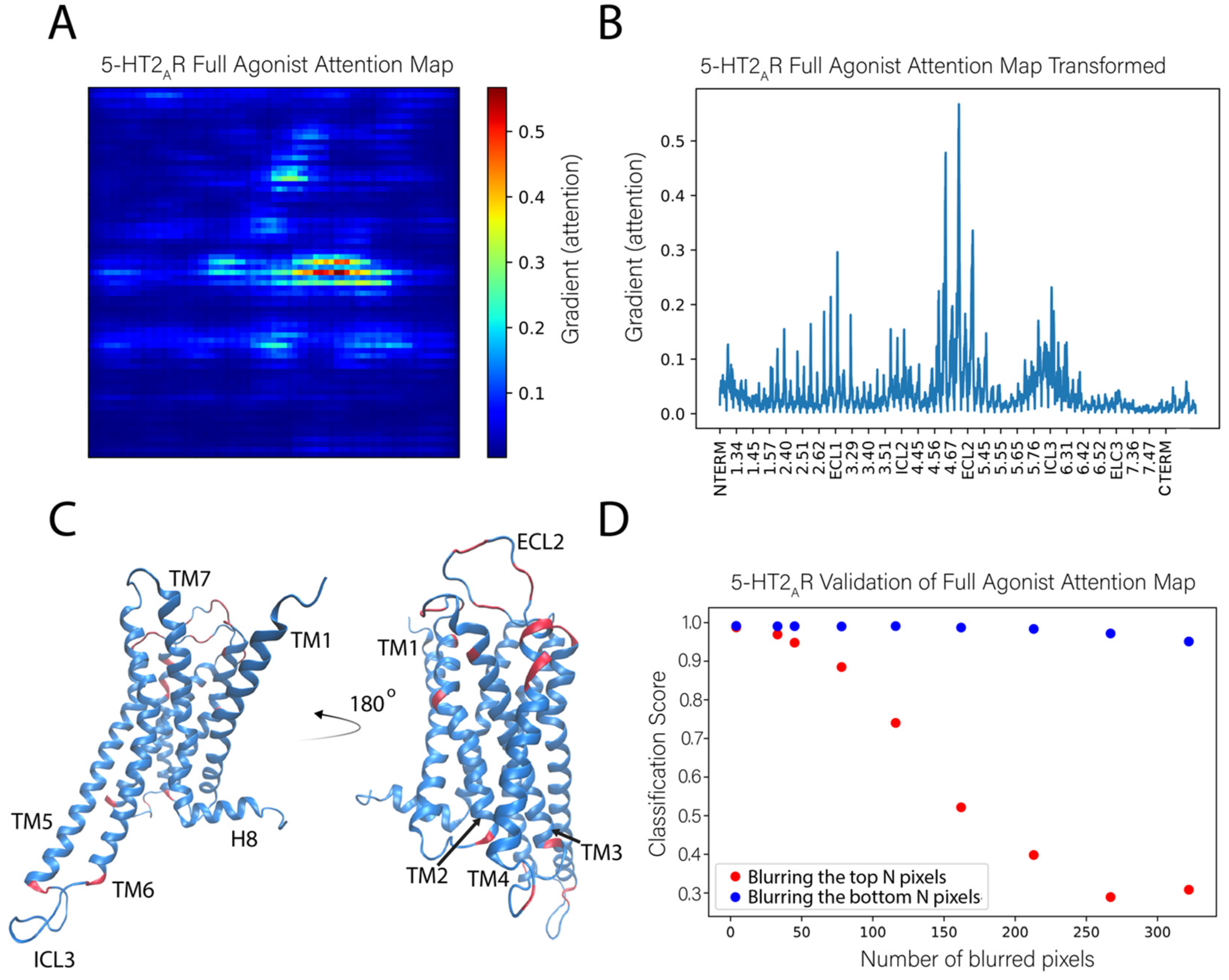
2.3. Identifying the Molecular Determinants of the Classification
2.3.1. General Protocol
2.3.2. Application of the Sensitivity Analysis to the 5-HT2AR Classifications
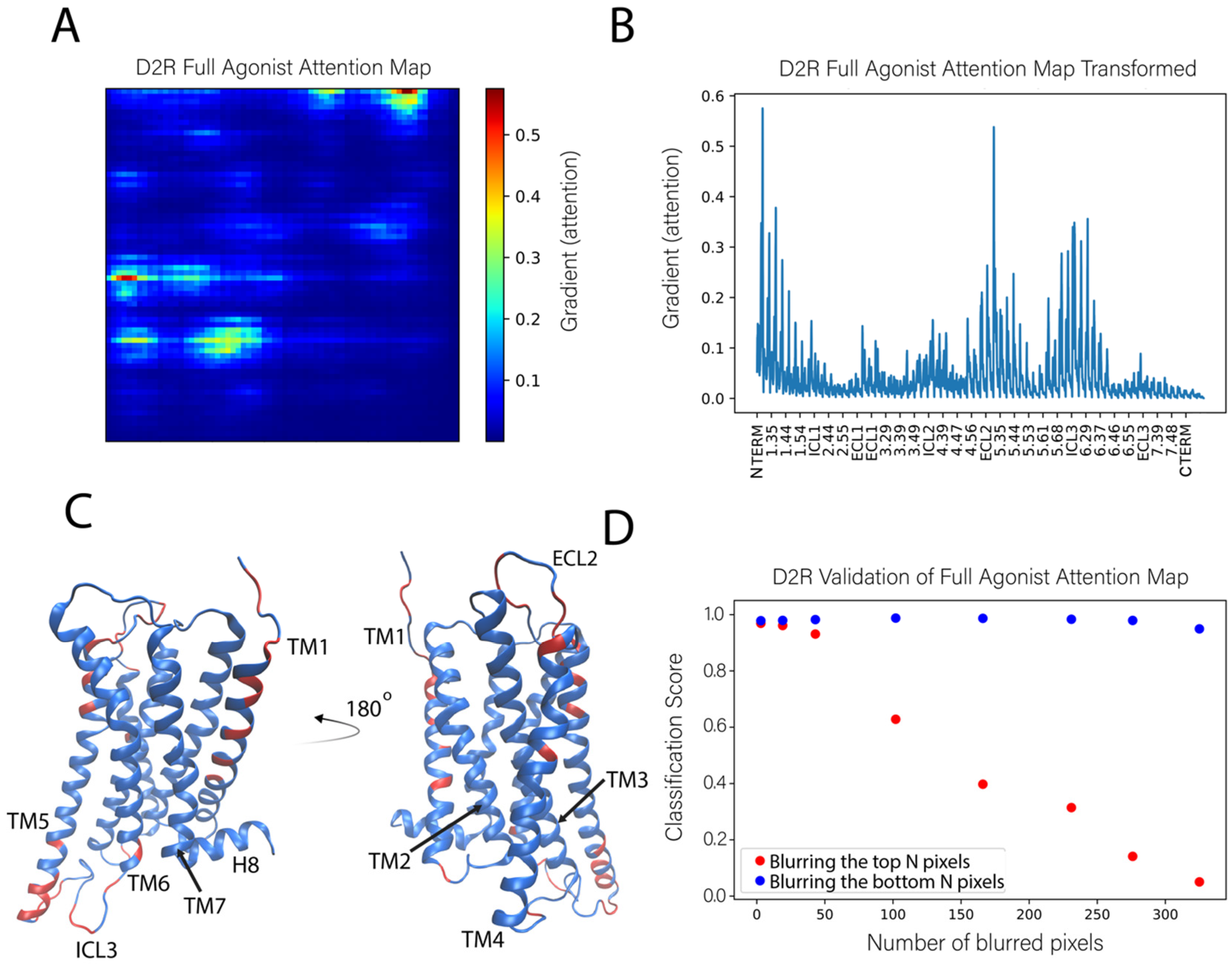
2.3.3. Sensitivity Analysis Applied to the Classification of the D2R Complexes
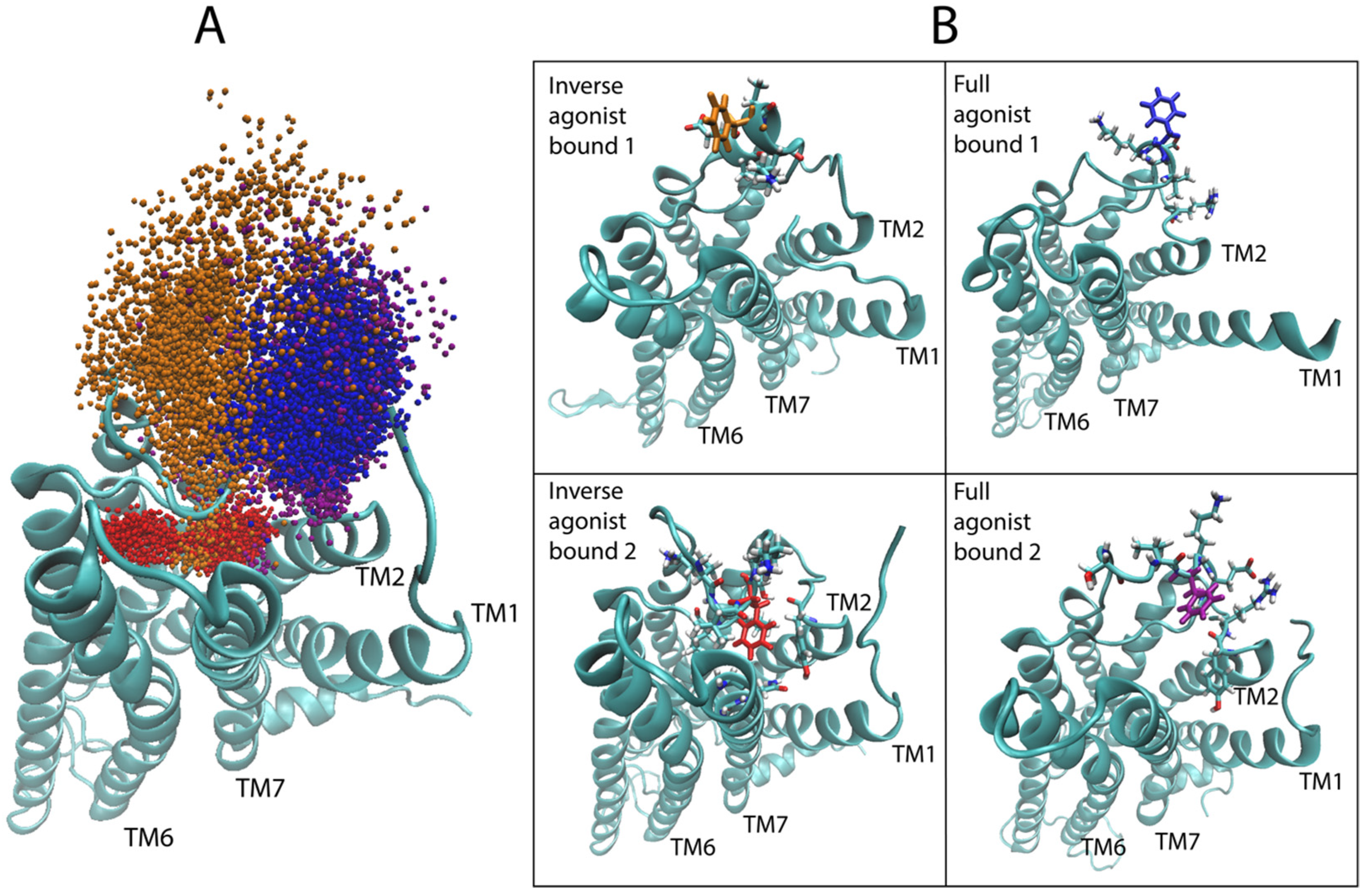
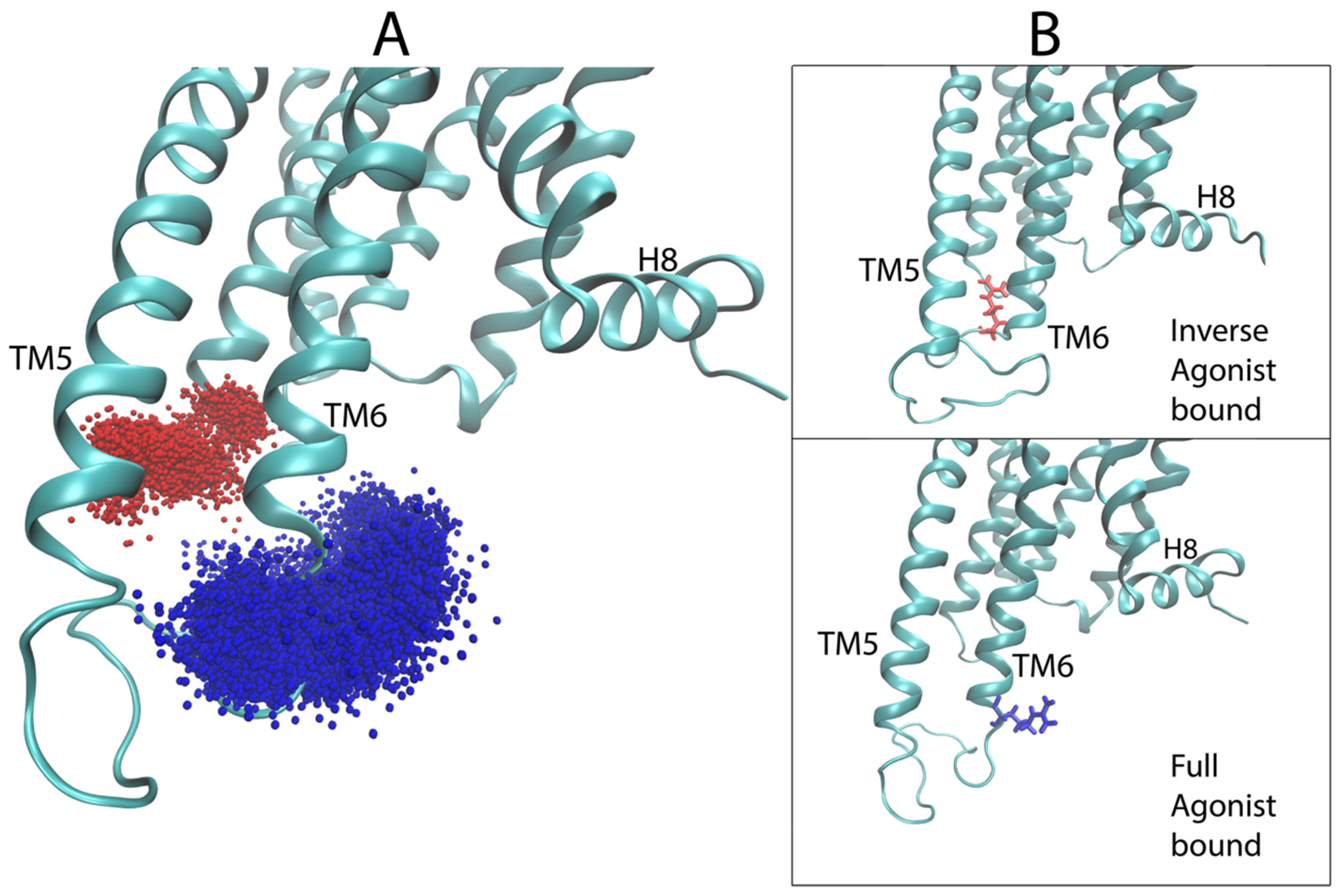
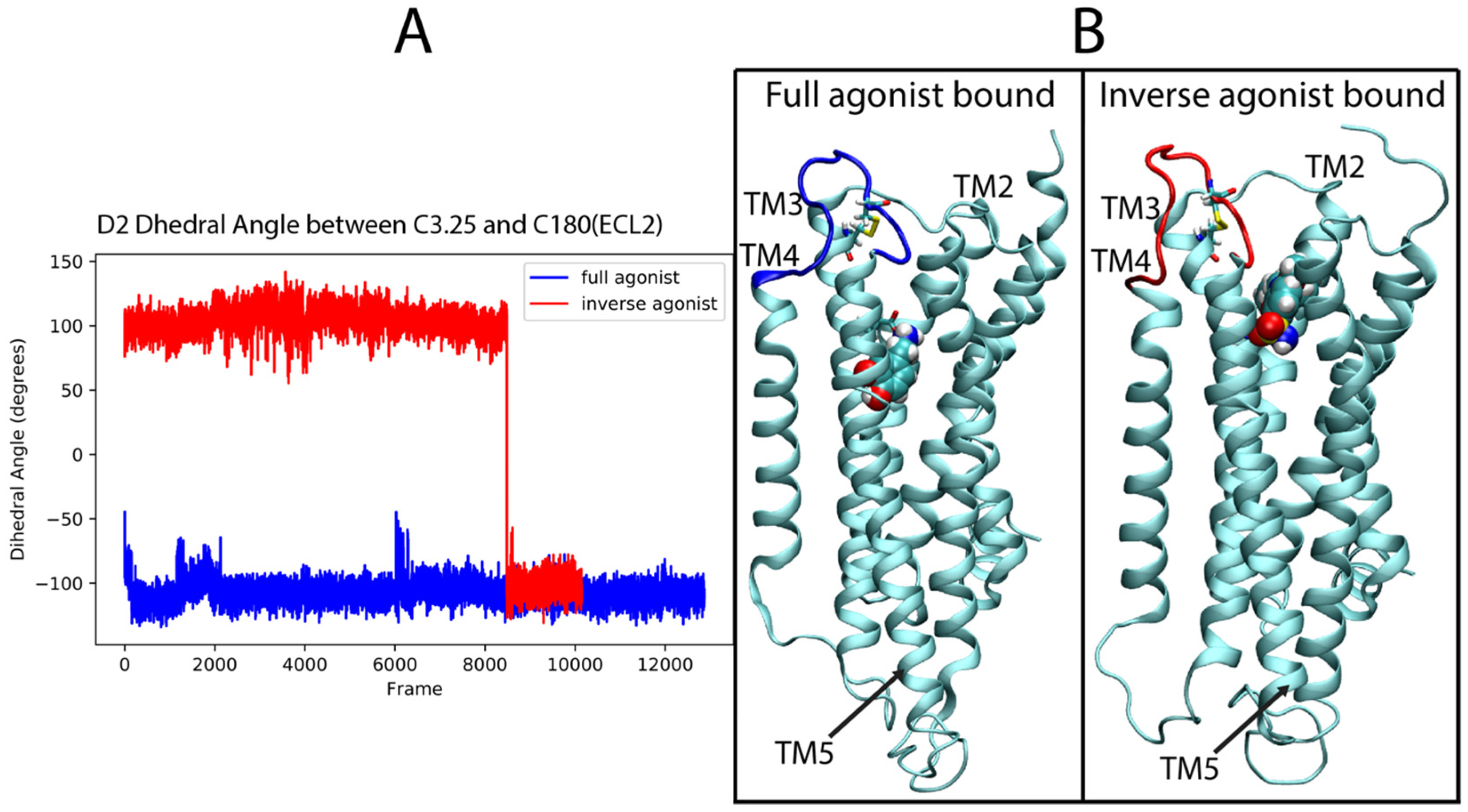
2.4. Dynamic Differences Discriminated by the DNN in the Regions Most Important for the Classification Decisions of Ligand-Bound GPCRs
2.4.1. 5-HT2AR Complexes
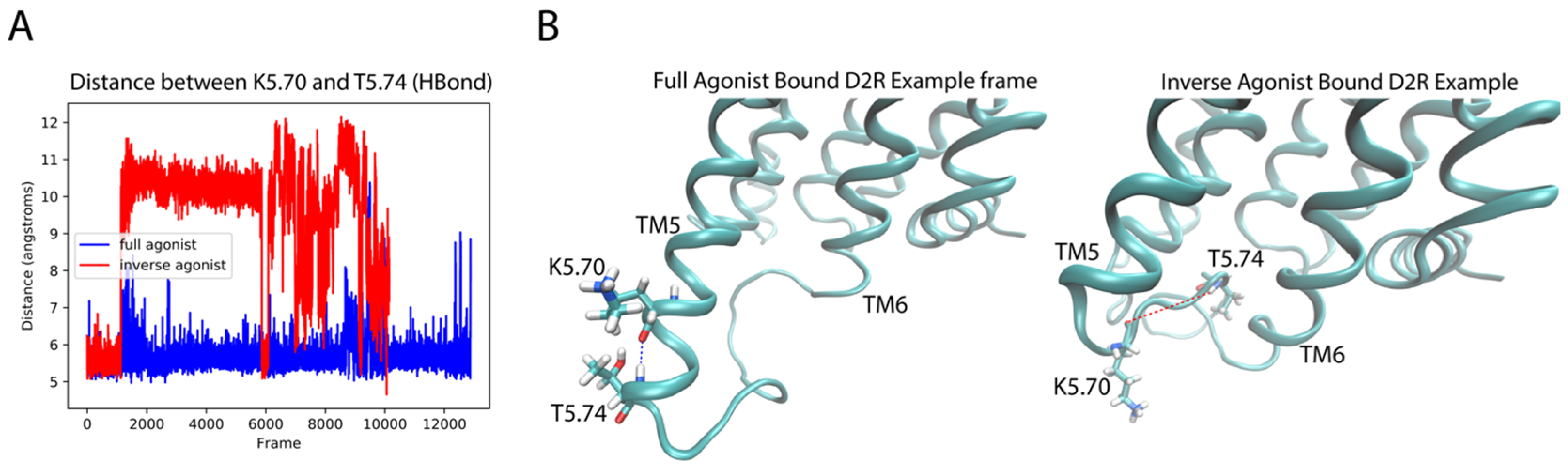
2.4.2. D2R Complexes
3. Discussion
4. Materials and Methods
4.1. Building Homology Model of the GPCRs
4.2. Molecular Construct for D2R
4.3. Parametrization and Docking of the Molecular Models
4.4. Molecular Dynamics Simulations
4.5. Machine Learning Procedures
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Smith, J.S.; Lefkowitz, R.J.; Rajagopal, S. Biased signalling: From simple switches to allosteric microprocessors. Nat. Rev. Drug Discov. 2018, 17, 243–260. [Google Scholar] [CrossRef] [PubMed]
- Berg, K.A.; Maayani, S.; Goldfarb, J.; Scaramellini, C.; Leff, P.; Clarke, W.P. Effector pathway-dependent relative efficacy at serotonin type 2A and 2C receptors: Evidence for agonist-directed trafficking of receptor stimulus. Mol. Pharm. 1998, 54, 94–104. [Google Scholar] [CrossRef]
- Urban, J.D.; Clarke, W.P.; von Zastrow, M.; Nichols, D.E.; Kobilka, B.; Weinstein, H.; Javitch, J.A.; Roth, B.L.; Christopoulos, A.; Sexton, P.M.; et al. Functional selectivity and classical concepts of quantitative pharmacology. J. Pharm. Exp. Ther. 2007, 320, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, H. Hallucinogen actions on 5-HT receptors reveal distinct mechanisms of activation and signaling by G protein-coupled receptors. Aaps J. 2005, 7, E871–E884. [Google Scholar] [CrossRef]
- Shan, J.F.; Khelashvili, G.; Mondal, S.; Mehler, E.L.; Weinstein, H. Ligand-Dependent Conformations and Dynamics of the Serotonin 5-HT2A Receptor Determine Its Activation and Membrane-Driven Oligomerization Properties. PLoS Comput. Biol. 2012, 8, e1002473. [Google Scholar] [CrossRef]
- Wingler, L.M.; Elgeti, M.; Hilger, D.; Latorraca, N.R.; Lerch, M.T.; Staus, D.P.; Dror, R.O.; Kobilka, B.K.; Hubbell, W.L.; Lefkowitz, R.J. Angiotensin Analogs with Divergent Bias Stabilize Distinct Receptor Conformations. Cell 2019, 176, 468–478. [Google Scholar] [CrossRef]
- Perez-Aguilar, J.M.; Shan, J.F.; LeVine, M.V.; Khelashvili, G.; Weinstein, H. A Functional Selectivity Mechanism at the Serotonin-2A GPCR Involves Ligand-Dependent Conformations of Intracellular Loop 2. J. ACS 2014, 136, 16044–16054. [Google Scholar] [CrossRef]
- Hollingsworth, S.A.; Dror, R.O. Molecular Dynamics Simulation for All. Neuron 2018, 99, 1129–1143. [Google Scholar] [CrossRef]
- Razavi, A.M.; Khelashvili, G.; Weinstein, H. A Markov State-based Quantitative Kinetic Model of Sodium Release from the Dopamine Transporter. Sci. Rep. 2017, 7, 40076. [Google Scholar] [CrossRef]
- Song, X.; Jensen, M.O.; Jogini, V.; Stein, R.A.; Lee, C.H.; McHaourab, H.S.; Shaw, D.E.; Gouaux, E. Mechanism of NMDA receptor channel block by MK-801 and memantine. Nature 2018, 556, 515–519. [Google Scholar] [CrossRef]
- Shaw, D.E.; Dror, R.O.; Salmon, J.K.; Grossman, J.P.; Mackenzie, K.M.; Bank, J.A.; Young, C.; Deneroff, M.M.; Batson, B.; Bowers, K.J.; et al. Millisecond-Scale Molecular Dynamics Simulations on Anton. In Proceedings of the Conference on High. Performance Computing Networking, Storage and Analysis, Portland, OR, USA, 14–20 November 2009; IEEE: New York, NY, USA, 2009. Article No. 39. [Google Scholar] [CrossRef]
- Motlagh, H.N.; Wrabl, J.O.; Li, J.; Hilser, V.J. The ensemble nature of allostery. Nature 2014, 508, 331–339. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Mao, S.W.; Liu, Y.H. Big Data: A Survey. Mobile Netw. Appl. 2014, 19, 171–209. [Google Scholar] [CrossRef]
- Frankel, F.; Reid, R. Big data: Distilling meaning from data. Nature 2008, 455, 30. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; van der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; IEEE: New York, NY, USA, 2017; pp. 2261–2269. [Google Scholar] [CrossRef]
- Khelashvili, G.; Stanley, N.; Sahai, M.A.; Medina, J.; LeVine, M.V.; Shi, L.; De Fabritiis, G.; Weinstein, H. Spontaneous inward opening of the dopamine transporter is triggered by PIP2-regulated dynamics of the N-terminus. ACS Chem. Neurosci. 2015, 6, 1825–1837. [Google Scholar] [CrossRef] [PubMed]
- LeVine, M.V.; Weinstein, H. NbIT—A New Information Theory-Based Analysis of Allosteric Mechanisms Reveals Residues that Underlie Function in the Leucine Transporter LeuT. PLoS Comput. Biol. 2014, 10, e1003603. [Google Scholar] [CrossRef] [PubMed]
- Pleiss, G.; Chen, D.; Huang, G.; Li, T.; van der Maaten, L.; Weinberger, K.Q. Memory-efficient implementation of densenets. arXiv preprint 2017, arXiv:170706990. (accessed on 2 March 2018). [Google Scholar]
- Simonyan, K.; Vedaldi, A.; Zisserman, A. Deep Inside Convolutional Networks: Visualising Image Classification Models and Saliency Maps. ICLR 2013. (accessed on 15 March 2018). [Google Scholar]
- Ballesteros, J.; Weinstein, H. Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods Neurosci. 1995, 25, 366–428. [Google Scholar]
- Venkatakrishnan, A.J.; Deupi, X.; Lebon, G.; Tate, C.G.; Schertler, G.F.; Babu, M.M. Molecular signatures of G-protein-coupled receptors. Nature 2013, 494, 185–194. [Google Scholar] [CrossRef]
- Yu, I.; Feig, M.; Sugita, Y. High-Performance Data Analysis on the Big Trajectory Data of Cellular Scale All-atom Molecular Dynamics Simulations. J. Phys. Conf. Ser. 2018, 1036, 012009. [Google Scholar] [CrossRef]
- Noé, F. Machine Learning for Molecular Dynamics on Long Timescales. arXiv 2018. [Google Scholar]
- Wheatley, M.; Wootten, D.; Conner, M.T.; Simms, J.; Kendrick, R.; Logan, R.T.; Poyner, D.R.; Barwell, J. Lifting the lid on GPCRs: The role of extracellular loops. Br. J. Pharm. 2012, 165, 1688–1703. [Google Scholar] [CrossRef]
- Peng, Y.; McCorvy, J.D.; Harpsoe, K.; Lansu, K.; Yuan, S.; Popov, P.; Qu, L.; Pu, M.; Che, T.; Nikolajsen, L.F.; et al. 5-HT2C Receptor Structures Reveal the Structural Basis of GPCR Polypharmacology. Cell 2018, 172, 719–730.e14. [Google Scholar] [CrossRef] [PubMed]
- Wacker, D.; Wang, S.; McCorvy, J.D.; Betz, R.M.; Venkatakrishnan, A.J.; Levit, A.; Lansu, K.; Schools, Z.L.; Che, T.; Nichols, D.E.; et al. Crystal Structure of an LSD-Bound Human Serotonin Receptor. Cell 2017, 168, 377–389.e12. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Che, T.; Levit, A.; Shoichet, B.K.; Wacker, D.; Roth, B.L. Structure of the D2 dopamine receptor bound to the atypical antipsychotic drug risperidone. Nature 2018, 555, 269–273. [Google Scholar] [CrossRef] [PubMed]
- Flock, T.; Hauser, A.S.; Lund, N.; Gloriam, D.E.; Balaji, S.; Babu, M.M. Selectivity determinants of GPCR-G-protein binding. Nature 2017, 545, 317. [Google Scholar] [CrossRef]
- Katritch, V.; Cherezov, V.; Stevens, R.C. Structure-function of the G protein-coupled receptor superfamily. Annu Rev. Pharm. Toxicol. 2013, 53, 531–556. [Google Scholar] [CrossRef]
- Webb, B.; Sali, A. Protein Structure Modeling with MODELLER. Methods Mol. Biol. 2017, 1654, 39–54. [Google Scholar]
- Harding, S.D.; Sharman, J.L.; Faccenda, E.; Southan, C.; Pawson, A.J.; Ireland, S.; Gray, A.J.G.; Bruce, L.; Alexander, S.P.H.; Anderton, S.; et al. The IUPHAR/BPS Guide to PHARMACOLOGY in 2018: Updates and expansion to encompass the new guide to IMMUNOPHARMACOLOGY. Nucleic Acids Res. 2018, 46, D1091–D1106. [Google Scholar] [CrossRef]
- Mysinger, M.M.; Carchia, M.; Irwin, J.J.; Shoichet, B.K. Directory of useful decoys, enhanced (DUD-E): Better ligands and decoys for better benchmarking. J. Med. Chem. 2012, 55, 6582–6594. [Google Scholar] [CrossRef]
- Sastry, G.M.; Adzhigirey, M.; Day, T.; Annabhimoju, R.; Sherman, W. Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 2013, 27, 221–234. [Google Scholar] [CrossRef] [PubMed]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [CrossRef] [PubMed]
- Irwin, J.J.; Sterling, T.; Mysinger, M.M.; Bolstad, E.S.; Coleman, R.G. ZINC: A free tool to discover chemistry for biology. J. Chem. Inf. Model. 2012, 52, 1757–1768. [Google Scholar] [CrossRef] [PubMed]
- Sherman, W.; Beard, H.S.; Farid, R. Use of an induced fit receptor structure in virtual screening. Chem. Biol. Drug Des. 2006, 67, 83–84. [Google Scholar] [CrossRef] [PubMed]
- Sherman, W.; Day, T.; Jacobson, M.P.; Friesner, R.A.; Farid, R. Novel procedure for modeling ligand/receptor induced fit effects. J. Med. Chem. 2006, 49, 534–553. [Google Scholar] [CrossRef] [PubMed]
- Farid, R.; Day, T.; Friesner, R.A.; Pearlstein, R.A. New insights about HERG blockade obtained from protein modeling, potential energy mapping, and docking studies. Bioorgan. Med. Chem. 2006, 14, 3160–3173. [Google Scholar] [CrossRef] [PubMed]
- Braden, M.R.; Parrish, J.C.; Naylor, J.C.; Nichols, D.E. Molecular interaction of serotonin 5-HT2A receptor residues Phe339((6.51)) and Phe340((6.52)) with superpotent N-benzyl phenethylamine agonists. Mol. Pharmacol. 2006, 70, 1956–1964. [Google Scholar] [CrossRef]
- Almaula, N.; Ebersole, B.J.; Zhang, D.Q.; Weinstein, H.; Sealfon, S.C. Mapping the binding site pocket of the serotonin 5-hydroxytryptamine(2A) receptor—Ser(3.36) provides a second interaction site for the protonated amine of serotonin but not of lysergic acid diethylamide or bufotenin. J. Biol. Chem. 1996, 271, 14672–14675. [Google Scholar] [CrossRef]
- Sealfon, S.C.; Chi, L.; Ebersole, B.J.; Rodic, V.; Zhang, D.; Ballesteros, J.A.; Weinstein, H. Related Contribution of Specific Helix-2 and Helix-7 Residues to Conformational Activation of the Serotonin 5-Ht2a Receptor. J. Biol. Chem. 1995, 270, 16683–16688. [Google Scholar] [CrossRef]
- Michino, M.; Free, R.B.; Doyle, T.B.; Sibley, D.R.; Shi, L. Structural basis for Na+-sensitivity in dopamine D2 and D3 receptors. Chem. Commun. 2015, 51, 8618–8621. [Google Scholar] [CrossRef]
- Kling, R.C.; Tschammer, N.; Lanig, H.; Clark, T.; Gmeiner, P. Active-State Model of a Dopamine D-2 Receptor—G alpha(i) Complex Stabilized by Aripiprazole-Type Partial Agonists. PLoS ONE 2014, 9, e100069. [Google Scholar] [CrossRef] [PubMed]
- Almaula, N.; Ebersole, B.J.; Ballesteros, J.A.; Weinstein, H.; Sealfon, S.C. Contribution of a helix 5 locus to selectivity of hallucinogenic and nonhallucinogenic ligands for the human 5-hydroxytryptamine2A and 5-hydroxytryptamine2C receptors: Direct and indirect effects on ligand affinity mediated by the same locus. Mol. Pharm. 1996, 50, 34–42. [Google Scholar]
- Shah, U.H.; Gaitonde, S.A.; Moreno, J.L.; Glennon, R.A.; Dukat, M.; Gonzalez-Maeso, J. A revised pharmacophore model for 5-HT2A receptor antagonists derived from the atypical antipsychotic agent risperidone. ACS Chem. Neurosci. 2019. [Google Scholar] [CrossRef] [PubMed]
- Chien, E.Y.; Liu, W.; Zhao, Q.; Katritch, V.; Han, G.W.; Hanson, M.A.; Shi, L.; Newman, A.H.; Javitch, J.A.; Cherezov, V.; et al. Structure of the human dopamine D3 receptor in complex with a D2/D3 selective antagonist. Science 2010, 330, 1091–1095. [Google Scholar] [CrossRef] [PubMed]
- Mansour, A.; Meng, F.; Meador-Woodruff, J.H.; Taylor, L.P.; Civelli, O.; Akil, H. Site-directed mutagenesis of the human dopamine D2 receptor. Eur. J. Pharm. 1992, 227, 205–214. [Google Scholar] [CrossRef]
- Vanommeslaeghe, K.; Hatcher, E.; Acharya, C.; Kundu, S.; Zhong, S.; Shim, J.; Darian, E.; Guvench, O.; Lopes, P.; Vorobyov, I.; et al. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 2010, 31, 671–690. [Google Scholar] [CrossRef]
- Vanommeslaeghe, K.; MacKerell, A.D., Jr. Automation of the CHARMM General Force Field (CGenFF) I: Bond perception and atom typing. J. Chem. Inf. Model. 2012, 52, 3144–3154. [Google Scholar] [CrossRef]
- Vanommeslaeghe, K.; Raman, E.P.; MacKerell, A.D., Jr. Automation of the CHARMM General Force Field (CGenFF) II: Assignment of bonded parameters and partial atomic charges. J. Chem. Inf. Model. 2012, 52, 3155–3168. [Google Scholar] [CrossRef]
- Mayne, C.G.; Saam, J.; Schulten, K.; Tajkhorshid, E.; Gumbart, J.C. Rapid Parameterization of Small Molecules Using the Force Field Toolkit. J. Comput. Chem. 2013, 34, 2757–2770. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 09, Revision B.01. G09, Revision B.01 ed; Gaussian, Inc.: Pittsburgh, PA, USA, 2009. [Google Scholar]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kale, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Cheng, X.; Jo, S.; MacKerell, A.D.; Klauda, J.B.; Im, W. CHARMM-GUI Input Generator for NAMD, Gromacs, Amber, Openmm, and CHARMM/OpenMM Simulations using the CHARMM36 Additive Force Field. J. Chem. Theory Comput. 2016, 110, 641a-a. [Google Scholar]
- Shi, L.; Quick, M.; Zhao, Y.; Weinstein, H.; Javitch, J.A. The mechanism of a neurotransmitter:sodium symporter--inward release of Na+ and substrate is triggered by substrate in a second binding site. Mol. Cell 2008, 30, 667–677. [Google Scholar] [CrossRef] [PubMed]
- Harvey, M.J.; Giupponi, G.; De Fabritiis, G. ACEMD: Accelerating Biomolecular Dynamics in the Microsecond Time Scale. J. Chem. Theory Comput. 2009, 5, 1632–1639. [Google Scholar] [CrossRef] [PubMed]
- Chollet, F. Keras, Github. 2015. Available online: https://github.com/keras-team/keras (accessed on 3 October 2018).
- Abadi, M.; Agarwal, A.; Barham, P.; Brevdo, E.; Chen, Z.; Citro, C.; Corrado, G.S.; Davis, A.; Dean, J.; Devin, M.; et al. TensorFlow: Large-Scale Machine Learning on Heterogeneous Systems. 2015. Available online: https://arxiv.org/abs/1603.04467 (accessed on 10 January 2019).
- Yu, F.F. DenseNet-Keras. Github. 2017. Available online: https://github.com/flyyufelix/DenseNet-Keras (accessed on 9 October 2018).
- Kotikalapudi, R.A.C. Keras-Vis. GitHub. 2017. Available online: https://github.com/raghakot/keras-vis. (accessed on 20 February 2019).



© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Plante, A.; Shore, D.M.; Morra, G.; Khelashvili, G.; Weinstein, H. A Machine Learning Approach for the Discovery of Ligand-Specific Functional Mechanisms of GPCRs. Molecules 2019, 24, 2097. https://doi.org/10.3390/molecules24112097
Plante A, Shore DM, Morra G, Khelashvili G, Weinstein H. A Machine Learning Approach for the Discovery of Ligand-Specific Functional Mechanisms of GPCRs. Molecules. 2019; 24(11):2097. https://doi.org/10.3390/molecules24112097
Chicago/Turabian StylePlante, Ambrose, Derek M. Shore, Giulia Morra, George Khelashvili, and Harel Weinstein. 2019. "A Machine Learning Approach for the Discovery of Ligand-Specific Functional Mechanisms of GPCRs" Molecules 24, no. 11: 2097. https://doi.org/10.3390/molecules24112097
APA StylePlante, A., Shore, D. M., Morra, G., Khelashvili, G., & Weinstein, H. (2019). A Machine Learning Approach for the Discovery of Ligand-Specific Functional Mechanisms of GPCRs. Molecules, 24(11), 2097. https://doi.org/10.3390/molecules24112097





