Sequencing and Analysis of Chrysanthemum carinatum Schousb and Kalimeris indica. The Complete Chloroplast Genomes Reveal Two Inversions and rbcL as Barcoding of the Vegetable
Abstract
1. Introduction
2. Results
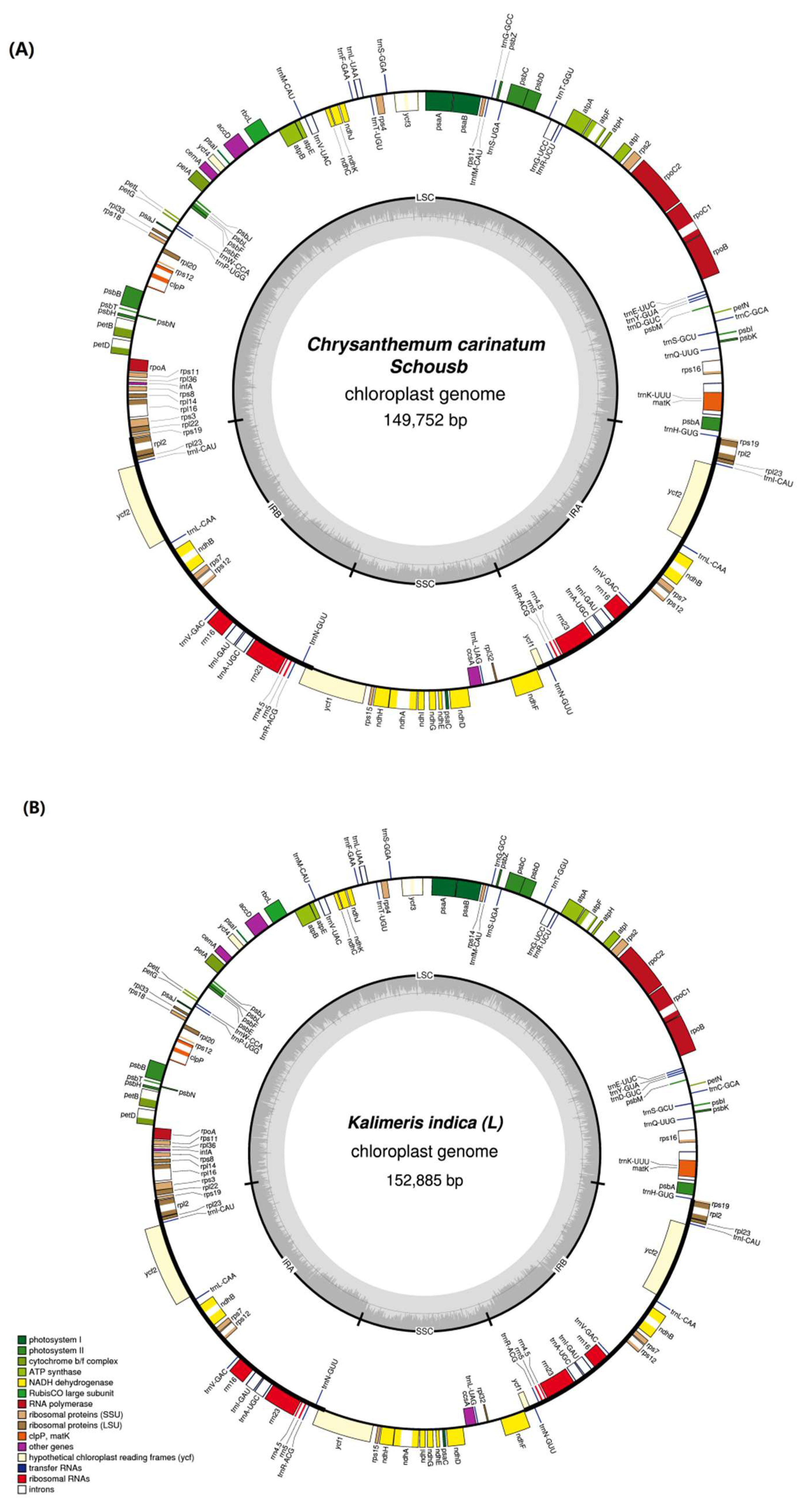
2.1. Features of the C. carinatum Schousb and K. indica cp Genome
2.2. Functional Genes of the C. carinatum Schousb and K. indica cp Genome
2.3. Codon Usage of the K. indica and C. carinatum Schousb cp Genome
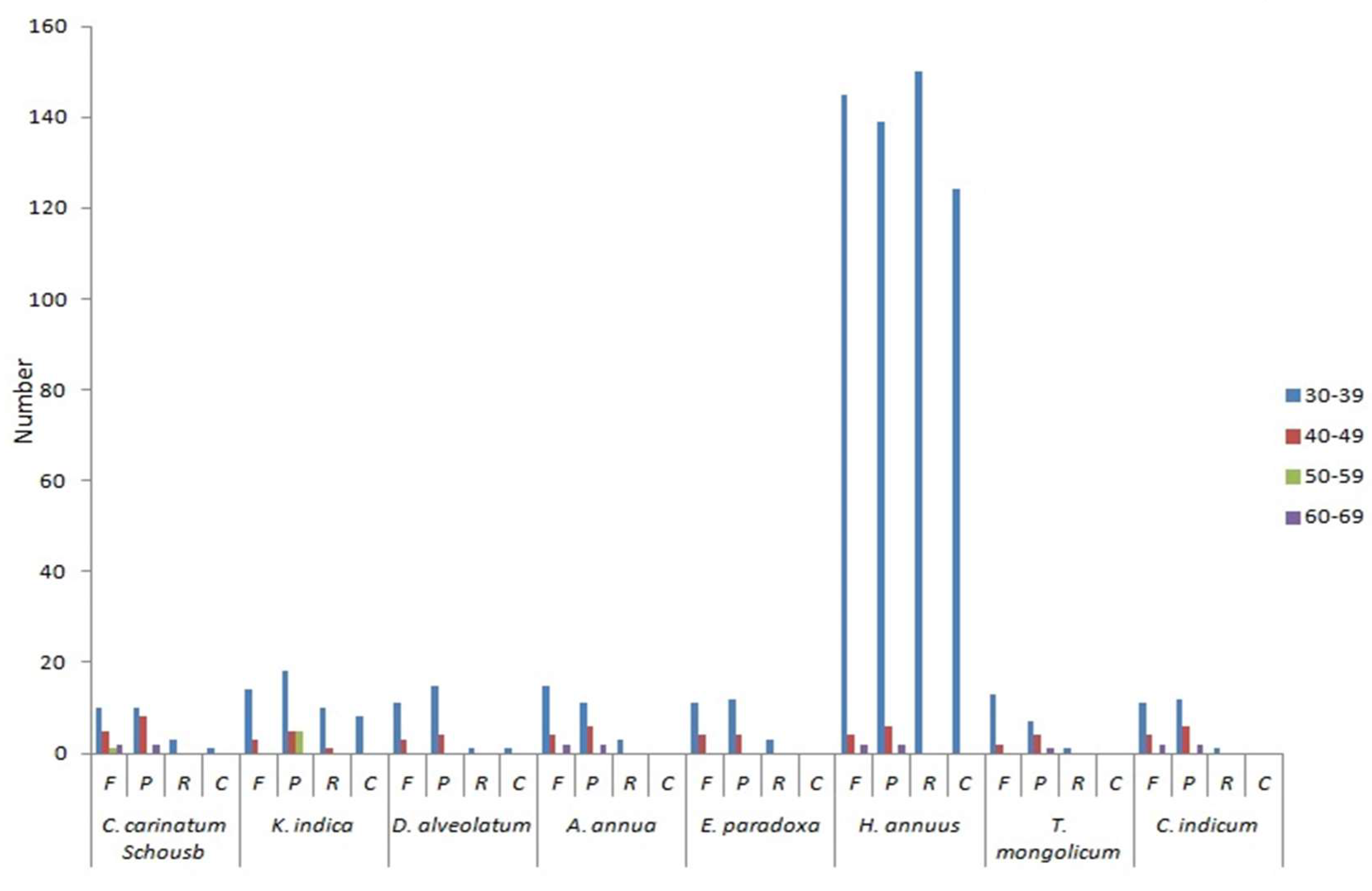
2.4. Repeats Structure and SSRs
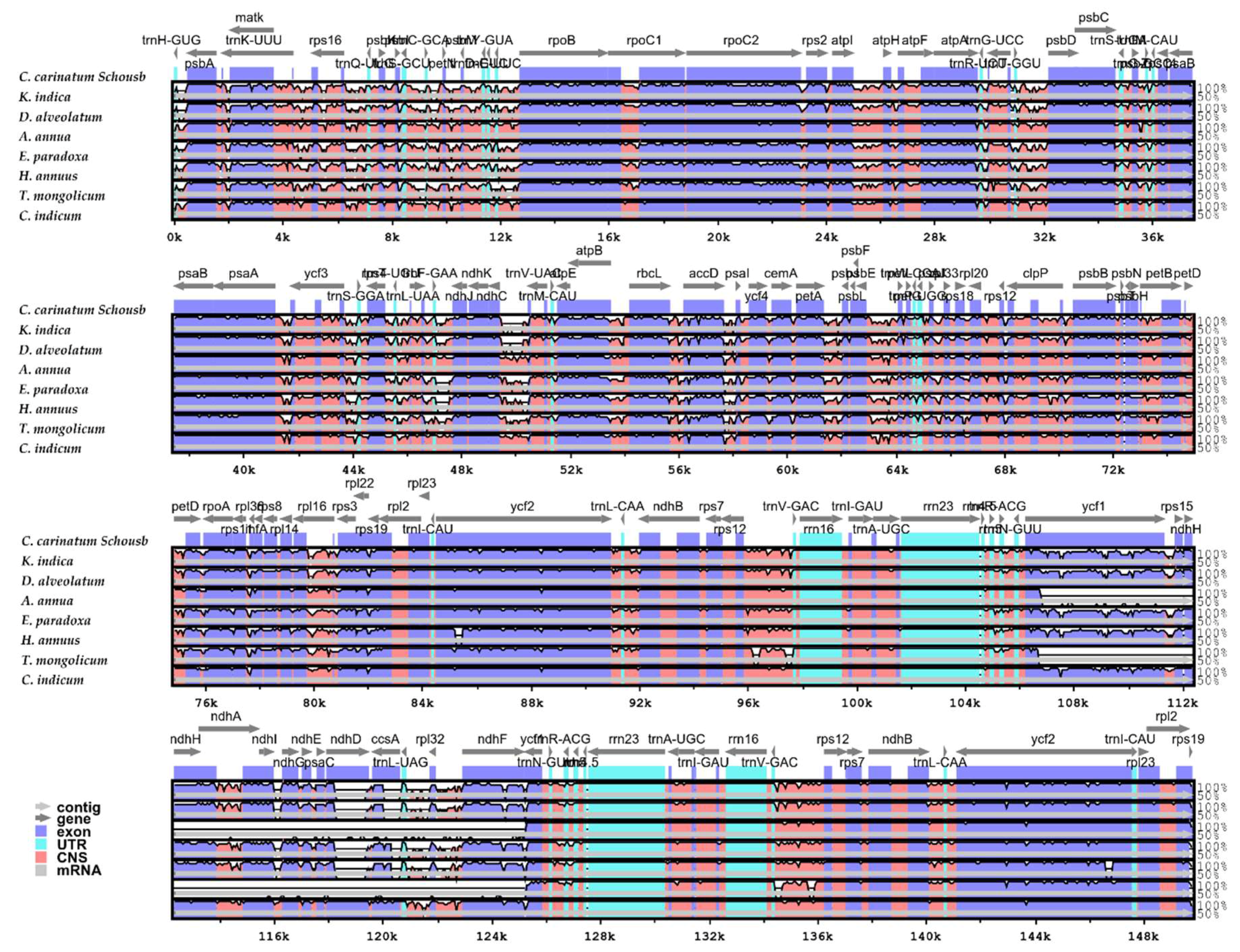
2.5. Comparison of the Whole cp Genome of Asteraceae
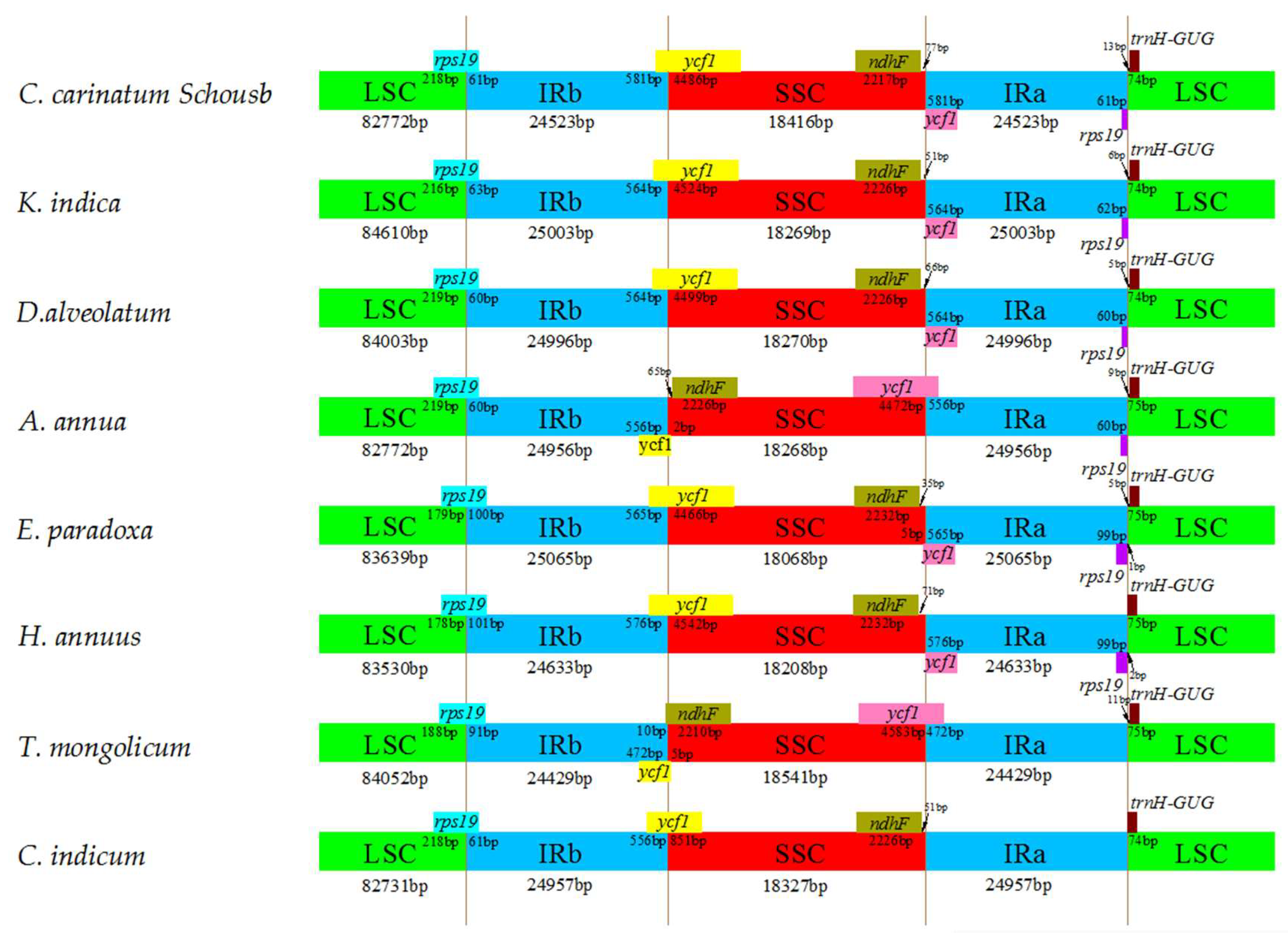
2.6. IR Contraction and Expansion
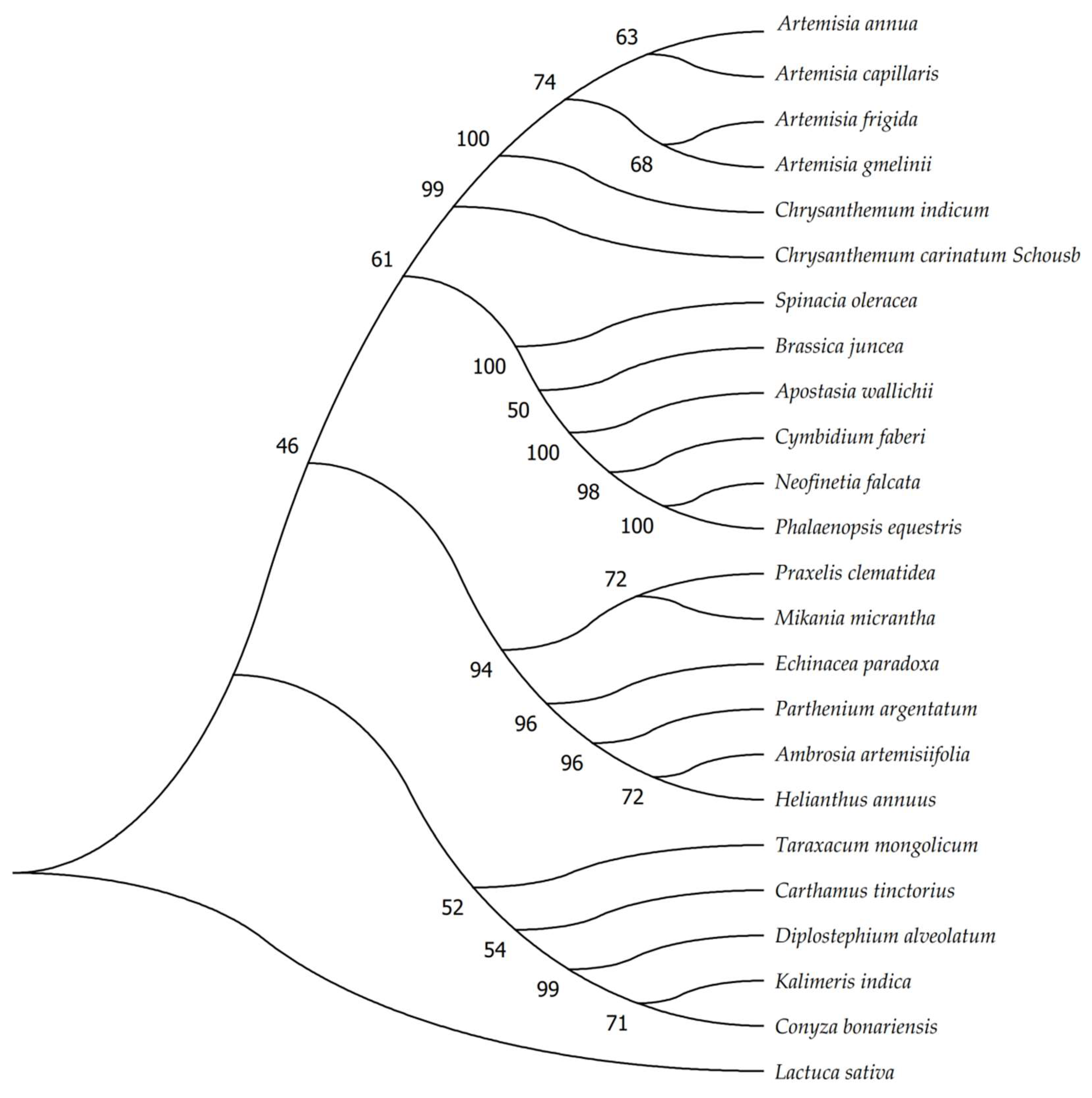
2.7. Phylogenetic Analysis and Barcoding
3. Discussion
4. Materials and Methods
4.1. DNA Extraction and Sequencing
4.2. Genome Assembly
4.3. Gene Annotation and Codon Usage Analysis
4.4. Repeat Structure and Single Sequence Repeats (SSRs) Analysis
4.5. Comparative Genome Analysis of the C. carinatum Schousb and K. indica Genomes
4.6. Phylogenetic Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cazzonelli, C.I. Carotenoids in nature: Insights from plants and beyond. Funct. Plant Biol. 2011, 38, 833–847. [Google Scholar] [CrossRef]
- Bobik, K.; Burch-Smith, T.M. Chloroplast signaling within, between and beyond cells. Front. Plant Sci. 2015, 6, 781. [Google Scholar] [CrossRef] [PubMed]
- Daniell, H.; Lin, C.; Yu, M.; Chang, W. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016, 17, 134. [Google Scholar] [CrossRef] [PubMed]
- Baczkiewicz, A.; Szczecinska, M.; Sawicki, J.; Stebel, A.; Buczkowska, K. DNA barcoding, ecology and geography of the cryptic species of Aneura pinguis and their relationships with Aneura maxima and Aneura mirabilis (Metzgeriales, Marchantiophyta). PLoS ONE 2017, 12, e0188837. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Deng, Z.; Zang, R.; Long, W. DNA barcoding analysis and phylogenetic relationships of tree species in tropical cloud forests. Sci. Rep. 2017, 7, 12564. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Chen, Y.; Lv, J.; Xu, J.; Zhu, S.; Li, M.; Chen, N. Development of Chloroplast Genomic Resources for Oryza Species Discrimination. Front. Plant Sci. 2017, 8, 1854. [Google Scholar] [CrossRef] [PubMed]
- Stegemann, S.; Keuthe, M.; Greiner, S.; Bock, R. Horizontal transfer of chloroplast genomes between plant species. Proc. Natl. Acad. Sci. USA 2012, 109, 2434–2438. [Google Scholar] [CrossRef] [PubMed]
- Rogalski, M.; Vieira, L.D.N.; Fraga, H.P.; Guerra, M.P. Plastid genomics in horticultural species: Importance and applications for plant population genetics, evolution, and biotechnology. Front. Plant Sci. 2015, 6, 586. [Google Scholar] [CrossRef] [PubMed]
- Ruck, E.C.; Linard, S.R.; Nakov, T.; Theriot, E.C.; Alverson, A.J. Hoarding and horizontal transfer led to an expanded gene and intron repertoire in the plastid genome of the diatom, Toxarium undulatum (Bacillariophyta). Curr. Genet. 2017, 63, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Kawabe, A.; Nukii, H.; Furihata, H.Y. Exploring the History of Chloroplast Capture in Arabis Using Whole Chloroplast Genome Sequencing. Int. J. Mol. Sci. 2018, 19, 602. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Peng, H.; Kilian, N. Molecular Phylogeny of the Lactuca Alliance (Cichorieae Subtribe Lactucinae, Asteraceae) with Focus on Their Chinese Centre of Diversity Detects Potential Events of Reticulation and Chloroplast Capture. PLoS ONE 2013, 8, e82692. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Cui, L.; Feng, K.; Deng, P.; Du, X.; Wan, F.; Song, W.; Nie, X. Comparative Analysis of Asteraceae Chloroplast Genomes: Structural Organization, RNA Editing and Evolution. Plant. Mol. Biol. Rep. 2015, 33, 1526–1538. [Google Scholar] [CrossRef]
- Jansen, R.K.; Palmer, J.D. A chloroplast DNA inversion marks an ancient evolutionary split in the sunflower family (Asteraceae). Proc. Natl. Acad. Sci. USA 1987, 84, 5818–5822. [Google Scholar] [CrossRef] [PubMed]
- Jansen, R.K.; Palmer, J.D. Chloroplast DNA from lettuce and Barnadesia (Asteraceae): Structure, gene localization, and characterization of a large inversion. Curr. Genet. 1987, 11, 553–564. [Google Scholar] [CrossRef]
- Kim, K.J.; Choi, K.S.; Jansen, R.K. Two chloroplast DNA inversions originated simultaneously during the early evolution of the sunflower family (Asteraceae). Mol. Biol. Evol. 2005, 22, 1783–1792. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Castillo-Ramirez, S.; Gonzalez, V.; Bustos, P.; Fernandez-Vazquez, J.L.; Santamaria, R.I.; Arellano, J.; Cevallos, M.A.; Davila, G. Rapid evolutionary change of common bean (Phaseolus vulgaris L.) plastome, and the genomic diversification of legume chloroplasts. BMC Genom. 2007, 8, 228. [Google Scholar] [CrossRef] [PubMed]
- Walker, J.F.; Zanis, M.J.; Emery, N.C. Comparative analysis of complete chloroplast genomw sequence and inversion variation in lasthenia burkei (Madieae, Asteraceae). Am. J. Bot 2014, 101, 722–729. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.S.; Park, S. The complete chloroplast genome sequence of Aster spathuhfolius (Asteraceae); genomic features and relationship with Asteraceae. Gene 2015, 572, 214–221. [Google Scholar] [CrossRef] [PubMed]
- Curci, P.L.; De Paola, D.; Danzi, D.; Vendramin, G.G.; Sonnante, G. Complete Chloroplast Genome of the Multifunctional Crop Globe Artichoke and Comparison with Other Asteraceae. PLoS ONE 2015, 10, e0120589. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, E.N.; Ruhlman, T.A.; Sabir, J.S.M.; Hajrah, N.H.; Alharbi, N.S.; Al-Malki, A.L.; Bailey, C.D.; Jansen, R.K. Plastid genome sequences of legumes reveal parallel inversions and multiple losses of rps16 in papilionoids. J. Syst. Evol. 2015, 53, 458–468. [Google Scholar] [CrossRef]
- Hernandez, L.F.; Rosetti, M.V. Is the abaxial palisade parenchyma in phyllaries of the sunflower (Helianthus annuus L.) capitulum a missing trait in modern genotypes? Phyton Int. J. Exp. Bot. 2016, 85, 291–296. [Google Scholar]
- Raman, G.; Park, S. The Complete Chloroplast Genome Sequence of Ampelopsis: Gene Organization, Comparative Analysis, and Phylogenetic Relationships to Other Angiosperms. Front. Plant Sci. 2016, 7, 341. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Wu, M.; Liao, B.; Liu, Z.; Bai, R.; Xiao, S.; Li, X.; Zhang, B.; Xu, J.; Chen, S. Complete Chloroplast Genome Sequence and Phylogenetic Analysis of the Medicinal Plant Artemisia annua. Molecules 2017, 22, 1330. [Google Scholar] [CrossRef] [PubMed]
- Xie, Q.; Shen, K.; Hao, X.; Phan, N.N.; Bui, T.N.H.; Chen, C.; Zhu, C.; Lin, Y.; Hsiao, C. The complete chloroplast genome of Tianshan Snow Lotus (Saussurea involucrata), a famous traditional Chinese medicinal plant of the family Asteraceae. Mitochondrial DNA Part A 2017, 28, 294–295. [Google Scholar] [CrossRef] [PubMed]
- Salih, R.H.M.; Majesky, L.; Schwarzacher, T.; Gornall, R.; Heslop-Harrison, P. Complete chloroplast genomes from apomictic Taraxacum (Asteraceae): Identity and variation between three microspecies. PLoS ONE 2017, 12, e168008. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Park, J.Y.; Lee, Y.S.; Woo, S.M.; Park, H.; Lee, S.; Kang, J.H.; Lee, T.J.; Sung, S.H.; Yang, T. The complete chloroplast genomes of two Taraxacum species, T. platycarpum Dahlst. and T. mongolicum Hand.-Mazz. (Asteraceae). Mitochondrial DNA PART B Res. 2016, 1, 412. [Google Scholar] [CrossRef]
- Smith, D.R.; Keeling, P.J. Mitochondrial and plastid genome architecture: Reoccurring themes, but significant differences at the extremes. Proc. Natl. Acad. Sci. USA 2015, 112, 10177–10184. [Google Scholar] [CrossRef] [PubMed]
- Chase, M.W.; Reveal, J.L. A phylogenetic classification of the land plants to accompany APG III. Bot. J. Linn. Soc. 2009, 161, 122–127. [Google Scholar] [CrossRef]
- Bremer, B.; Bremer, K.; Chase, M.W.; Fay, M.F.; Reveal, J.L.; Soltis, D.E.; Soltis, P.S.; Stevens, P.F.; Anderberg, A.A.; Moore, M.J.; et al. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Bot. J. Linn. Soc. 2009, 161, 105–121. [Google Scholar]
- Byng, J.W.; Chase, M.W.; Christenhusz, M.J.M.; Fay, M.F.; Judd, W.S.; Mabberley, D.J.; Sennikov, A.N.; Soltis, D.E.; Soltis, P.S.; Stevens, P.F.; et al. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 2016, 181, 1–20. [Google Scholar]
- Xu, W.; Gong, X.; Zhou, X.; Zhao, C.; Chen, H. Chemical constituents and bioactivity of Kalimeris indica. China J. Chin. Mater. Med. 2010, 35, 3172–3174. [Google Scholar]
- Wang, G.; Liu, J.; Zhang, C.; Wang, Z.; Cai, B.; Wang, G. Study on Chemical Constituents of Kalimeris indica. J. Chin. Med. Mater. 2015, 38, 81–84. [Google Scholar]
- Fan, G.; Kim, S.; Han, B.H.; Han, Y.N. Glyceroglycolipids, a novel class of platelet-activating factor antagonists from Kalimeris indica. Phytochem. Lett. 2008, 1, 207–210. [Google Scholar] [CrossRef]
- Zhong, R.; Xu, G.; Wang, Z.; Wang, A.; Guan, H.; Li, J.; He, X.; Liu, J.; Zhou, M.; Li, Y.; et al. Identification of anti-inflammatory constituents from Kalimeris indica with UHPLC-ESI-Q-TOF-MS/MS and GC-MS. J. Ethnopharmacol. 2015, 165, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Yu, Y.; Wang, Z.; Cai, B.; Zhou, Z.; Wang, G.; Liu, J. Two new terpenoids from Kalimeris indica. Nat. Prod. Res. 2017, 31, 2348–2353. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Choi, J.N.; Ku, K.M.; Kang, D.; Kim, J.S.; Park, J.H.Y.; Lee, C.H. A Correlation between Antioxidant Activity and Metabolite Release during the Blanching of Chrysanthemum coronarium L. Biosci. Biotechnol. Biochem. 2011, 75, 674–680. [Google Scholar] [CrossRef] [PubMed]
- Polatoglu, K.; Karakoc, O.C.; Demirci, B.; Baser, K.H.C. Chemical composition and insecticidal activity of edible garland (Chrysanthemum coronarium L.) essential oil against the granary pest Sitophilus granarius L. (Coleoptera). J. Essent. Oil Res. 2018, 30, 120–130. [Google Scholar] [CrossRef]
- Flamini, G.; Cioni, P.L.; Morelli, I. Differences in the fragrances of pollen, leaves, and floral parts of garland (Chrysanthemum coronarium) and composition of the essential oils from flowerheads and leaves. J. Agric. Food Chem. 2003, 51, 2267–2271. [Google Scholar] [CrossRef] [PubMed]
- Abd-Alla, H.I.; Albalawy, M.A.; Aly, H.F.; Shalaby, N.M.M.; Shaker, K.H. Flavone Composition and Antihypercholesterolemic and Antihyperglycemic Activities of Chrysanthemum coronarium L. Z Naturforsch. C 2014, 69, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Song, M.; Yang, H.; Jeong, T.; Kim, K.; Baek, N. Heterocyclic compounds from Chrysanthemum coronarium L. and their inhibitory activity on hACAT-1, hACAT-2, and LDL-oxidation. Arch. Pharm. Res. 2008, 31, 573–578. [Google Scholar] [CrossRef] [PubMed]
- Vargas, O.M.; Ortiz, E.M.; Simpson, B.B. Conflicting phylogenomic signals reveal a pattern of reticulate evolution in a recent high-Andean diversification (Asteraceae: Astereae: Diplostephium). New Phytol. 2017, 214, 1736–1750. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.; Jiao, B.; Nie, B.; Zhang, G.; Gao, T. A comprehensive generic-level phylogeny of the sunflower family: Implications for the systematics of Chinese Asteraceae. J. Syst. Evol. 2016, 54, 416–437. [Google Scholar] [CrossRef]
- Muse, S.V. Examining rates and patterns of nucleotide substitution in plants. Plant Mol. Biol. 2000, 42, 25–43. [Google Scholar] [CrossRef] [PubMed]
- Muse, S.V.; Gaut, B.S. Comparing patterns of nucleotide substitution rates among chloroplast loci using the relative ratio test. Genetics 1997, 146, 393–399. [Google Scholar] [PubMed]
- Wakeley, J. Substitution-rate variation among sites and the estimation of transition bias. Mol. Biol. Evol. 1994, 11, 436–442. [Google Scholar] [PubMed]
- Morton, B.R. Codon use and the rate of divergence of land plant chloroplast genes. Mol. Biol. Evol. 1994, 11, 231–238. [Google Scholar] [PubMed]
- Liu, X.; Yang, H.; Zhao, J.; Zhou, B.; Li, T.; Xiang, B. The complete chloroplast genome sequence of the folk medicinal and vegetable plant purslane (Portulaca oleracea L.). J. Hortic. Sci. Biotechnol. 2017, 1–10. [Google Scholar] [CrossRef]
- Xia Liu, Y.L.H.Y. Chloroplast Genome of the Folk Medicine and Vegetable Plant Talinum paniculatum (Jacq.) Gaertn.: Gene Organization, Comparative and Phylogenetic Analysis. Molecules 2018, 23, 857. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Cui, Y.; Chen, X.; Li, Y.; Xu, Z.; Duan, B.; Li, Y.; Song, J.; Yao, H. Complete Chloroplast Genomes of Papaver rhoeas and Papaver orientale: Molecular Structures, Comparative Analysis, and Phylogenetic Analysis. Molecules 2018, 23, 437. [Google Scholar] [CrossRef] [PubMed]
- Zuo, L.; Shang, A.; Zhang, S.; Yu, X.; Ren, Y.; Yang, M.; Wang, J. The first complete chloroplast genome sequences of Ulmus species by de novo sequencing: Genome comparative and taxonomic position analysis. PLoS ONE 2017, 12, e0171264. [Google Scholar] [CrossRef] [PubMed]
- Jian, H.; Zhang, Y.; Yan, H.; Qiu, X.; Wang, Q.; Li, S.; Zhang, S. The Complete Chloroplast Genome of a Key Ancestor of Modern Roses, Rosa chinensis var. spontanea, and a Comparison with Congeneric Species. Molecules 2018, 23, 389. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Saina, J.K.; Gichira, A.W.; Kyalo, C.M.; Wang, Q.; Chen, J. Comparative Genomics of the Balsaminaceae Sister Genera Hydrocera triflora and Impatiens pinfanensis. Int. J. Mol. Sci. 2018, 19, 319. [Google Scholar] [CrossRef] [PubMed]
- Ni, L.; Zhao, Z.; Xu, H.; Chen, S.; Dorje, G. The complete chloroplast genome of Gentiana straminea (Gentianaceae), an endemic species to the Sino-Himalayan subregion. Gene 2016, 577, 281–288. [Google Scholar] [CrossRef] [PubMed]
- Khakhlova, O.; Bock, R. Elimination of deleterious mutations in plastid genomes by gene conversion. Plant J. 2006, 46, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Moore, A.J.; Bartoli, A.; Tortosa, R.D.; Baldwin, B.G. Phylogeny, biogeography, and chromosome evolution of the amphitropical genus Grindelia (Asteraceae) inferred from nuclear ribosomal and chloroplast sequence data. Taxon 2012, 61, 211–230. [Google Scholar]
- Urbatsch, L.E.; Roberts, R.P.; Karaman, V. Phylogenetic evaluation of Xylothamia, Gundlachia, and related genera (Asteraceae, Astereae) based on ETS and ITS nrDNA sequence data. Am. J. Bot. 2003, 90, 634–649. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.Q.; Gao, T.G.; Chen, Z.D.; Lu, A.M. Molecular phylogeny and biogeography of the Qinghai-Tibet Plateau endemic Nannoglottis (Asteraceae). Mol. Phylogenet. Evol. 2002, 23, 307–325. [Google Scholar] [CrossRef]
- Bayer, R.J.; Cross, E.W. A reassessment of tribal affinities of the enigmatic genera Printzia and Isoetopsis (Asteraceae), based on three chloroplast sequences. Aust. J. Bot. 2002, 50, 677–686. [Google Scholar] [CrossRef]
- Eldenas, P.; Kallersjo, M.; Anderberg, A.A. Phylogenetic placement and circumscription of tribes Inuleae s. str. and Plucheeae (Asteraceae): Evidence from sequences of chloroplast gene ndhF. Mol. Phylogenet. Evol. 1999, 13, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Dempewolf, H.; Kane, N.C.; Ostevik, K.L.; Geleta, M.; Barker, M.S.; Lai, Z.; Stewart, M.L.; Bekele, E.; Engels, J.M.M.; Cronk, Q.C.B.; et al. Establishing genomic tools and resources for Guizotia abyssinica (L.f.) Cass.-the development of a library of expressed sequence tags, microsatellite loci, and the sequencing of its chloroplast genome. Mol. Ecol. Resour. 2010, 10, 1048–1058. [Google Scholar] [CrossRef] [PubMed]
- Bock, D.G.; Kane, N.C.; Ebert, D.P.; Rieseberg, L.H. Genome skimming reveals the origin of the Jerusalem Artichoke tuber crop species: Neither from Jerusalem nor an artichoke. New Phytol. 2014, 201, 1021–1030. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Erickson, D.L.; Ramachandran, P.; Ottesen, A.R.; Timme, R.E.; Funk, V.A.; Luo, Y.; Handy, S.M. An analysis of Echinacea chloroplast genomes: Implications for future botanical identification. Sci. Rep. 2017, 7, 216. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Park, J.; Lee, H.; Sohn, S.; Lee, J. Complete chloroplast genomic sequence of Citrus platymamma determined by combined analysis of Sanger and NGS data. Hortic. Environ. Biotechnol. 2015, 56, 704–711. [Google Scholar] [CrossRef]
- Su, H.; Hogenhout, S.A.; Al-Sadi, A.M.; Kuo, C. Complete Chloroplast Genome Sequence of Omani Lime (Citrus aurantiifolia) and Comparative Analysis within the Rosids. PLoS ONE 2014, 9, e113049. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhu, J.; Feng, L.; Zhou, T.; Bei, G.; Yang, J.; Zhao, G. Plastid Genome Comparative and Phylogenetic Analyses of the Key Genera in Fagaceae: Highlighting the Effect of Codon Composition Bias in Phylogenetic Inference. Front. Plant Sci. 2018, 9, 82. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Guo, L.; Zhao, W.; Xu, J.; Li, Y.; Zhang, X.; Shen, X.; Wu, M.; Hou, X. Complete Chloroplast Genome Sequence and Phylogenetic Analysis of Paeonia ostii. Molecules 2018, 23, 246. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Yu, H.; Wang, J.; Lei, W.; Gao, J.; Qiu, X.; Wang, J. The Complete Chloroplast Genome Sequences of the Medicinal Plant Forsythia suspensa (Oleaceae). Int. J. Mol. Sci. 2017, 18, 2288. [Google Scholar] [CrossRef] [PubMed]
- Saina, J.K.; Gichira, A.W.; Li, Z.; Hu, G.; Wang, Q.; Liao, K. The complete chloroplast genome sequence of Dodonaea viscosa: Comparative and phylogenetic analyses. Genetica 2018, 146, 101–113. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Chen, X.; Cui, Y.; Sun, W.; Li, Y.; Wang, Y.; Song, J.; Yao, H. Molecular Structure and Phylogenetic Analyses of Complete Chloroplast Genomes of Two Aristolochia Medicinal Species. Int. J. Mol. Sci. 2017, 18, 1839. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Chen, C.; Wei, Y.; Chang, Y.; Bai, G.; Li, Z.; Kanwal, N.; Zhao, G. Comparative Transcriptome and Chloroplast Genome Analyses of Two Related Dipteronia Species. Front. Plant Sci. 2016, 7, 1512. [Google Scholar] [CrossRef] [PubMed]
- Mosa, K.A.; Soliman, S.; El-Keblawy, A.; Ali, M.A.; Hassan, H.A.; Tamim, A.A.B.; Al-Ali, M.M. Using DNA barcoding to detect adulteration in different herbal plant-based products in the United Arab Emirates: Proof of concept and validation. Recent Pat. Food Nutr. Agric. 2018. [Google Scholar] [CrossRef] [PubMed]
- Mishra, P.; Shukla, A.K.; Sundaresan, V. Candidate DNA Barcode Tags Combined with High Resolution Melting (Bar-HRM) Curve Analysis for Authentication of Senna alexandrina Mill. with Validation in Crude Drugs. Front. Plant Sci. 2018, 9, 283. [Google Scholar] [CrossRef] [PubMed]
- Viljoen, E.; Odeny, D.A.; Coetzee, M.P.A.; Berger, D.K.; Rees, D.J.G. Application of Chloroplast Phylogenomics to Resolve Species Relationships Within the Plant Genus Amaranthus. J. Mol. Evol. 2018, 86, 216–239. [Google Scholar] [CrossRef] [PubMed]
- Allen, G.C.; Flores-Vergara, M.A.; Krasynanski, S.; Kumar, S.; Thompson, W.F. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat. Protoc. 2006, 1, 2320–2325. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST plus: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Wyman, S.K.; Jansen, R.K.; Boore, J.L. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 2004, 20, 3252–3255. [Google Scholar] [CrossRef] [PubMed]
- Schattner, P.; Brooks, A.N.; Lowe, T.M. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005, 33, W686–W689. [Google Scholar] [CrossRef] [PubMed]
- Lohse, M.; Drechsel, O.; Bock, R. OrganellarGenomeDRAW (OGDRAW): A tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet. 2007, 52, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, S.; Choudhuri, J.V.; Ohlebusch, E.; Schleiermacher, C.; Stoye, J.; Giegerich, R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001, 29, 4633–4642. [Google Scholar] [CrossRef] [PubMed]
- Beier, S.; Thiel, T.; Muench, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, S.; Phillippy, A.; Delcher, A.L.; Smoot, M.; Shumway, M.; Antonescu, C.; Salzberg, S.L. Versatile and open software for comparing large genomes. Genome Biol. 2004, 5. [Google Scholar]
- Frazer, K.A.; Pachter, L.; Poliakov, A.; Rubin, E.M.; Dubchak, I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004, 322, W273–W279. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of C. carinatum Schousb and K. indica are available from the authors. |
| Region | C. carinatum Schousb | K. indica | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| T(U) (%) | C (%) | A (%) | G (%) | Length (bp) | T(U) (%) | C (%) | A (%) | G (%) | Length (bp) | |
| LSC | 32.4 | 17.6 | 32.0 | 18.1 | 82,290 | 32.6 | 17.3 | 32.2 | 17.9 | 84,610 |
| SSC | 35.1 | 14.7 | 34.1 | 16.1 | 18,416 | 34.9 | 14.8 | 33.9 | 16.4 | 18,269 |
| IRa | 28.3 | 22.3 | 28.6 | 20.8 | 24,523 | 28.3 | 22.2 | 28.7 | 20.8 | 25,003 |
| IRb | 28.6 | 20.8 | 28.3 | 22.3 | 24,523 | 28.7 | 20.8 | 28.3 | 22.2 | 25,003 |
| Total | 31.8 | 19.0 | 30.7 | 18.5 | 149,752 | 31.8 | 19.0 | 30.7 | 18.5 | 152,885 |
| CDS | 31.5 | 17.7 | 30.7 | 20.1 | 77,289 | 31.5 | 17.7 | 30.6 | 20.3 | 78,372 |
| 1st position | 23.7 | 18.9 | 30.6 | 26.7 | 25,763 | 23.9 | 18.8 | 30.6 | 26.8 | 26,124 |
| 2nd position | 32.6 | 20.4 | 29.3 | 17.7 | 25,763 | 32.6 | 20.3 | 29.3 | 17.8 | 26,124 |
| 3rd position | 38.3 | 13.7 | 32.1 | 15.9 | 25,763 | 38.0 | 13.9 | 31.8 | 16.3 | 26,124 |
| Group of Genes | C. carinatum Schousb Gene Names | K. indica Gene Names |
|---|---|---|
| Photosystem I | psaA, psaB, psaC, psaI, psaJ | psaA, psaB, psaC, psaI, psaJ |
| Photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ |
| Cytochrome b/f complex | petA, petB *, petD *, petG, petL, petN | petA, petB *, petD *, petG, petL, petN |
| ATP synthase | atpA, atpB, atpE, atpF *, atpH, atpI | atpA, atpB, atpE, atpF *, atpH, atpI |
| NADH dehydrogenase | ndhA *, ndhB * (×2), ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | ndhA *, ndhB * (×2), ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK |
| RuBisCO large subunit | rbcL | rbcL |
| RNA polymerase | rpoA, rpoB, rpoC1 *, rpoC2 | rpoA, rpoB, rpoC1 *, rpoC2 |
| Ribosomal proteins (SSU) | rps2, rps3, rps4, rps7 (×2), rps8, rps11, rps12 ** (×2), rps14, rps15, rps16 *, rps18, rps19 | rps2, rps3, rps4, rps7 (×2), rps8, rps11, rps12 ** (×2), rps14, rps15, rps16 *, rps18, rps19 |
| Ribosomal proteins (LSU) | rpl2 * (×2), rpl14, rpl16 *, rpl20, rpl22, rpl23 (×2), rpl32, rpl33, rpl36 | rpl2 * (×2), rpl14, rpl16 *, rpl20, rpl22, rpl23 (×2), rpl32, rpl33, rpl36 |
| Miscellaneous proteins | accD, clpP **, matK, ccsA, cemA, infA | accD, clpP **, matK, ccsA, cemA, infA |
| Hypothetical chloroplast reading frames (ycf) | ycf1, ycf2 (×2), ycf3 **, ycf4 | ycf1, ycf2 (×2), ycf3 **, ycf4 |
| Transfer RNAs | trnA-UGC (×2), trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnG-GCC, trnG-UCC, trnH-GUG, trnI-CAU (×2), trnI-GAU (×2), trnK-UUU, trnL-CAA(×2), trnL-UAA, trnL-UAG, trnfM-CAU, trnM-CAU, trnN-GUU (×2), trnP-UGG, trnQ-UUG, trnR-ACG (×2), trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC (×2), trnV-UAC, trnW-CCA, trnY-GUA | trnA-UGC (×2), trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnG-GCC, trnG-UCC, trnH-GUG, trnI-CAU (×2), trnI-GAU (×2), trnK-UUU, trnL-CAA(×2), trnL-UAA, trnL-UAG, trnfM-CAU, trnM-CAU, trnN-GUU (×2), trnP-UGG, trnQ-UUG, trnR-ACG (×2), trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC (×2), trnV-UAC, trnW-CCA, trnY-GUA |
| Ribosomal RNAs | rrn4.5 (×2), rrn5 (×2), rrn16 (×2), rrn23 (×2) | rrn4.5 (×2), rrn5 (×2), rrn16 (×2), rrn23 (×2) |
| Pseudogenes | ycf1, rps19 | ycf1, rps19 |
| Total | 132 | 132 |
| C. Gene | Location | Exon I (bp) | Intron I (bp) | Exon II (bp) | Intron II (bp) | Exon III (bp) | K. Gene | Location | Exon I (bp) | Intron I (bp) | Exon II (bp) | Intron II (bp) | Exon III (bp) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| atpF | LSC | 145 | 699 | 410 | atpF | LSC | 145 | 709 | 410 | ||||
| clpP | LSC | 71 | 796 | 291 | 611 | 229 | clpP | LSC | 71 | 814 | 291 | 615 | 229 |
| ndhA | SSC | 539 | 1045 | 553 | ndhA | SSC | 553 | 1064 | 539 | ||||
| ndhB | IR | 777 | 670 | 756 | ndhB | IR | 777 | 674 | 756 | ||||
| petB | LSC | 6 | 751 | 642 | petB | LSC | 6 | 823 | 642 | ||||
| petD | LSC | 8 | 675 | 475 | petD | LSC | 8 | 809 | 475 | ||||
| rpl16 | LSC | 9 | 1029 | 399 | rpl16 | LSC | 9 | 1098 | 399 | ||||
| rpl2 | IR | 391 | 664 | 434 | rpl2 | IR | 391 | 671 | 434 | ||||
| rpoC1 | LSC | 432 | 733 | 1641 | rpoC1 | LSC | 432 | 742 | 1638 | ||||
| rps12 * | LSC | 114 | - | 232 | 536 | 26 | rps12 * | LSC | 114 | 232 | 535 | 26 | |
| rps16 | LSC | 41 | 891 | 184 | rps16 | LSC | 41 | 876 | 226 | ||||
| trnA-UGC | IR | 38 | 812 | 35 | trnA-UGC | IR | 35 | 820 | 38 | ||||
| trnG-UCC | LSC | 23 | 722 | 47 | trnG-UCC | LSC | 23 | 732 | 48 | ||||
| trnI-GAU | IR | 43 | 775 | 35 | trnI-GAU | IR | 38 | 780 | 35 | ||||
| trnK-UUU | LSC | 37 | 2562 | 30 | trnK-UUU | LSC | 37 | 2539 | 35 | ||||
| trnL-UAA | LSC | 37 | 422 | 50 | trnL-UAA | LSC | 37 | 438 | 50 | ||||
| trnV-UAC | LSC | 38 | 573 | 37 | trnV-UAC | LSC | 38 | 573 | 37 | ||||
| ycf3 | LSC | 125 | 698 | 229 | 735 | 153 | ycf3 | LSC | 125 | 702 | 229 | 739 | 153 |
| Amino Acid | Codon | No. | RSCU | tRNA | Amino Acid | Codon | No. | RSCU | tRNA |
|---|---|---|---|---|---|---|---|---|---|
| Phe | UUU | 956 | 1.32 | Stop | UAA | 49 | 1.73 | ||
| Phe | UUC | 496 | 0.68 | trnF-GAA | Stop | UAG | 21 | 0.74 | |
| Leu | UUA | 874 | 1.89 | trnL-UAA | Stop | UGA | 15 | 0.53 | |
| Leu | UUG | 553 | 1.19 | trnL-CAA | His | CAU | 452 | 1.52 | |
| Leu | CUU | 620 | 1.34 | His | CAC | 143 | 0.48 | trnH-GUG | |
| Leu | CUC | 183 | 0.40 | Gln | CAA | 703 | 1.51 | trnQ-UUG | |
| Leu | CUA | 358 | 0.77 | trnL-UAG | Gln | CAG | 227 | 0.49 | |
| Leu | CUG | 190 | 0.41 | Asn | AAU | 978 | 1.56 | ||
| Ile | AUU | 1075 | 1.47 | Asn | AAC | 277 | 0.44 | trnN-GUU | |
| Ile | AUC | 428 | 0.58 | trnI-GAU | Lys | AAA | 1026 | 1.50 | trnK-UUU |
| Ile | AUA | 692 | 0.95 | trnI-CAU | Lys | AAG | 345 | 0.50 | |
| Met | AUG | 613 | 1.00 | trn(f)M-CAU | Asp | GAU | 831 | 1.59 | |
| Val | GUU | 490 | 1.44 | Asp | GAC | 214 | 0.41 | trnD-GUC | |
| Val | GUC | 166 | 0.49 | trnV-GAC | Glu | GAA | 986 | 1.50 | trnE-UUC |
| Val | GUA | 523 | 1.54 | trnV-UAC | Glu | GAG | 331 | 0.50 | |
| Val | GUG | 178 | 0.52 | Cys | UGU | 198 | 1.38 | ||
| Ser | UCU | 580 | 1.77 | Cys | UGC | 88 | 0.62 | trnC-GCA | |
| Ser | UCC | 313 | 0.96 | trnS-GGA | Trp | UGG | 448 | 1.00 | trnW-CCA |
| Ser | UCA | 394 | 1.20 | trnS-UGA | Arg | CGU | 336 | 1.32 | trnR-ACG |
| Ser | UCG | 154 | 0.47 | Arg | CGC | 100 | 0.39 | ||
| Ser | AGA | 474 | 1.86 | Arg | CGA | 339 | 1.33 | ||
| Ser | AGG | 165 | 0.65 | trnS-GCU | Arg | CGG | 118 | 0.46 | |
| Tyr | UAU | 799 | 1.64 | Arg | AGU | 406 | 1.24 | trnR-UCU | |
| Tyr | UAC | 175 | 0.36 | trnY-GUA | Arg | AGC | 118 | 0.36 | |
| Pro | CCA | 322 | 1.17 | trnP-UGG | Gly | GGU | 581 | 1.32 | |
| Pro | CCG | 159 | 0.58 | Gly | GGC | 188 | 0.43 | trnG-GCC | |
| Pro | CCU | 434 | 1.58 | Gly | GGA | 686 | 1.56 | trnG-UCC | |
| Pro | CCC | 184 | 0.67 | Gly | GGG | 305 | 0.69 | ||
| Thr | ACU | 529 | 1.64 | Ala | GCA | 416 | 1.18 | trnA-UGC | |
| Thr | ACC | 236 | 0.73 | trnT-GGU | Ala | GCG | 162 | 0.46 | |
| Thr | ACA | 404 | 1.25 | trnT-UGU | Ala | GCU | 611 | 1.73 | |
| Thr | ACG | 125 | 0.39 | Ala | GCC | 223 | 0.63 |
| Amino Acid | Codon | No. | RSCU | tRNA | Amino Acid | Codon | No. | RSCU | tRNA |
|---|---|---|---|---|---|---|---|---|---|
| Phe | UUU | 982 | 1.31 | Stop | UAA | 49 | 1.73 | ||
| Phe | UUC | 515 | 0.69 | trnF-GAA | Stop | UAG | 21 | 0.74 | |
| Leu | UUA | 870 | 1.87 | trnL-UAA | Stop | UGA | 15 | 0.53 | |
| Leu | UUG | 578 | 1.24 | trnL-CAA | His | CAU | 452 | 1.51 | |
| Leu | CUU | 607 | 1.30 | His | CAC | 148 | 0.49 | trnH-GUG | |
| Leu | CUC | 186 | 0.4 | Gln | CAA | 723 | 1.53 | trnQ-UUG | |
| Leu | CUA | 375 | 0.81 | trnL-UAG | Gln | CAG | 220 | 0.47 | |
| Leu | CUG | 179 | 0.38 | Asn | AAU | 969 | 1.53 | ||
| Ile | AUU | 1072 | 1.47 | Asn | AAC | 295 | 0.47 | trnN-GUU | |
| Ile | AUC | 427 | 0.58 | trnI-GAU | Lys | AAA | 1036 | 1.48 | trnK-UUU |
| Ile | AUA | 691 | 0.95 | trnI-CAU | Lys | AAG | 360 | 0.52 | |
| Met | AUG | 633 | 1 | trn(f)M-CAU | Asp | GAU | 847 | 1.61 | |
| Val | GUU | 503 | 1.45 | Asp | GAC | 205 | 0.39 | trnD-GUC | |
| Val | GUC | 180 | 0.52 | trnV-GAC | Glu | GAA | 987 | 1.47 | trnE-UUC |
| Val | GUA | 517 | 1.49 | trnV-UAC | Glu | GAG | 359 | 0.53 | |
| Val | GUG | 190 | 0.55 | Cys | UGU | 209 | 1.42 | ||
| Ser | UCU | 585 | 1.76 | Cys | UGC | 85 | 0.58 | trnC-GCA | |
| Ser | UCC | 308 | 0.93 | trnS-GGA | Trp | UGG | 463 | 1 | trnW-CCA |
| Ser | UCA | 401 | 1.21 | trnS-UGA | Arg | CGU | 351 | 1.33 | trnR-ACG |
| Ser | AGA | 488 | 1.85 | Arg | CGC | 108 | 0.41 | ||
| Ser | AGG | 177 | 0.67 | trnS-GCU | Arg | CGA | 346 | 1.31 | |
| Ser | UCG | 172 | 0.52 | Arg | CGG | 114 | 0.43 | ||
| Tyr | UAU | 804 | 1.64 | Arg | AGU | 405 | 1.22 | trnR-UCU | |
| Tyr | UAC | 175 | 0.36 | trnY-GUA | Arg | AGC | 121 | 0.36 | |
| Pro | CCU | 414 | 1.50 | Gly | GGU | 571 | 1.28 | ||
| Pro | CCC | 209 | 0.76 | Gly | GGC | 199 | 0.45 | trnG-GCC | |
| Pro | CCA | 314 | 1.14 | trnP-UGG | Gly | GGA | 681 | 1.53 | trnG-UCC |
| Pro | CCG | 167 | 0.61 | Gly | GGG | 328 | 0.74 | ||
| Thr | ACU | 531 | 1.62 | Ala | GCU | 622 | 1.74 | ||
| Thr | ACC | 239 | 0.73 | trnT-GGU | Ala | GCC | 233 | 0.65 | |
| Thr | ACA | 405 | 1.23 | trnT-UGU | Ala | GCA | 410 | 1.15 | trnA-UGC |
| Thr | ACG | 137 | 0.42 | Ala | GCG | 161 | 0.45 |
| No. | Size (bp) | Type | Repeat 1 Location | Repeat 2 Location | Region | No. | Size (bp) | Type | Repeat 1 Location | Repeat 2 Location | Region |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 60 | F | ycf2 (CDS) | ycf2 (CDS) | IRb | 22 | 31 | R | IGS (trnT-GGU, psbD) | IGS (trnT-GGU, psbD) | LSC |
| 2 | 60 | P | ycf2 (CDS) | ycf2 (CDS) | IRb, IRa | 23 | 30 | P | IGS (psbI, trnS-GUC) | trnS-GGA (CDS) | LSC |
| 3 | 60 | P | ycf2 (CDS) | ycf2 (CDS) | IRb, IRa | 24 | 30 | F | ycf2 (CDS) | ycf2 (CDS) | IRb |
| 4 | 60 | F | ycf2 (CDS) | ycf2 (CDS) | IRa, IRa | 25 | 30 | P | ycf2 (CDS) | ycf2 (CDS) | IRb, IRa |
| 5 | 51 | F | IGS (rps11, rpl36) | IGS (rps11, rpl36) | LSC | 26 | 30 | P | ycf2 (CDS) | ycf2 (CDS) | IRb, IRa |
| 6 | 48 | P | IGS (psbT, psbN) | IGS (psbT, psbN) | LSC | 27 | 35 | F | psaB (CDS) | psaA (CDS) | LSC |
| 7 | 46 | F | IGS (accD, psaI) | IGS (accD, psaI) | LSC | 28 | 35 | F | ycf3 (intron2) | ndhB (intron) | LSC, IRb |
| 8 | 45 | F | ycf2 (CDS) | ycf2 (CDS) | IRb | 29 | 35 | P | ycf3 (intron2) | ndhB (intron) | LSC, IRa |
| 9 | 45 | P | ycf2 (CDS) | ycf2 (CDS) | IRb, IRa | 30 | 32 | P | IGS (trnT-GGU, psbD) | IGS (trnT-GGU, psbD) | LSC |
| 10 | 45 | P | ycf2 (CDS) | ycf2 (CDS) | IRb, IRa | 31 | 31 | F | IGS (psbI, trnS-GUC) | IGS (psbI, trnS-GUC) | LSC |
| 11 | 46 | P | IGS (petN, psbM) | IGS (petN, psbM) | LSC | 32 | 30 | P | IGS (psbC, trnS-UGA) | trnS-GGA (CDS) | LSC |
| 12 | 39 | P | IGS (rps12, trnV-GAC) | ndhA (intron) | IRb, SSC | 33 | 30 | F | rbcL (CDS) | IGS (rbcL, accD) | LSC |
| 13 | 39 | F | ndhA (intron) | IGS (trnV-GAC, rps12) | SSC, IRa | 34 | 32 | F | IGS (psbI, trnS-GUC) | IGS (psbC, trnS-UGA) | LSC |
| 14 | 41 | F | ycf3 (intron2) | IGS (rps12, trnV-GAC) | LSC, IRb | 35 | 31 | R | IGS (trnL-UAG, rpl32) | IGS (trnL-UAG, rpl32) | SSC |
| 15 | 41 | P | ycf3 (intron2) | IGS (trnV-GAC, rps12) | LSC, IRa | 36 | 30 | C | IGS (trnT-GGU, psbD) | IGS (trnT-GGU, psbD) | LSC |
| 16 | 39 | P | ycf3 (intron2) | ndhA (intron) | LSC, SSC | 37 | 30 | F | psaB (CDS) | psaA (CDS) | LSC |
| 17 | 42 | F | ycf2 (CDS) | ycf2 (CDS) | IRb | 38 | 30 | F | ycf2 (CDS) | ycf2 (CDS) | IRb, IRa |
| 18 | 42 | P | ycf2 (CDS) | ycf2 (CDS) | IRb, IRa | 39 | 30 | P | ycf2 (CDS) | ycf2 (CDS) | IRb |
| 19 | 42 | P | ycf2 (CDS) | ycf2 (CDS) | IRb, IRa | 40 | 30 | F | IGS (trnL-UAG, rpl32) | IGS (trnL-UAG, rpl32) | SSC |
| 20 | 42 | F | ycf2 (CDS) | ycf2 (CDS) | IRa | 41 | 30 | R | IGS (trnL-UAG, rpl32) | IGS (trnL-UAG, rpl32) | SSC |
| 21 | 41 | P | IGS (ndhE, psaC) | IGS (psaC, ndhD) | SSC | 42 | 30 | P | ycf2 (CDS) | ycf2 (CDS) | IRa |
| No. | Size (bp) | Type | Repeat 1 Location | Repeat 2 Location | Region | No. | Size (bp) | Type | Repeat 1 Location | Repeat 2 Location | Region |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 48 | P | IGS (psbT, psbN) | IGS (psbT, psbN) | LSC | 31 | 32 | F | IGS (rrn5, rrn4.5) | IGS (rrn5, rrn4.5) | IRa |
| 2 | 39 | P | ycf3 (intron2) | ndhA (intron) | LSC, SSC | 32 | 34 | R | IGS (trnT-GGU, psbD) | IGS (trnT-GGU, psbD) | LSC |
| 3 | 41 | F | ycf3 (intron2) | IGS (rps12, trnV-GAC) | LSC, IRb | 33 | 31 | R | IGS (ndhC, trnV-UAC) | petB (intron) | LSC |
| 4 | 41 | P | ycf3 (intron2) | IGS (trnV-GAC, rps12) | LSC, IRa | 34 | 33 | C | IGS (accD, psaI) | petD (CDS) | LSC |
| 5 | 39 | P | IGS (rps12, trnV-GAC) | ndhA (intron) | IRb, SSC | 35 | 33 | C | IGS (accD, psaI) | IGS (accD, psaI) | LSC |
| 6 | 39 | F | ndhA (intron) | IGS (trnV-GAC, rps12) | SSC, IRa | 36 | 30 | P | IGS (psbC, trnS-UGA) | trnS-GGA (CDS) | LSC |
| 7 | 42 | F | ycf2 (CDS) | ycf2 (CDS) | IRb | 37 | 32 | C | IGS (trnH-GUG, psbA) | IGS (accD, psaI) | LSC |
| 8 | 42 | P | ycf2 (CDS) | ycf2 (CDS) | IRb, IRa | 38 | 32 | F | IGS (psbI, trnS-GCU) | IGS (psbC, trnS-UGA) | LSC |
| 9 | 42 | P | ycf2 (CDS) | ycf2 (CDS) | IRb, IRa | 39 | 32 | P | IGS (petN, psbM) | IGS (petN, psbM) | LSC |
| 10 | 42 | F | ycf2 (CDS) | ycf2 (CDS) | IRa | 40 | 32 | C | IGS (trnG-UCC, trnT-GGU) | petD (intron) | LSC |
| 11 | 41 | R | petD (intron) | petD (intron) | LSC | 41 | 32 | F | psaB (CDS) | psaA (CDS) | LSC |
| 12 | 43 | P | IGS (psaC, ndhD) | IGS (psaC, ndhD) | SSC | 42 | 32 | R | IGS (accD, psaI) | IGS (accD, psaI) | LSC |
| 13 | 39 | R | IGS (accD, psaI) | IGS (accD, psaI) | LSC | 43 | 31 | P | IGS (trnG-UCC, trnT-GGU) | IGS (ndhC, trnV-UAC) | LSC |
| 14 | 31 | F | IGS (rps12, trnV-GAC) | IGS (rps12, trnV-GAC) | IRb | 44 | 31 | P | IGS (trnG-UCC, trnT-GGU) | IGS (trnG-UCC, trnT-GGU) | LSC |
| 15 | 31 | P | IGS (rps12, trnV-GAC) | IGS (trnV-GAC, rps12) | IRb, IRa | 45 | 31 | P | IGS (trnT-GGU, psbD) | IGS (trnT-UGU, trnL-UAA) | LSC |
| 16 | 31 | P | IGS (rps12, trnV-GAC) | IGS (trnV-GAC, rps12) | IRb, IRa | 46 | 31 | F | IGS (psaA, ycf3) | IGS (psaA, ycf3) | LSC |
| 17 | 31 | F | IGS (trnV-GAC, rps12) | IGS (trnV-GAC, rps12) | IRa | 47 | 31 | R | IGS (accD, psaI) | IGS (accD, psaI) | LSC |
| 18 | 37 | P | IGS (rpl22, rps19) | IGS (rpl22, rps19) | LSC | 48 | 31 | F | IGS (accD, psaI) | petD (intron) | LSC |
| 19 | 31 | R | petD (intron) | petD (intron) | LSC | 49 | 31 | C | petD (intron) | petD (intron) | LSC |
| 20 | 30 | P | IGS (psbI, trnS-GCU) | trnS-GGA (CDS) | LSC | 50 | 31 | F | ndhF (CDS) | IGS (ndhF, ycf1) | SSC |
| 21 | 30 | F | ycf2 (CDS) | ycf2 (CDS) | IRb | 51 | 30 | C | IGS (trnG-UCC, trnT-GGU) | IGS (trnT-GGU, psbD) | LSC |
| 22 | 30 | P | ycf2 (CDS) | ycf2 (CDS) | IRb, IRa | 52 | 30 | R | IGS (trnT-GGU, psbD) | IGS (trnT-GGU, psbD) | LSC |
| 23 | 30 | P | ycf2 (CDS) | ycf2 (CDS) | IRb, IRa | 53 | 30 | C | ycf3 (intron2) | IGS (ndhC, trnV-UAC) | LSC |
| 24 | 32 | P | IGS (trnG-UCC, trnT-GGU) | IGS (trnG-UCC, trnT-GGU) | LSC | 54 | 30 | C | IGS (ndhC, trnV-UAC) | IGS (accD, psaI) | LSC |
| 25 | 32 | P | IGS (psbZ, trnG-GCC) | IGS (psbZ, trnG-GCC) | LSC | 55 | 30 | F | IGS (ndhC, trnV-UAC) | IGS (ndhC, trnV-UAC) | LSC |
| 26 | 32 | P | IGS (accD, psaI) | petD (CDS) | LSC | 56 | 30 | F | IGS (accD, psaI) | IGS (accD, psaI) | LSC |
| 27 | 32 | R | petD (CDS) | petD (CDS) | LSC | 57 | 30 | R | IGS (accD, psaI) | IGS (accD, psaI) | LSC |
| 28 | 32 | F | IGS (rrn4.5, rrn5) | IGS (rrn4.5, rrn5) | IRb | 58 | 30 | R | IGS (accD, psaI) | petD (intron) | LSC |
| 29 | 32 | P | IGS (rrn4.5, rrn5) | IGS (rrn5, rrn4.5) | IRb, IRa | 59 | 30 | F | petD (intron) | petD (intron) | LSC |
| 30 | 32 | P | IGS (rrn4.5, rrn5) | IGS (rrn5, rrn4.5) | IRb, IRa |
| Unit | Length | No. | Location | Region | Unit | Length | No. | Location | Region | Unit | Length | No. | Location | Region |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | 19 | 1 | IGS (rpl32, ndhF) | SSC | T | 22 | 1 | IGS (atpF, atpA) | LSC | TA | 14 | 1 | IGS (trnH-GUG, psbA) | LSC |
| 13 | 1 | IGS (psbE, petL) | LSC | 16 | 1 | rps16 (intron) | LSC | 12 | 2 | IGS (trnT-GGU, psbD) | LSC | |||
| 12 | 1 | IGS (ycf1, rps15) | SSC | 14 | 3 | IGS (trnE-UUC, rpoB) | LSC | IGS (rpl33, rps18) | LSC | |||||
| 11 | 6 | IGS (trnK-UUU, rps16) | LSC | IGS (atpB, rbcL) | LSC | 10 | 1 | rpoC1 (exonII) | LSC | |||||
| IGS (trnR-UCU, trnG-UCC) | LSC | IGS (rps8, rpl14) | LSC | TG | 10 | 1 | IGS (rps16, trnQ-UUG) | LSC | ||||||
| IGS (psbZ, trnG-GCC) | LSC | 13 | 3 | IGS (ndhC, trnV-UAC) | LSC | ATT | 12 | 2 | IGS (cemA, petA) | LSC | ||||
| IGS (psbE, petL) | LSC | IGS (rbcL, accD) | LSC | IGS (trnL-UAG, rpl32) | SSC | |||||||||
| IGS (rps18, rpl20) | LSC | IGS (psbB, psbT) | LSC | GAA | 15 | 1 | ycf1 | SSC | ||||||
| IGS (petD, rpoA) | LSC | 12 | 5 | IGS (trnH-GUG, psbA) | LSC | TTA | 12 | 1 | ndhA (intron) | SSC | ||||
| 10 | 10 | trnk-UUU (intron) | LSC | IGS (trnD-GUC, trnY-GUA) | LSC | TTC | 12 | 1 | psbC | LSC | ||||
| rpoB | LSC | ycf3 (intronII) | LSC | AATA | 12 | 2 | IGS (trnK-UUU, rps16) | LSC | ||||||
| rpoC1 (intron) | LSC | clpP (intronI) | LSC | ndhA (intron) | SSC | |||||||||
| rpoC1 (exonII) | LSC | ycf1 | LSC | ATAC | 12 | 1 | IGS (trnF-GAA, ndhJ) | LSC | ||||||
| rpoC1 (exonII) | LSC | 11 | 1 | IGS (petA, psbJ) | LSC | ATTG | 12 | 1 | ycf2 | IRa | ||||
| IGS (psaA, ycf3) | LSC | 10 | 9 | trnk-UUU (intron) | LSC | ATTT | 22 | 1 | IGS (atpF, atpA) | LSC | ||||
| IGS (ycf3, trnS-GGA) | LSC | IGS (psbM, trnD-GUC) | LSC | CAAT | 12 | 1 | IGS (ycf2, trnL-CAA) | IRb | ||||||
| IGS (trnT-UGU, rnL-UAA) | LSC | rpoC1 (intron) | LSC | GATT | 12 | 1 | ndhA (intron) | SSC | ||||||
| psbT | LSC | IGS (atpI, atpH) | LSC | TAGA | 12 | 1 | IGS (rbcL, accD) | LSC | ||||||
| IGS (rrn5, trnR-ACG) | IRb | IGS (atpI, atpH) | LSC | TAAA | 12 | 1 | IGS (atpI, atpH) | LSC | ||||||
| C | 12 | 1 | rps16 (intron) | LSC | IGS (atpA, trnR-UCU) | LSC | TAAT | 12 | 1 | IGS (petN, psbM) | LSC | |||
| AT | 14 | 1 | rps16 (intron) | LSC | rpoA | LSC | TATT | 12 | 2 | IGS (rpl33, rps18) | LSC | |||
| 12 | 1 | IGS (psbZ, trnG-GCC) | LSC | IGS (rpl14, rpl16) | LSC | ndhD | SSC | |||||||
| 10 | 1 | rpoC2 | LSC | IGS (trnR-ACG, rrn5) | IRa | TTTC | 16 | 1 | rpl16 (intron) | LSC | ||||
| AATTT | 15 | 1 | IGS (ccsA, trnL-UAG) | SSC |
| Unit | Length | No. | Location | Region | Unit | Length | No. | Location | Region | Unit | Length | No. | Location | Region |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | 18 | 1 | IGS (trnS-UGA, psbZ) | LSC | AT | 16 | 1 | IGS (psbZ, trnG-GCC) | LSC | GAA | 15 | 1 | ycf1 | SSC |
| 12 | 1 | IGS (psbZ, trnG-GCC) | LSC | 10 | 7 | rps16 (intron) | LSC | 12 | 1 | ycf1 | SSC | |||
| 11 | 3 | IGS (trnQ-UUG, psbK) | LSC | 10 | rpoC2 | LSC | TAT | 21 | 1 | IGS (rps12, trnV-GAC) | IRb | |||
| 11 | IGS (psbM, trnD-GUC) | LSC | 10 | IGS (psbZ, trnG-GCC) | LSC | 12 | 2 | IGS (trnT-UGU, trnL-UAA) | LSC | |||||
| 11 | IGS (ycf4, cemA) | LSC | 10 | IGS (psbZ, trnG-GCC) | LSC | 12 | IGS (trnF-GAA, ndhJ) | LSC | ||||||
| 10 | 7 | rpoB | LSC | 10 | IGS (psbE, petL) | LSC | TTA | 15 | 2 | IGS (ndhC, trnV-UAC) | LSC | |||
| 10 | rpoC1 (exonII) | LSC | 10 | IGS (petD, rpoA) | LSC | 15 | IGS (clpP, psbB) | LSC | ||||||
| 10 | IGS (petB, petD) | LSC | 10 | rpl16 (intron) | LSC | 12 | 2 | petB (intron) | LSC | |||||
| 10 | ndhB (intron) | IRb | TA | 23 | 1 | petD (intron) | LSC | 12 | petD (intron) | LSC | ||||
| 10 | ndhA (intron) | SSC | 20 | 1 | IGS (accD, psaI) | LSC | 12 | 1 | psbC | LSC | ||||
| 10 | IGS (trnL-UAG, rpl32) | SSC | 18 | 1 | petD (intron) | LSC | 12 | 1 | IGS (trnK-UUU, rps16) | LSC | ||||
| 10 | IGS (rpl2, rps19) | IRa | 14 | 1 | IGS (accD, psaI) | LSC | 12 | 1 | IGS (trnG-UCC, trnT-GGU) | LSC | ||||
| T | 18 | 1 | IGS (trnE-UUC, rpoB) | LSC | 12 | 3 | IGS (trnT-GGU, psbD) | LSC | 12 | 1 | IGS (trnE-UUC, rpoB) | LSC | ||
| 17 | 1 | rpoA | LSC | 12 | petD (intron) | LSC | 12 | 1 | IGS (clpP, psbB) | LSC | ||||
| 14 | 2 | IGS (rpl20, rps12) | LSC | 12 | IGS (rpl22, rps19) | LSC | 12 | 1 | IGS (trnS-GCU, trnC-GCA) | LSC | ||||
| 14 | ndhA (intron) | SSC | 10 | 3 | rpoC1 (exonII) | LSC | 12 | 1 | IGS (ndhI, ndhG) | SSC | ||||
| 13 | 1 | IGS (psbE, petL) | LSC | 10 | IGS (psaJ, rpl33) | LSC | TATC | 12 | 1 | IGS (psbA, trnK-UUU) | LSC | |||
| 12 | 2 | IGS (atpF, atpA) | LSC | 10 | ndhA (intron) | SSC | TATT | 12 | 2 | IGS (rpl33, rps18) | LSC | |||
| 12 | clpP (intronII) | LSC | AAT | 12 | 2 | IGS (trnR-UCU, trnG-UCC) | LSC | 12 | IGS (psaC, ndhD) | SSC | ||||
| 11 | 2 | IGS (atpB, rbcL) | LSC | 12 | IGS (trnT-GGU, psbD) | LSC | TCTA | 12 | 1 | ndhA (intron) | SSC | |||
| 11 | IGS (psaI, ycf4) | LSC | ATA | 21 | 1 | IGS (trnV-GAC, rps12) | IRa | TTCT | 12 | 1 | IGS (trnG-UCC, trnT-GGU) | LSC | ||
| 10 | 9 | IGS (trnC-GCA, petN) | LSC | 15 | 3 | ycf3 (intron) | LSC | TTTA | 12 | 1 | petD (intron) | LSC | ||
| 10 | IGS (rpoC2, rps2) | LSC | 15 | IGS (trnP-UGG, psaJ) | LSC | TTTC | 12 | 1 | trnK-UUU (intron) | LSC | ||||
| 10 | IGS (atpI, atpH) | LSC | 15 | IGS (trnL-UAG, rpl32) | SSC | ATTAG | 15 | 1 | IGS (rbcL, accD) | LSC | ||||
| 10 | IGS (ndhC, trnV-UAC) | LSC | 12 | 2 | IGS (trnG-UCC, trnT-GGU) | LSC | TATAT | 23 | 1 | petD (intron) | LSC | |||
| 10 | clpP (intronI) | LSC | 12 | rpl16 (intron) | LSC | 20 | 1 | IGS (accD, psaI) | LSC | |||||
| 10 | IGS (rps8, rpl14) | LSC | ATT | 15 | 1 | IGS (trnG-UCC, trnT-GGU) | LSC | 15 | 1 | IGS (accD, psaI) | LSC | |||
| 10 | IGS (rps19, rpl2) | IRb | TAA | 15 | 1 | IGS (accD, psaI) | LSC | TATTA | 21 | 2 | IGS (rps12, trnV-GAC) | IRb | ||
| 10 | IGS (psaC, ndhD) | SSC | 12 | 2 | IGS (trnE-UUC, rpoB) | LSC | 21 | IGS (trnV-GAC, rps12) | IRa | |||||
| 10 | ndhB (intron) | IRa | 12 | IGS (trnT-GGU, psbD) | LSC | TCCTA | 15 | 1 | IGS (rps4, trnT-UGU) | LSC |
| Taxon | Genome Size (bp) | GC (%) | SSR Type | CDS | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mono | Di | Tri | Tetra | Penta | Hexa | Total | % a | No. b | % c | |||
| C. carinatum Schousb | 149,752 | 37.47 | 43 | 8 | 5 | 13 | 1 | 0 | 70 | 51.6 | 12 | 17.1 |
| K. indica | 152,885 | 37.25 | 30 | 18 | 22 | 13 | 7 | 0 | 90 | 51.3 | 9 | 10 |
| D. alveolatum | 152,265 | 37.37 | 34 | 14 | 11 | 14 | 2 | 0 | 75 | 51.4 | 8 | 10.7 |
| A. annua | 150,952 | 37.48 | 39 | 10 | 4 | 15 | 3 | 0 | 71 | 52.1 | 10 | 14.1 |
| E. paradoxa | 151,837 | 37.59 | 37 | 4 | 4 | 5 | 0 | 1 | 51 | 51.5 | 14 | 27.5 |
| H. annuus | 151,104 | 37.62 | 40 | 4 | 4 | 4 | 0 | 0 | 52 | 51.2 | 14 | 26.9 |
| T. mongolicum | 151,451 | 37.67 | 14 | 5 | 3 | 6 | 1 | 1 | 30 | 51.3 | 11 | 36.7 |
| C. indicum | 150,972 | 37.48 | 38 | 10 | 4 | 14 | 1 | 0 | 67 | 48.8 | 7 | 10.4 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, X.; Zhou, B.; Yang, H.; Li, Y.; Yang, Q.; Lu, Y.; Gao, Y. Sequencing and Analysis of Chrysanthemum carinatum Schousb and Kalimeris indica. The Complete Chloroplast Genomes Reveal Two Inversions and rbcL as Barcoding of the Vegetable. Molecules 2018, 23, 1358. https://doi.org/10.3390/molecules23061358
Liu X, Zhou B, Yang H, Li Y, Yang Q, Lu Y, Gao Y. Sequencing and Analysis of Chrysanthemum carinatum Schousb and Kalimeris indica. The Complete Chloroplast Genomes Reveal Two Inversions and rbcL as Barcoding of the Vegetable. Molecules. 2018; 23(6):1358. https://doi.org/10.3390/molecules23061358
Chicago/Turabian StyleLiu, Xia, Boyang Zhou, Hongyuan Yang, Yuan Li, Qian Yang, Yuzhuo Lu, and Yu Gao. 2018. "Sequencing and Analysis of Chrysanthemum carinatum Schousb and Kalimeris indica. The Complete Chloroplast Genomes Reveal Two Inversions and rbcL as Barcoding of the Vegetable" Molecules 23, no. 6: 1358. https://doi.org/10.3390/molecules23061358
APA StyleLiu, X., Zhou, B., Yang, H., Li, Y., Yang, Q., Lu, Y., & Gao, Y. (2018). Sequencing and Analysis of Chrysanthemum carinatum Schousb and Kalimeris indica. The Complete Chloroplast Genomes Reveal Two Inversions and rbcL as Barcoding of the Vegetable. Molecules, 23(6), 1358. https://doi.org/10.3390/molecules23061358






