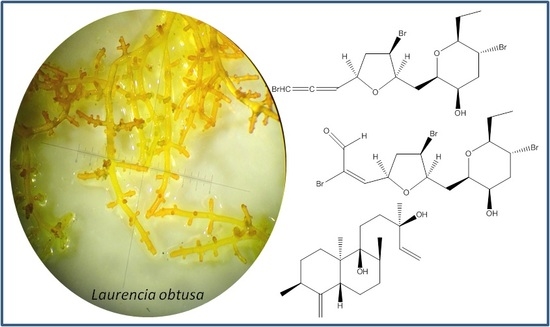
New Metabolites Isolated from a Laurencia obtusa Population Collected in Corsica
Abstract
:1. Introduction
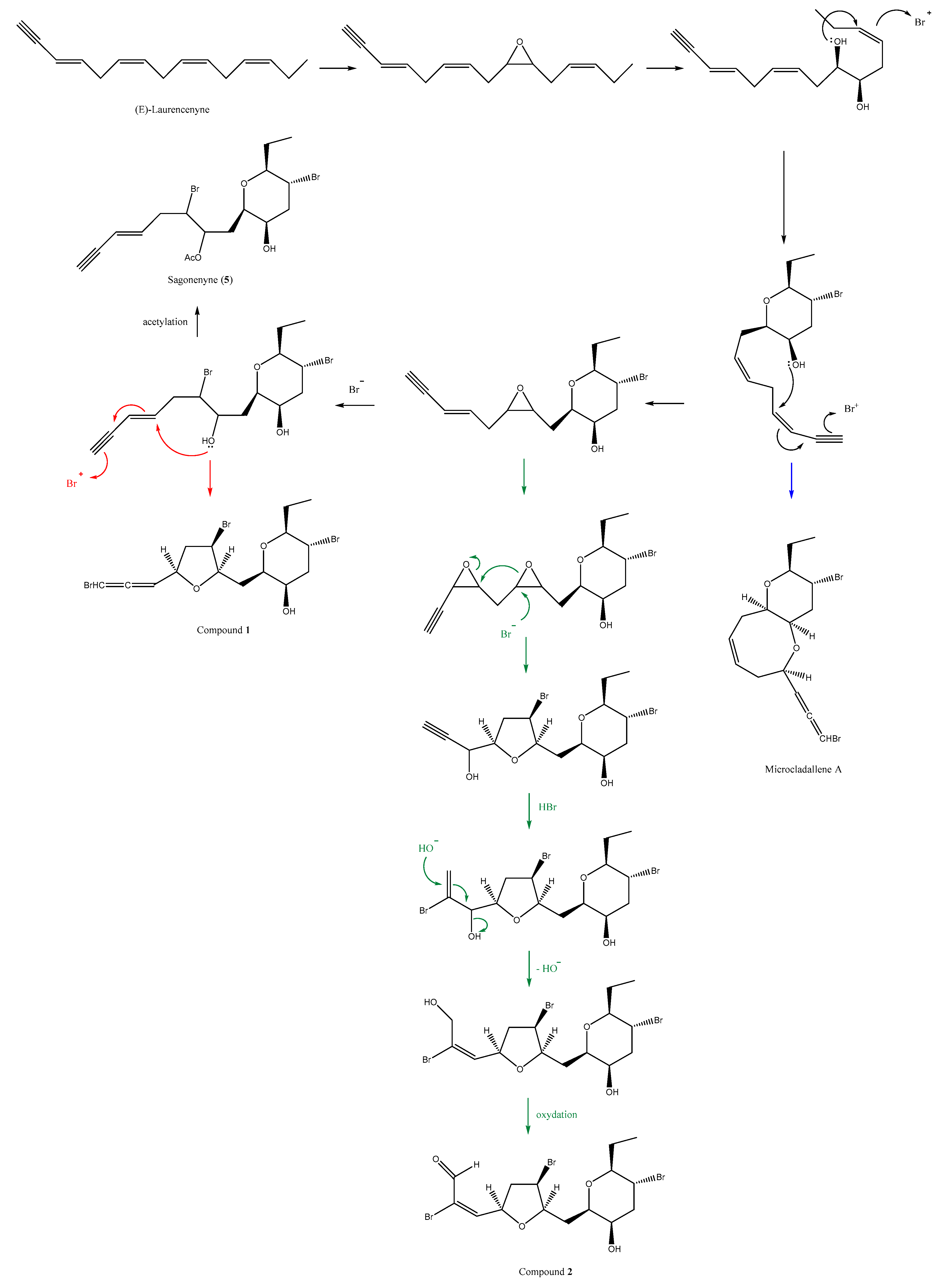
2. Results
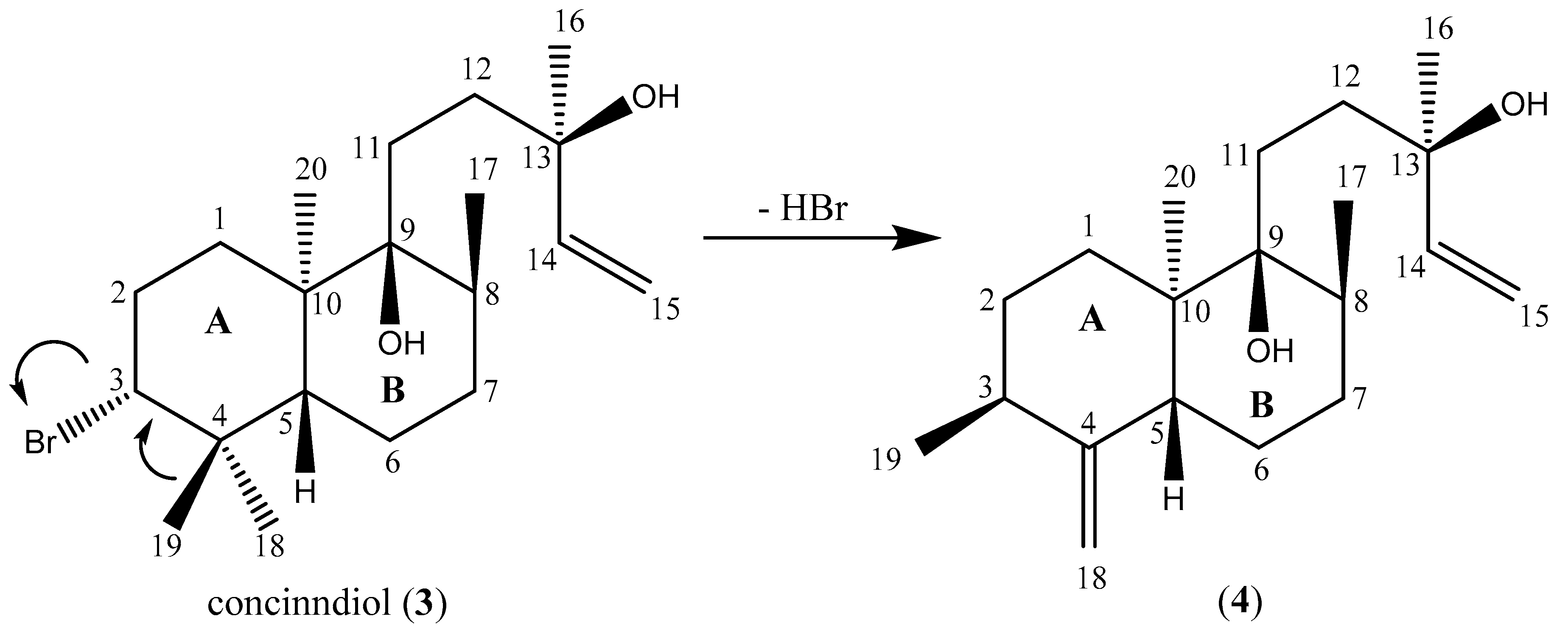
Structure Elucidation of Compounds 1–4
3. Discussion
4. Materials and Methods
4.1. General Experimental Procedures
4.2. Plant Material
4.3. Extraction and Isolation
4.3.1. Compound 1
4.3.2. Compound 2
4.3.3. Concinndiol (Compound 3)
4.3.4. Compound 4
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Al Massarani, S.M. Phytochemical and biological properties of sesquiterpene constituents from the marine red seaweed Laurencia: A review. Nat. Prod. Chem. Res. 2014, 2, 1–13. [Google Scholar]
- De Carvalho, L.R.; Farias, J.N.; Riul, P.; Fujii, M.T. An overview of global distribution of the diterpenes synthesized by the red algae Laurencia complex (Ceremiales, Rhodomelaceae). In Marine Algae Extracts: Processes, Products, and Applications, 1st ed.; Kim, S.K., Chojnacka, K., Eds.; Wiley-VCH Verlag GmbH & Co. KGaA: Winheim, Germany, 2015; Volume 2, pp. 245–266. ISBN 978-3-527-33708-8. [Google Scholar]
- Wang, B.G.; Gloer, J.B.; Ji, N.Y.; Zhao, J.C. Halogenated organic molecules of rhodomelaceae origin: Chemistry and biology. Chem. Rev. 2013, 113, 3632–3685. [Google Scholar] [CrossRef] [PubMed]
- Dembitsky, V.M.; Tolstikov, A.G.; Tolstikov, G.A. Natural halogenated non-terpenic C15-acetogenins of sea organisms. Chem. Sustain. Dev. 2003, 11, 329–339. [Google Scholar]
- Wanke, T.; Phillippus, A.C.; Zatelli, G.A.; Vieira, L.F.O.; Lhullier, C.; Falkenberg, M. C15-acetogenins from the Laurencia complex: 50 years of research—An overview. Rev. Bras. Farmacogn. 2015, 25, 569–587. [Google Scholar] [CrossRef]
- Kornprobst, J.M. Substances Naturelles d’Origine Marine; Lavoisier: Paris, France, 2005; pp. 358–369. ISBN 2-7430-0721-4. [Google Scholar]
- AlgaeBase. Available online: http://www.algaebase.org/ (accessed on 22 January 2018).
- Sutour, S.; Esselin, H.; Bighelli, A.; Casanova, J.; Le Gall, L.; Tomi, F. Discrimination and characterization of two Mediterranean species from the Laurencia complex (Rhodomelaceae) using an NMR-based metabolomic approach. Chem. Biodivers. 2017, 14, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Esselin, H.; Sutour, S.; Liberal, J.; Cruz, M.T.; Salgueiro, L.; Siegler, B.; Freuze, I.; Castola, V.; Paoli, M.; Bighelli, A.; et al. Chemical composition of Laurencia obtusa extract and isolation of a new C15-acetogenin. Molecules 2017, 22, 779. [Google Scholar] [CrossRef] [PubMed]
- Norte, M.; Fernandez, J.J.; Ruano, J.Z. Three new bromo ethers from the red alga Laurencia obtusa. Tetrahedron 1989, 45, 5987–5994. [Google Scholar] [CrossRef]
- Howard, B.M.; Fenical, W. Isoconcinndiol, a brominated diterpenoid from Laurencia snyderae var. guadalupensis. Phytochemistry 1980, 19, 2774–2776. [Google Scholar] [CrossRef]
- Sims, J.J.; Lin, G.H.Y.; Wing, R.M.; Fenical, W. Marine natural products. Concinndiol, a bromo-diterpene alcohol from the red alga, Laurencia concinna. J. Chem. Soc. Chem. Commun. 1973, 470–471. [Google Scholar] [CrossRef]
- Corriero, G.; Madaio, A.; Mayol, L.; Piccialli, V.; Sica, D. Rotalin A and B, two novel diterpene metabolites from the encrusting Mediterranean sponge Mycale rotalis (Bowerbank). Tetrahedron 1989, 45, 277–288. [Google Scholar] [CrossRef]
- Harizani, M.; Ioannou, E.; Roussis, V. The Laurencia paradox: An endless source of chemiodiversity. Prog. Chem. Org. Nat. Prod. 2016, 102, 91–252. [Google Scholar] [PubMed]
- Giordano, F.; Mayol, L.; Notaro, G.; Piccialli, V.; Sica, D. Structure and absolute configuration of two new polybrominated C15 acetogenins from the sponge Mycale Rotalis. J. Chem. Soc. Chem. Commun. 1990, 1559–1561. [Google Scholar] [CrossRef]
- Imre, S.; Aydogmus, Z.; Güner, H.; Lotter, H. Polybrominated non-terpenoid C15 compounds from Laurencia paniculata and Laurencia obtusa. Z. Naturforsch. C 1995, 50, 743–747. [Google Scholar]
- Mihopoulos, N.; Vagias, C.; Scoullos, M.; Roussis, V. Laurencienyne B, a new acetylenic cyclic ether from the red alga Laurencia obtusa. Nat. Prod. Lett. 1999, 13, 151–156. [Google Scholar] [CrossRef]
- Erickson, K.L. Constituents of Laurencia. In Marine Natural Products: Chemical and Biological Perspectives; Academic Press: London, UK, 1983; pp. 131–257. [Google Scholar]
- Moore, R.E. Algal nonisoprenoids. In Marine Natural Products: Chemical and Biological Perspectives; Academic Press: London, UK, 1978; pp. 44–121. [Google Scholar]
- Suzuki, M.; Takahashi, Y.; Matsuo, Y.; Guiry, M.D.; Masuda, M. Scanlonenyne, a novel halogenated C15-acetogenin from the red alga Laurencia obtusa on Irish waters. Tetrahedron 1997, 12, 4271–4278. [Google Scholar] [CrossRef]
- Kigoshi, H.; Shizuri, Y.; Niwa, H.; Yamada, K. Four new C15 acetylenic polyenes of biogenetic significance from the red alga Laurencia okamurai: Structures and synthesis. Tetrahedron 1986, 42, 3781–3787. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |

| 1 | 2 | |||
|---|---|---|---|---|
| δH (J in Hz) | δC | δH (J in Hz) | δC | |
| 1 | 6.11 (dd 5.7, 1.4) | 73.94 | 9.22 (s) | 185.19 |
| 2 | - | 201.63 | - | 126.11 |
| 3 | 5.66 (dd 7.2, 5.7) | 102.01 | 7.54 (d 6.3) | 156.11 |
| 4 | 4.64 (dddd 9.0, 7.2, 4.4, 1.4) | 74.83 | 5.09 (ddd 10.1, 6.3, 3.8) | 76.56 |
| 5a | 2.97 (ddd 15.0, 9.0, 6.2) | 43.01 | 3.24 (ddd 15.0, 10.1, 5.6) | 44.12 |
| 5b | 2.46 (ddd 15.0, 4.4, 1.8) | 2.47 (ddd 15.0, 3.8, 1.1) | ||
| 6 | 4.46 (ddd 6.2, 3.5, 1.8) | 53.08 | 4.54 (ddd 5.6, 3.0, 1.1) | 53.68 |
| 7 | 3.89 (ddd 9.5, 3.5, 2.3) | 79.09 | 3.97 (ddd 9.2, 3.0, 2.8) | 79.93 |
| 8a | 2.01 (m) | 36.20 | 2.05 (m) | 36.24 |
| 8b | 1.85 (ddd 14.5, 9.5, 4.1) | 1.89 (ddd 14.7, 9.2, 3.7) | ||
| 9 | 3.71 (ddd 9.6, 4.1, 1.1) | 77.16 | 3.71 (ddd 10.0, 3.7, 1.1) | 77.22 |
| 10 | 3.75 (ddd 3.2, 2.8, 1.1) | 69.75 | 3.75 (ddd 3.2, 3.0, 1.1) | 69.82 |
| 11a | 2.60 (ddd 13.6, 4.6, 3.2) | 43.17 | 2.61 (ddd 13.8, 4.6, 3.2) | 43.23 |
| 11b | 2.13 (ddd 13.6, 12.3, 2.8) | 2.14 (m) | ||
| 12 | 4.03 (ddd 12.3, 10.2, 4.6) | 47.95 | 4.03 (ddd 12.4, 10.1, 4.6) | 47.58 |
| 13 | 3.38 (ddd 10.2, 8.8, 2.3) | 83.53 | 3.39 (ddd 10.1, 8.7, 2.4) | 83.63 |
| 14a | 2.06 (m) | 26.36 | 2.07 (m) | 26.32 |
| 14b | 1.51 (m) | 1.52 (m) | ||
| 15 | 0.97 (t 7.4) | 9.57 | 0.99 (t 7.4) | 9.58 |
| 3 | 4 | |||
|---|---|---|---|---|
| δH (J in Hz) | δC | δH (J in Hz) | δC | |
| 1a | 1.88 (m) | 34.55 | 2.14 (m) | 27.21 |
| 1b | 1.47 (m) | 1.30 (m) | ||
| 2 | 2.18 (m) | 32.00 | 1.75 (m) | 29.65 |
| 2b | 2.05 (m) | 1.43 (m) | ||
| 3 | 4.07 (dd 12.7, 4.2) | 71.35 | 2.47 (m) | 39.53 |
| 4 | - | 40.27 | - | 157.43 |
| 5 | 1.85 (m) | 47.54 | 2.69 (m) | 39.32 |
| 6a | 1.59 (m) | 24.08 | 1.37 (m) | 25.30 |
| 6b | 1.44 (m) | |||
| 7a | 1.42 m) | 32.18 | 1.52 (m) | 31.09 |
| 7b | 1.40 (m) | |||
| 8 | 1.72 (m) | 37.12 | 1.83 (m) | 36.00 |
| 9 | - | 76.87 | - | 76.51 |
| 10 | - | 44.33 | - | 45.97 |
| 11a | 1.67 (m) | 29.20 | 1.66 (m) | 29.85 |
| 11b | 1.58 (m) | |||
| 12 | 1.62 (m) | 38.47 | 1.64 (m) | 38.47 |
| 13 | - | 73.36 | - | 73.32 |
| 14 | 5.91 (dd 17.3, 10.7) | 147.17 | 5.92 (dd 17.3, 10.7) | 147.15 |
| 15a | 5.20 (dd 17.3, 1.9) | 111.35 | 5.21 (dd 17.3, 1.9) | 111.34 |
| 15b | 4.96 (dd 10.7, 1.9) | 4.97 (dd 10.7, 1.9) | ||
| 16 | 1.22 (s) | 28.31 | 1.23 (s) | 28.35 |
| 17 | 0.86 (d 6.6) | 16.81 | 0.90 (d 6.6) | 16.83 |
| 18a | 0.96 (s) | 18.74 | 4.77 (t 2.1) | 106.62 |
| 18b | 4.41 (t 2.1) | |||
| 19 | 1.05 (s) | 31.29 | 1.08 (d 7.2) | 20.08 |
| 20 | 0.99 (s) | 16.65 | 0.78 (s) | 15.49 |
| OH (C-9) | 3.04 (br s) | - | 3.00 (br s) | - |
| OH (C-13) | 3.71 (br s) | - | 3.64 (br s) | - |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Esselin, H.; Tomi, F.; Bighelli, A.; Sutour, S. New Metabolites Isolated from a Laurencia obtusa Population Collected in Corsica. Molecules 2018, 23, 720. https://doi.org/10.3390/molecules23040720
Esselin H, Tomi F, Bighelli A, Sutour S. New Metabolites Isolated from a Laurencia obtusa Population Collected in Corsica. Molecules. 2018; 23(4):720. https://doi.org/10.3390/molecules23040720
Chicago/Turabian StyleEsselin, Hélène, Félix Tomi, Ange Bighelli, and Sylvain Sutour. 2018. "New Metabolites Isolated from a Laurencia obtusa Population Collected in Corsica" Molecules 23, no. 4: 720. https://doi.org/10.3390/molecules23040720
APA StyleEsselin, H., Tomi, F., Bighelli, A., & Sutour, S. (2018). New Metabolites Isolated from a Laurencia obtusa Population Collected in Corsica. Molecules, 23(4), 720. https://doi.org/10.3390/molecules23040720