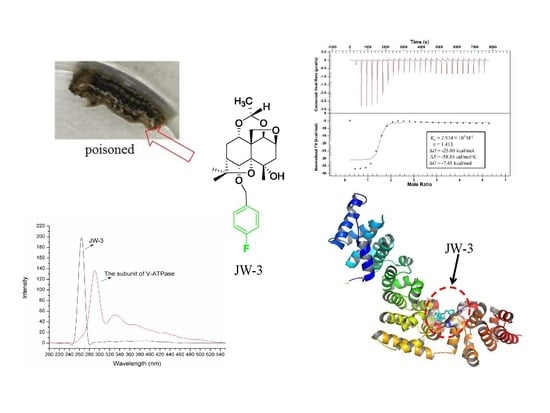
Molecular Insights into the Potential Insecticidal Interaction of β-Dihydroagarofuran Derivatives with the H Subunit of V-ATPase
Abstract
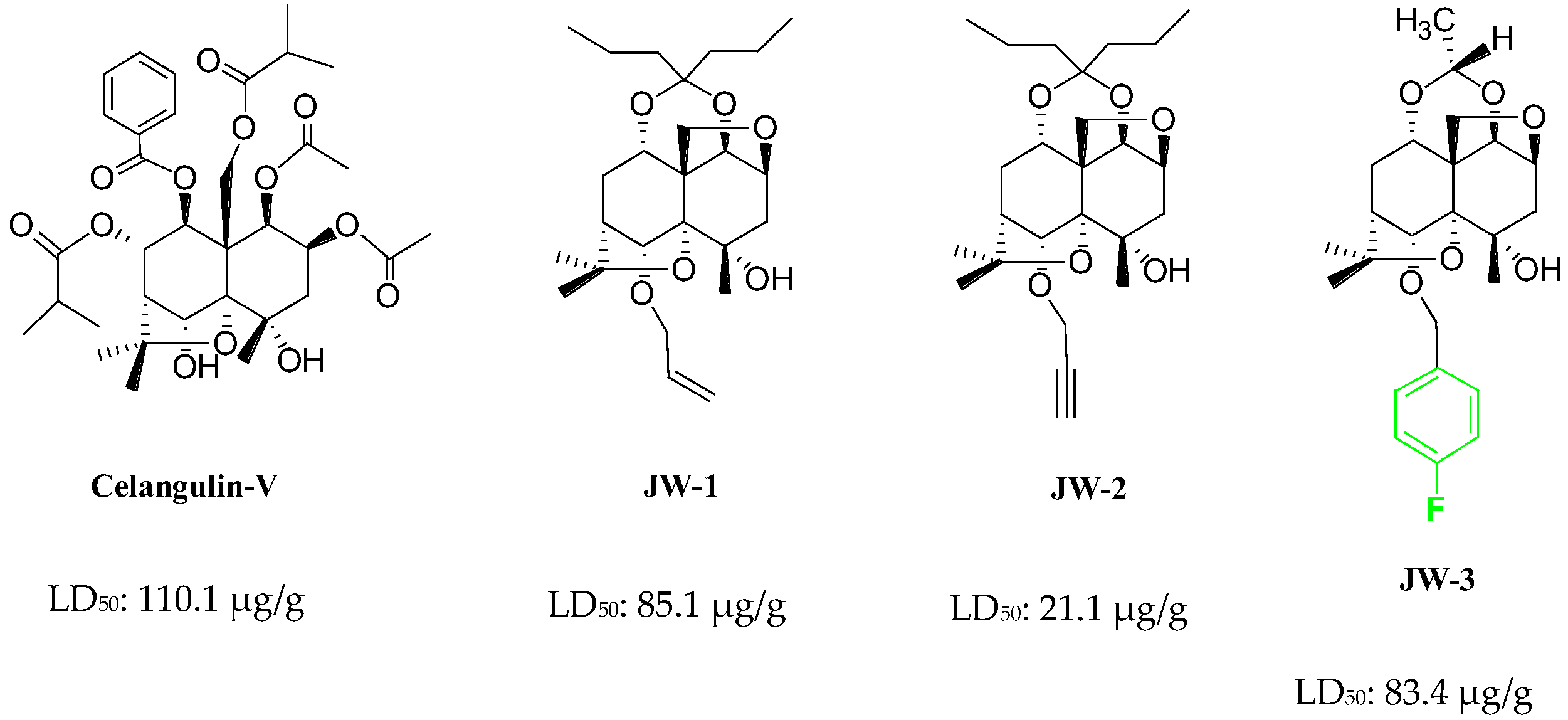
1. Introduction
2. Results
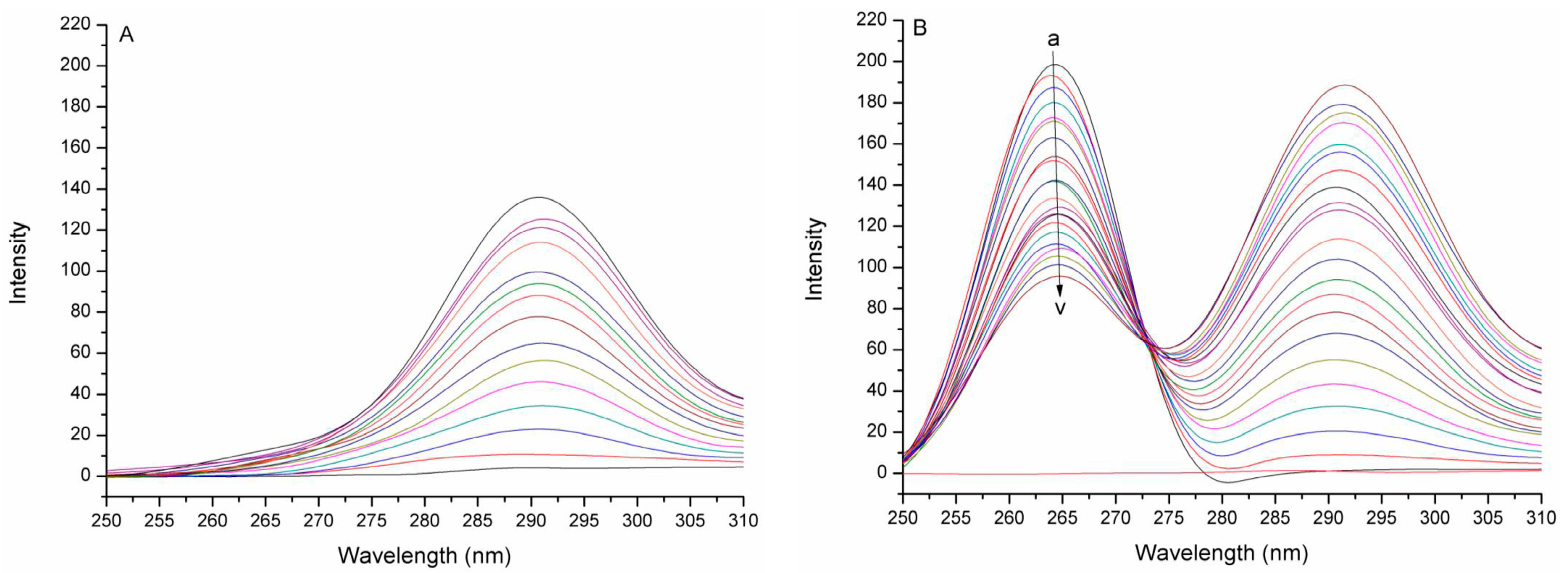
2.1. Synchronous Fluorescence Spectra Studies and Quenching Mechanism Analysis
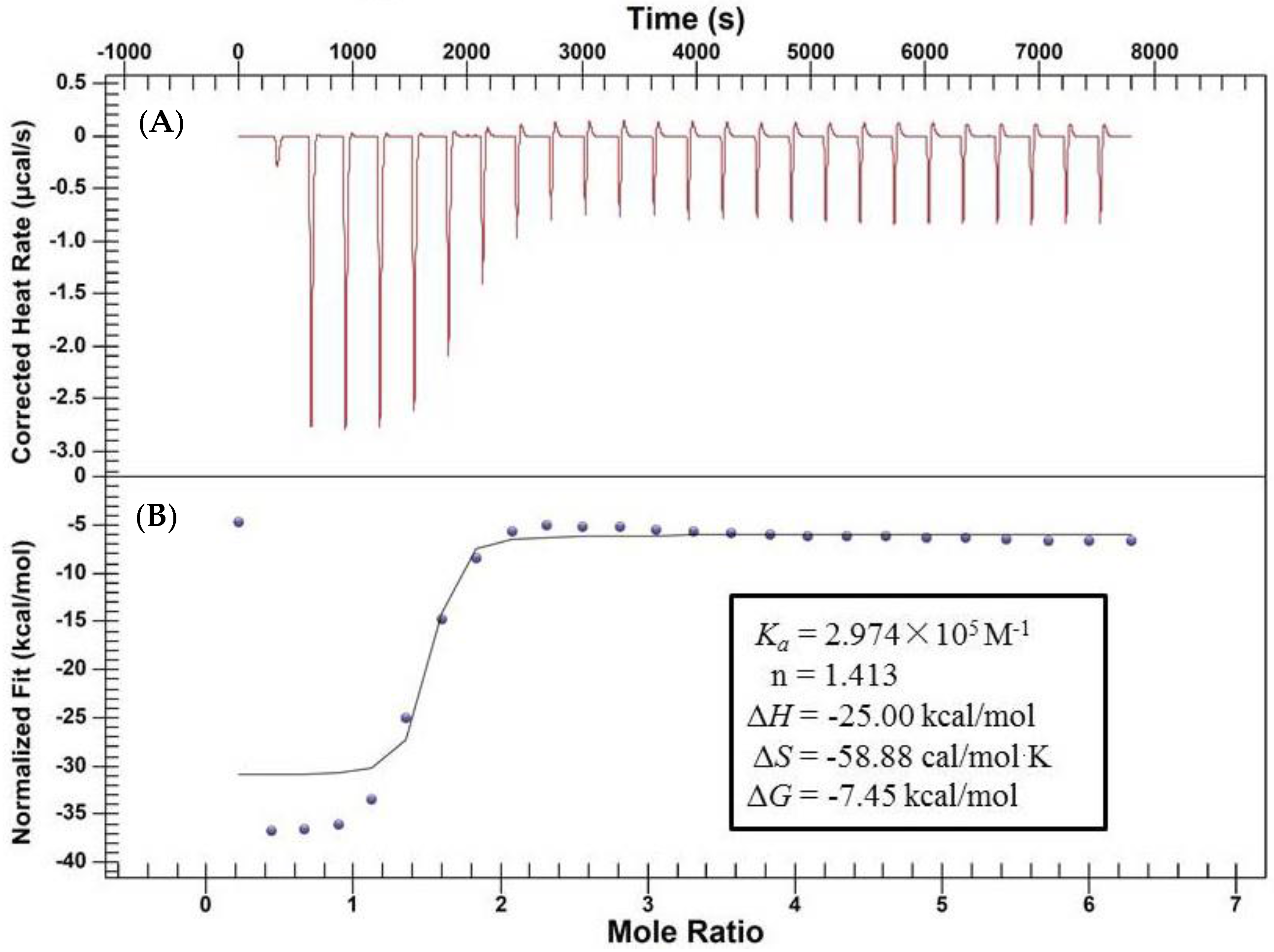
2.2. Characterization of the Binding Interaction between JW-3 and the Subunit H of V-ATPase by Isothermal Titration Calorimetry Measurements
2.3. Homology Modeling
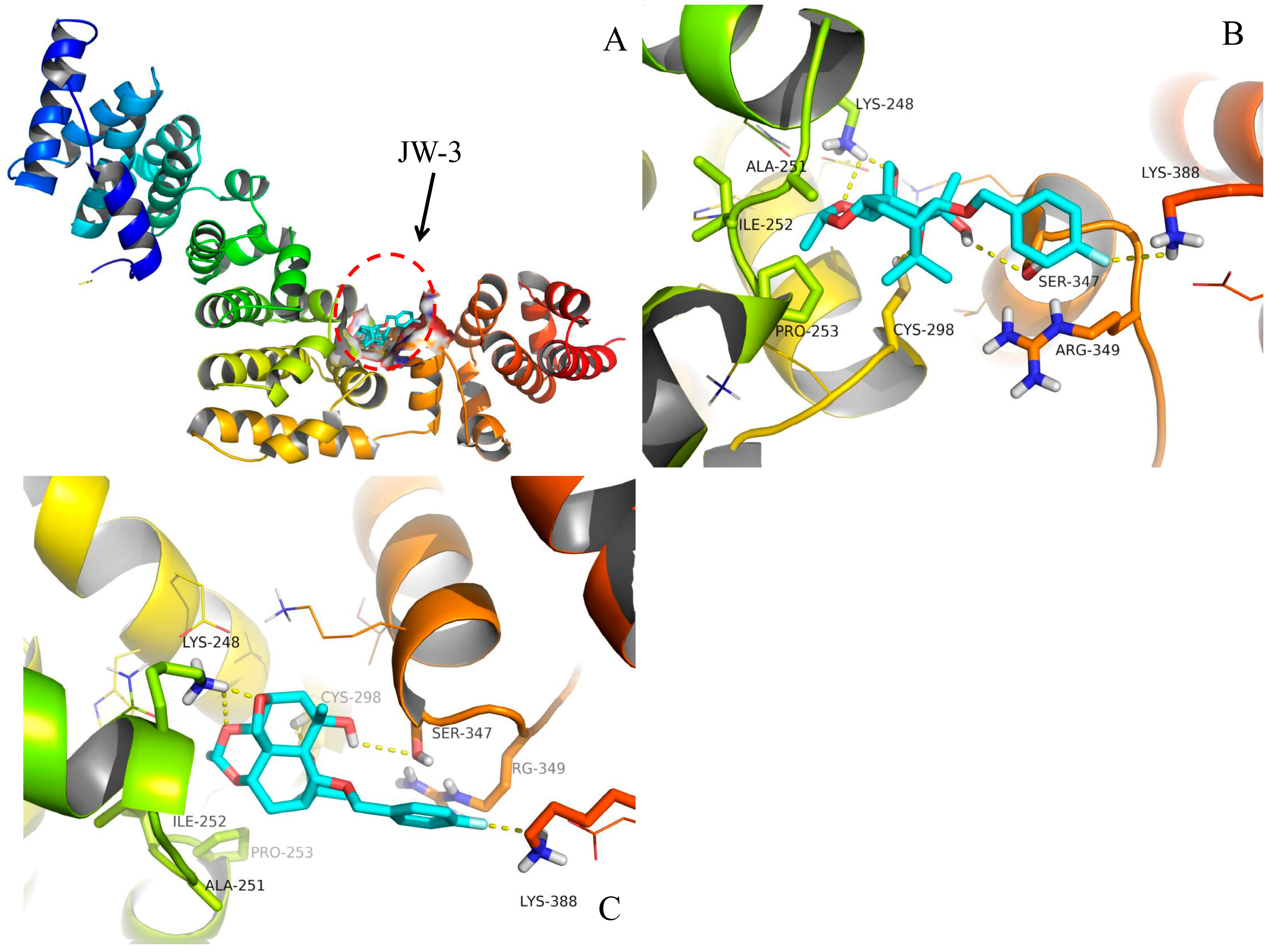
2.4. Molecular Docking
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Expression and Purification of the Subunit H of V-ATPase
4.3. Fluorescence and Synchronous Fluorescence Measurements
4.4. Isothermal Titration Calorimetry (ITC) Experiment
4.5. Data Analysis
4.6. Homology Modeling
4.7. Molecular Docking
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| CV | Celangulin V |
| V-ATPase | Vacuolar H+ATPase |
| TEM | Transmission electron microscopy |
| ITC | Isothermal titration calorimetry |
| SFS | Synchronous fluorescence spectra |
References
- Spivey, A.C.; Weston, M.; Woodhead, S. Celastraceae sesquiterpenoids: Biological activity and synthesis. Chem. Soc. Rev. 2002, 31, 43–59. [Google Scholar] [CrossRef] [PubMed]
- Wakabayashi, N.; Wu, W.; Waters, R.; Redfern, R.; Mills, G., Jr.; DeMilo, A.B.; Lusby, W.R.; Andrzejewski, D. Celangulin: A nonalkaloidal insect antifeedant from Chinese bittersweet, Celastrus angulatus. J. Nat. Prod. 1988, 51, 537–542. [Google Scholar] [CrossRef]
- Wu, W.; Tu, Y.; Liu, H.; Zhu, J. Celangulins II, III, and IV: New insecticidal sesquiterpenoids from Celastrus angulatus. J. Nat. Prod. 1992, 55, 1294–1298. [Google Scholar] [CrossRef]
- Wenjun, W.; Mingan, W.; Wenming, Z.; Jinbo, Z.; Zhiqing, J.; Zhaonong, H. Insecticidal sesquiterpene polyol esters from Celastrus angulatus. Phytochemistry 2001, 58, 1183–1187. [Google Scholar] [CrossRef]
- Gao, J.-M.; Wu, W.-J.; Zhang, J.-W.; Konishi, Y. The dihydro-[small beta]-agarofuran sesquiterpenoids. Nat. Prod. Rep. 2007, 24, 1153–1189. [Google Scholar] [CrossRef] [PubMed]
- Zhiqing, J.; Qidong, Z.; Baojun, S.; Shaopeng, W.; Mingan, W.; Wenjun, W. Three new insecticidal sesquiterpene pyridine alkaloids from Celastrus angulatus. Nat. Prod. Res. 2009, 23, 470–478. [Google Scholar] [CrossRef] [PubMed]
- Zhiqing, J.I.; Wenjun, W.; Hua, Y.; Baojun, S.; Mingan, W. Four novel insecticidal sesquiterpene esters from Celastrus angulatus. Nat. Prod. Res. 2007, 21, 334–342. [Google Scholar] [CrossRef] [PubMed]
- Qi, Z.; Shi, B.; Hu, Z.; Zhang, Y.; Wu, W. Ultrastructural effects of Celangulin V on midgut cells of the oriental armyworm, Mythimna separata walker (Lepidoptera: Noctuidae). Ecotoxicol. Environ. Saf. 2011, 74, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Hu, Z.; Li, J.; Li, L.; Wu, W.; Zhang, J. Synthesis and insecticidal activity of β-dihydroagarofuran ether analogues. Pest Manag. Sci. 2016, 72, 754–759. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Xi, X.; Hu, Z.; Wu, W.; Zhang, J. Exploration of Novel Botanical Insecticide Leads: Synthesis and Insecticidal Activity of β-Dihydroagarofuran Derivatives. J. Agric. Food Chem. 2016, 64, 1503–1508. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Qi, Z.; Li, Q.; Wu, W. Validation of the Target Protein of Insecticidal Dihydroagarofuran Sesquiterpene Polyesters. Toxins 2016, 8, 79. [Google Scholar] [CrossRef] [PubMed]
- Copping, L.G.; Duke, S.O. Natural products that have been used commercially as crop protection agents. Pest Manag. Sci. 2007, 63, 524–554. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Qi, Z.; Zhang, J.; Wu, W. Separation of binding protein of Celangulin V from the Midgut of Mythimna separata Walker by affinity chromatography. Toxins 2015, 7, 1738–1748. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Liao, X.; Li, D.; Feng, L.; Li, J.; Wang, X.; Jin, J.; Yi, F.; Zhou, L.; Wan, J. Study on the interaction between cyanobacteria FBP/SBPase and metal ions. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2012, 89, 337–344. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Liu, Y.; Zhou, S.; Fu, Y.; Li, C. Analysis of the interaction of Dp44mT with human serum albumin and calf thymus DNA using molecular docking and spectroscopic techniques. Int. J. Mol. Sci. 2016, 17, 1042. [Google Scholar] [CrossRef] [PubMed]
- Biasini, M.; Bienert, S.; Waterhouse, A.; Arnold, K.; Studer, G.; Schmidt, T.; Kiefer, F.; Cassarino, T.G.; Bertoni, M.; Bordoli, L. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014, 42, W252–W258. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Han, X.; Tu, Q.; Feng, L.; Wu, D.; Sun, Y.; Chen, H.; Li, Y.; Ren, Y.; Wan, J. Structure-based design and synthesis of novel dual-target inhibitors against cyanobacterial fructose-1, 6-bisphosphate aldolase and fructose-1,6-bisphosphatase. J. Agric. Food Chem. 2013, 61, 7453–7461. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Chi, B.; Wang, W.-W.; Gao, J.-M.; Wan, J. Exploring the possible binding mode of trisubstituted benzimidazoles analogues in silico for novel drug designtargeting Mtb FtsZ. Med. Chem. Res. 2017, 26, 153–169. [Google Scholar] [CrossRef]
- Lovell, S.C.; Davis, I.W.; Arendall, W.B.; de Bakker, P.I.; Word, J.M.; Prisant, M.G.; Richardson, J.S.; Richardson, D.C. Structure validation by Cα geometry: ϕ, ψ and Cβ deviation. Proteins Struct. Funct. Bioinf. 2003, 50, 437–450. [Google Scholar] [CrossRef] [PubMed]
- Bobone, S.; van de Weert, M.; Stella, L. A reassessment of synchronous fluorescence in the separation of Trp and Tyr contributions in protein emission and in the determination of conformational changes. J. Mol. Struct. 2014, 1077, 68–76. [Google Scholar] [CrossRef]
- Cui, M.; Liu, B.; Li, T.; Duan, S. Synchronous Fluorescence Characteristics on the Interaction of Cefotaxime Sodium and Transferrin. BioChem. Indian J. 2016, 10, 105. [Google Scholar]
- Lu, S.; Yu, X.; Yang, Y.; Li, X. Spectroscopic investigation on the intermolecular interaction between N-confused porphyrins-(3-methylisoxazole) diad and bovine serum albumin. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2012, 99, 116–121. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Huang, M.; Mi, C.; Wang, T.; Chen, D.; Teng, Y. Molecular Insights into the Potential Toxicological Interaction of 2-Mercaptothiazoline with the Antioxidant Enzyme—Catalase. Int. J. Mol. Sci. 2016, 17, 1330. [Google Scholar] [CrossRef] [PubMed]
- Perspicace, S.; Rufer, A.C.; Thoma, R.; Mueller, F.; Hennig, M.; Ceccarelli, S.; Schulz-Gasch, T.; Seelig, J. Isothermal titration calorimetry with micelles: Thermodynamics of inhibitor binding to carnitine palmitoyltransferase 2 membrane protein. FEBS Open Biol. 2013, 3, 204–211. [Google Scholar] [CrossRef] [PubMed]
- Sagermann, M.; Stevens, T.H.; Matthews, B.W. Crystal structure of the regulatory subunit H of the V-type ATPase of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2001, 98, 7134–7139. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Beyrakhova, K.; Liu, Y.; Alvarez, C.P.; Bueler, S.A.; Xu, L.; Xu, C.; Boniecki, M.T.; Kanelis, V.; Luo, Z.-Q.; et al. Molecular basis for the binding and modulation of V-ATPase by a bacterial effector protein. PLOS Pathog. 2017, 13, e1006394. [Google Scholar] [CrossRef] [PubMed]
- Oot, R.A.; Kane, P.M.; Berry, E.A.; Wilkens, S. Crystal structure of yeast V1-ATPase in the autoinhibited state. EMBO J. 2016, 35, 1694–1706. [Google Scholar] [CrossRef] [PubMed]
- Karonen, M.; Oraviita, M.; Mueller-Harvey, I.; Salminen, J.-P.; Green, R.J. Binding of an Oligomeric Ellagitannin Series to Bovine Serum Albumin (BSA): Analysis by Isothermal Titration Calorimetry (ITC). J. Agric. Food Chem. 2015, 63, 10647–10654. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- DeLano, W.L. The PyMOL Molecular Graphics System; System; DeLano Scientific: San Carlos, CA, USA, 2016; Available online: http://www.pymol.org/ (accessed on 10 October 2017).
Sample Availability: Samples of the compounds CV, JW1–3 are available from the authors. |

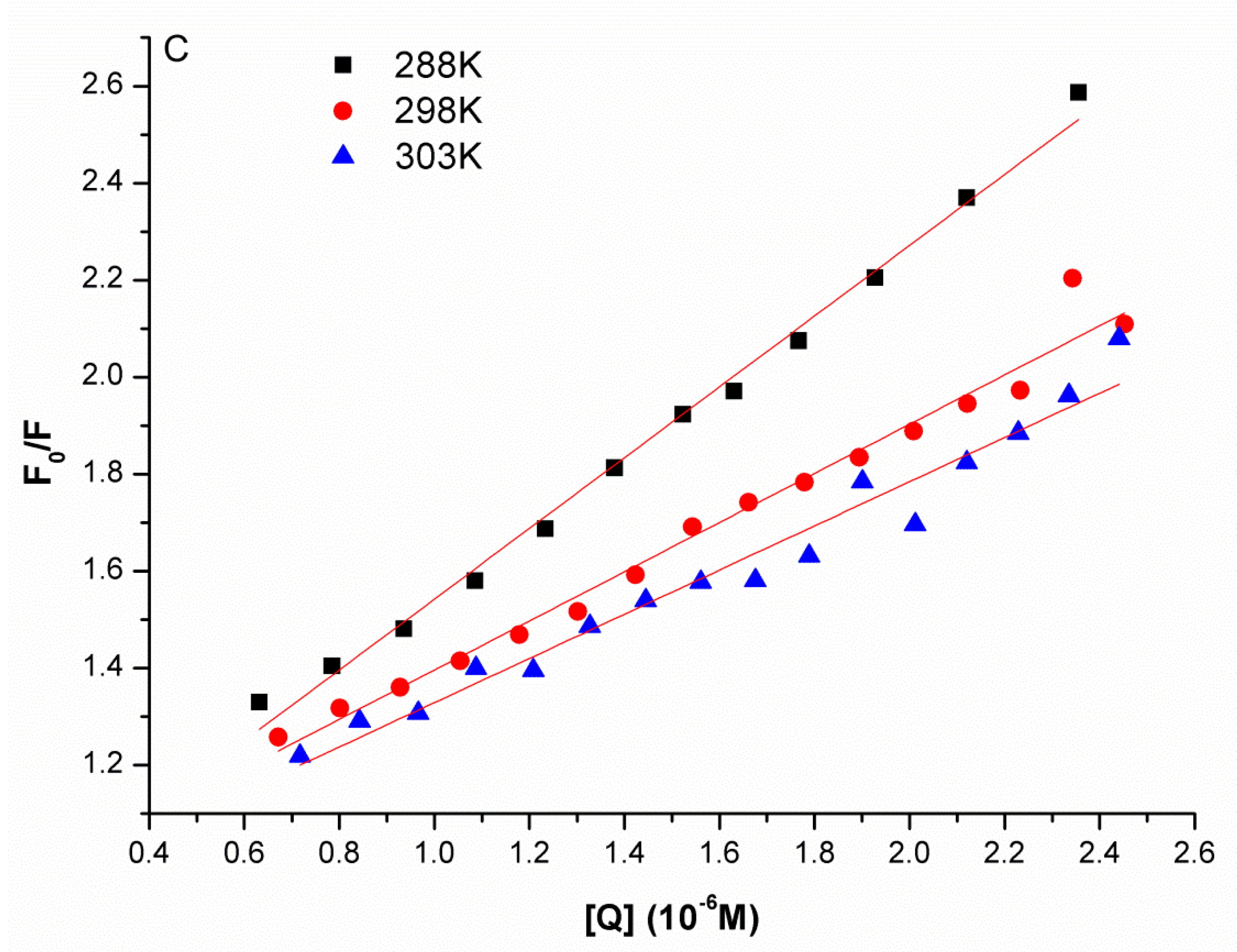
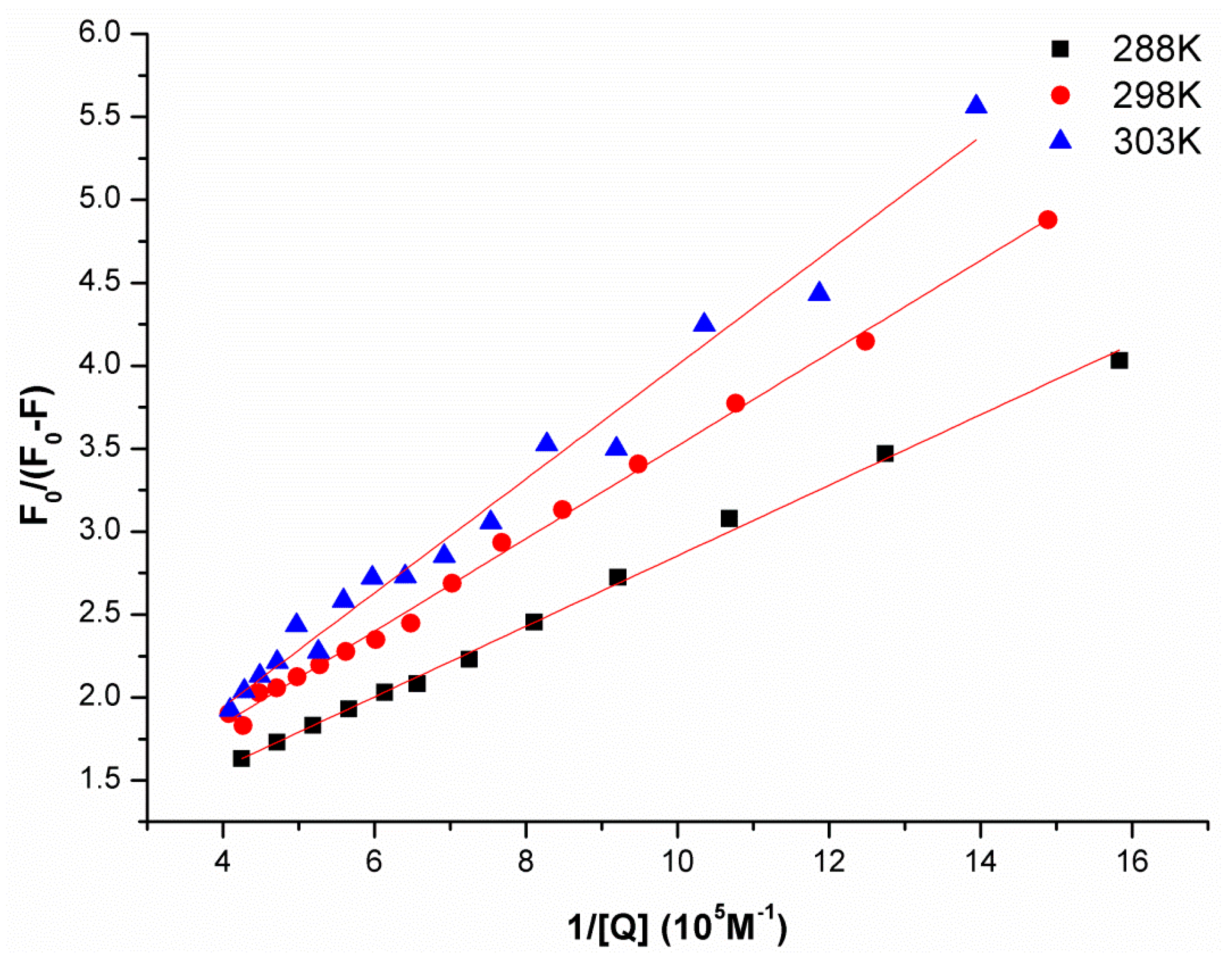

| pH | Temperature/K | KSV (M−1) | R2 |
|---|---|---|---|
| 288 | 7.295 × 105 | 0.9935 | |
| 6.8 | 298 | 5.070 × 105 | 0.9799 |
| 303 | 4.557 × 105 | 0.9662 |
| pH | Temperature/K | Ka (M−1) | R2 |
|---|---|---|---|
| 288 | 3.42 × 105 | 0.9974 | |
| 6.8 | 298 | 2.58 × 105 | 0.9970 |
| 303 | 1.65 × 105 | 0.9840 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, J.; Li, D.; Xi, X.; Liu, L.; Zhao, X.; Wu, W.; Zhang, J. Molecular Insights into the Potential Insecticidal Interaction of β-Dihydroagarofuran Derivatives with the H Subunit of V-ATPase. Molecules 2017, 22, 1701. https://doi.org/10.3390/molecules22101701
Wei J, Li D, Xi X, Liu L, Zhao X, Wu W, Zhang J. Molecular Insights into the Potential Insecticidal Interaction of β-Dihydroagarofuran Derivatives with the H Subunit of V-ATPase. Molecules. 2017; 22(10):1701. https://doi.org/10.3390/molecules22101701
Chicago/Turabian StyleWei, Jielu, Ding Li, Xin Xi, Lulu Liu, Ximei Zhao, Wenjun Wu, and Jiwen Zhang. 2017. "Molecular Insights into the Potential Insecticidal Interaction of β-Dihydroagarofuran Derivatives with the H Subunit of V-ATPase" Molecules 22, no. 10: 1701. https://doi.org/10.3390/molecules22101701
APA StyleWei, J., Li, D., Xi, X., Liu, L., Zhao, X., Wu, W., & Zhang, J. (2017). Molecular Insights into the Potential Insecticidal Interaction of β-Dihydroagarofuran Derivatives with the H Subunit of V-ATPase. Molecules, 22(10), 1701. https://doi.org/10.3390/molecules22101701