Dereplication of Known Nucleobase and Nucleoside Compounds in Natural Product Extracts by Capillary Electrophoresis-High Resolution Mass Spectrometry
Abstract
:1. Introduction
2. Results and Discussion
2.1. Optimization of CE-ESI-TOF/MS Methodology

2.2. Validation of the CE-TOF/MS Method
| Analytes | Precision RSD, %; n = 6 | Reproducibility RSD, %; n = 6 | LOD (µg/mL) | Error (ppm) | ||
|---|---|---|---|---|---|---|
| Intra-Day | Inter-Day | |||||
| 1 | Cytosine | 8.85 | 10.97 | 22.27 | 0.25 | 1.44 |
| 2 | Uracil | 9.87 | 10.14 | 22.63 | 0.25 | 2.18 |
| 3 | Thymine | 14.56 | 7.64 | 24.34 | 0.20 | 1.54 |
| 4 | Adenine | 9.60 | 8.60 | 22.15 | 0.03 | 0.21 |
| 5 | Hypoxanthine | 8.91 | 12.26 | 19.71 | 0.12 | −0.64 |
| 6 | Guanine | 10.93 | 10.56 | 23.42 | 0.20 | 1.41 |
| 7 | Xanthine | 11.45 | 11.25 | 18.84 | 0.25 | −1.32 |
| 8 | Thymidine | 15.74 | 11.72 | 23.33 | 0.50 | 1.54 |
| 9 | Cytidine | 14.75 | ND a | ND a | 1.00 | −1.22 |
| 10 | Uridine | 14.85 | 10.57 | 19.00 | 0.75 | 3.21 |
| 11 | Cordycepin | 13.34 | 6.87 | 9.38 | 0.25 | 0.33 |
| 12 | Adenosine | 13.96 | 14.04 | 17.48 | 1.00 | −1.23 |
| 13 | 2'-Deoxyguanosine | 12.57 | 8.87 | 19.47 | 0.50 | −1.23 |
| 14 | Inosine | 13.79 | 16.11 | 17.32 | 0.75 | 0.20 |
| 15 | Guanosine | 14.58 | 8.34 | 19.52 | 0.75 | −0.86 |
2.3. Dereplication of Nucleobase and Nucleoside Compounds in Real Natural Product Samples
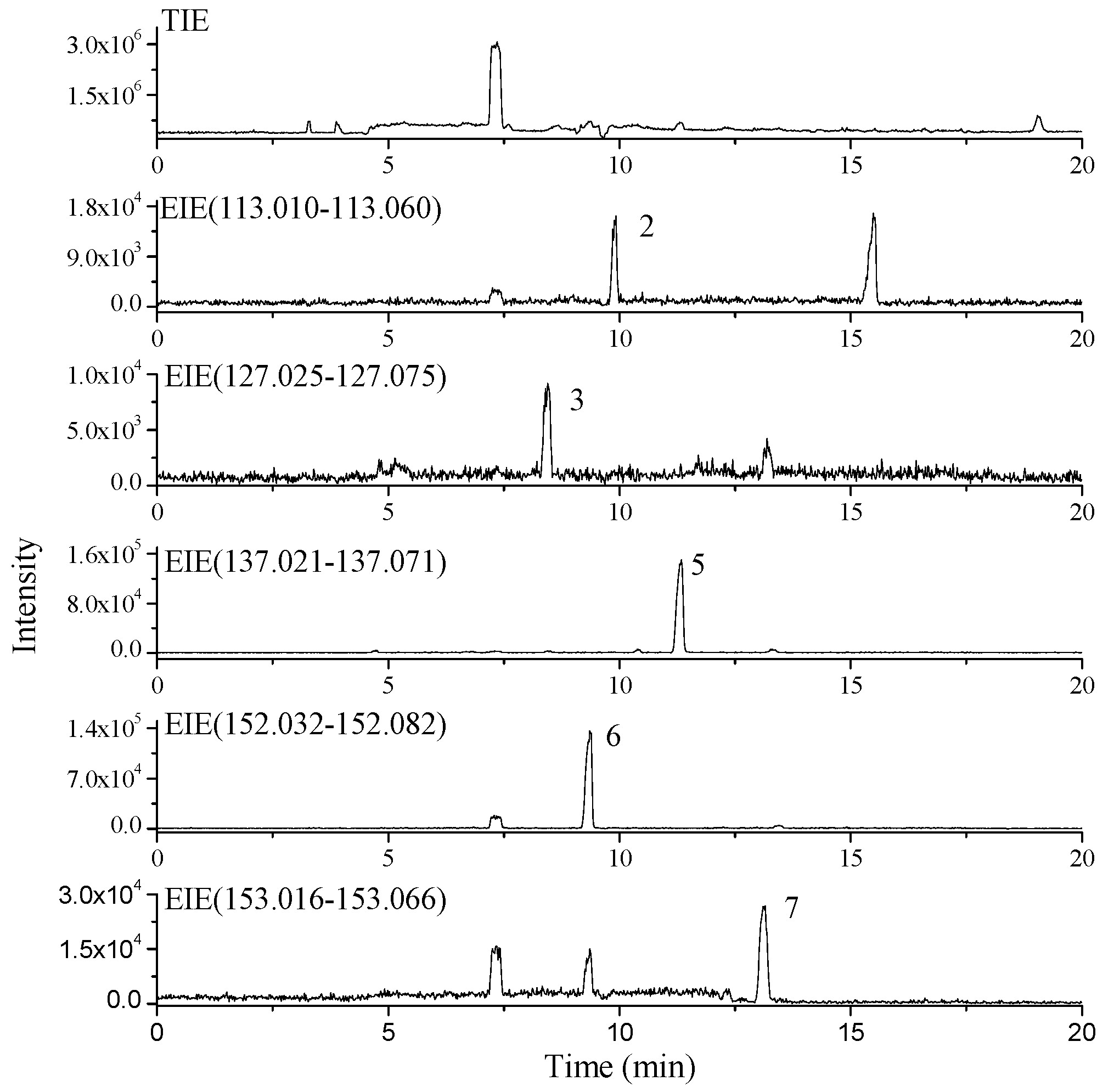
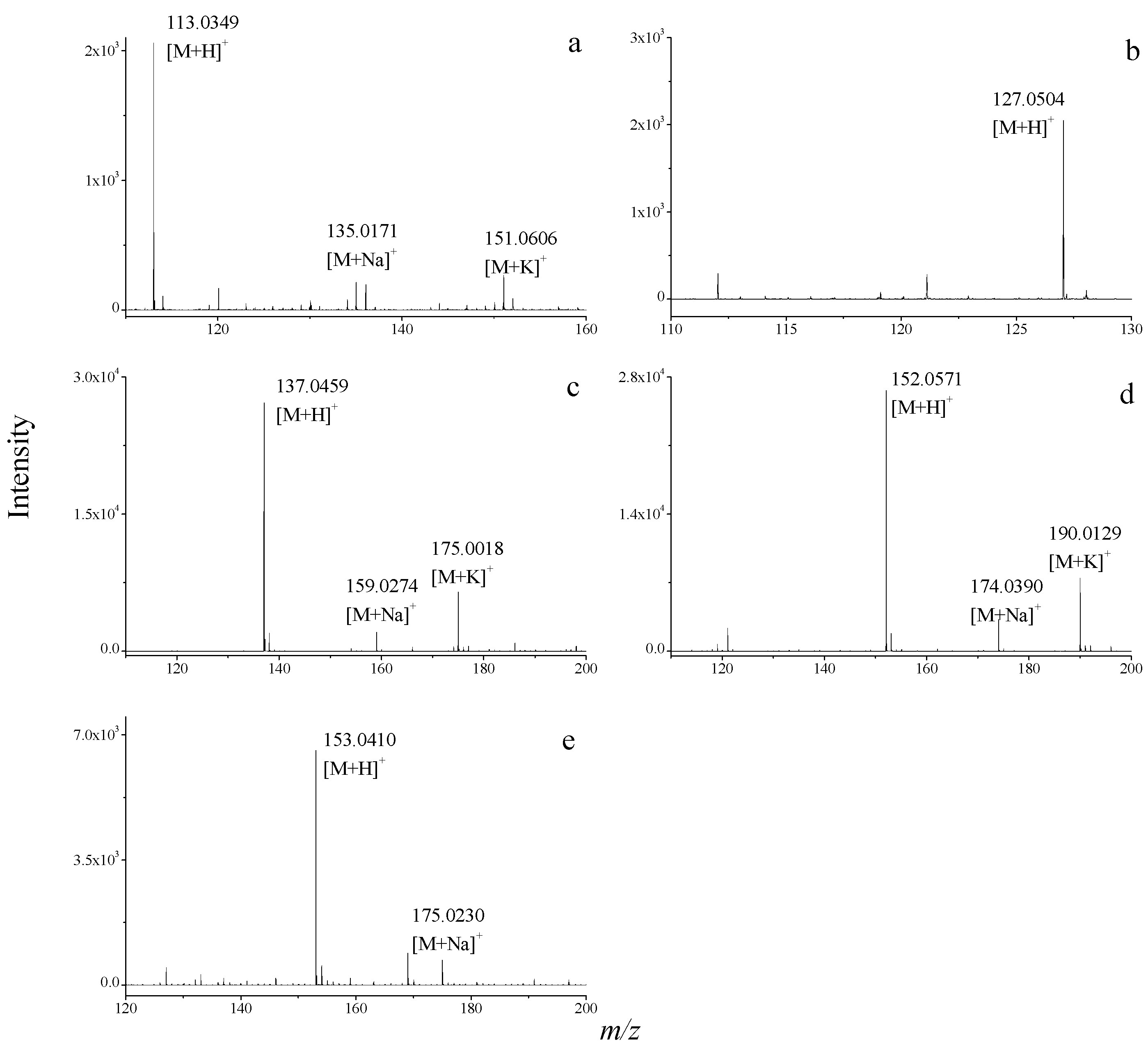
3. Experimental Section
3.1. Reagents, Materials and Standard Solutions
3.2. Sample Preparation
3.3. CE-ESI-TOF/MS Conditions
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Harvey, A.L.; Clark, R.L.; Mackay, S.P.; Johnston, B.F. Current strategies for drug discovery through natural products. Expert Opin. Drug Discov. 2010, 5, 559–568. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Liang, Q.; Wang, M.; Jeffries, C.; Smithson, D.; Tu, Y.; Boulos, N.; Jacob, M.R.; Shelat, A.A.; Wu, Y.S.; et al. UPLC-MS-ELSD-PDA as a powerful dereplication tool to facilitate compound identification from small-molecule natural product libraries. J. Nat. Prod. 2014, 77, 902–909. [Google Scholar] [CrossRef] [PubMed]
- Tchoumtchoua, J.; Njamen, D.; Mbanya, J.C.; Skaltsounis, A.L.; Halabalaki, M. Structure-oriented UHPLC-LTQ orbitrap-based approach as a dereplication strategy for the identification of isoflavonoids from Amphimas pterocarpoides crude extract. J. Mass. Spectrom. 2013, 48, 561–575. [Google Scholar] [CrossRef] [PubMed]
- El-Elimat, T.; Figueroa, M.; Ehrmann, B.M.; Cech, N.B.; Pearce, C.J.; Oberlies, N.H. High-resolution MS, MS/MS, and UV database of fungal secondary metabolites as a dereplication protocol for bioactive natural products. J. Nat. Prod. 2013, 76, 1709–1716. [Google Scholar] [CrossRef] [PubMed]
- Kildgaard, S.; Mansson, M.; Dosen, I.; Klitgaard, A.; Frisvad, J.C.; Larsen, T.O.; Nielsen, K.F. Accurate dereplication of bioactive secondary metabolites from marine-derived fungi by UHPLC-DAD-QTOFMS and a MS/HRMS library. Mar. Drugs 2014, 12, 3681–3705. [Google Scholar] [CrossRef] [PubMed]
- Abdelmohsen, U.R.; Cheng, C.; Viegelmann, C.; Zhang, T.; Grkovic, T.; Ahmed, S.; Quinn, R.J.; Hentschel, U.; Edrada-Ebel, R. Dereplication strategies for targeted isolation of new antitrypanosomal actinosporins A and B from a marine sponge associated-Actinokineospora. sp. EG49. Mar. Drugs 2014, 12, 1220–1244. [Google Scholar] [CrossRef] [PubMed]
- Blunt, J.; Munro, M.; Upjohn, M. The role of databases in marine natural products research. In Handbook of Marine Natural Products; Fattorusso, E., Gerwick, W.H., Taglialatela-Scafati, O., Eds.; Springer: Dordrecht, The Netherlands, 2012; pp. 389–421. [Google Scholar]
- Hostettmann, K.; Wolfender, J.L.; Terreaux, C. Modern screening techniques for plant extracts. Pharm. Biol. 2001, 39, 18–32. [Google Scholar] [CrossRef] [PubMed]
- Wolfender, J.L.; Ndjoko, K.; Hostettmann, K. Liquid chromatography with ultraviolet absorbance-mass spectrometric detection and with nuclear magnetic resonance spectrometry: A powerful combination for the on-line structural investigation of plant metabolites. J. Chromatogr. A 2003, 1000, 437–455. [Google Scholar] [CrossRef] [PubMed]
- Wolfender, J.L.; Queiroz, E.F.; Hostettmann, K.E.O. The importance of hyphenated techniques in the discovery of new lead compounds from nature. Expert Opin. Drug Discov. 2006, 1, 237–260. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, K.F.; Månsson, M.; Rank, C.; Frisvad, J.C.; Larsen, T.O. Dereplication of microbial natural products by LC-DAD-TOFMS. J. Nat. Prod. 2011, 74, 2338–2348. [Google Scholar] [CrossRef] [PubMed]
- Neto, F.C.; Siquitelli, C.D.; Pilon, A.C.; Silva, D.H.S.; Bolzani, V.S.; Castro-Gamboa, I. Dereplication of phenolic derivatives of Qualea grandiflora and Qualea. cordata (Vochysiaceae) using liquid chromatography coupled with ESI-QToF-MS/MS. J. Braz. Chem. Soc. 2013, 24, 758–764. [Google Scholar]
- Yang, J.Y.; Sanchez, L.M.; Rath, C.M.; Liu, X.T.; Boudreau, P.D.; Bruns, N.; Glukhov, E.; Wodtke, A.; de Felicio, R.; Fenner, A.; et al. Molecular networking as a dereplication strategy. J. Nat. Prod. 2013, 76, 1686–1699. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, K.A.; Jarvis, M.F.; Williams, M. Purine and pyrimidine (P2) receptors as drug targets. J. Med. Chem. 2002, 45, 4057–4093. [Google Scholar] [CrossRef] [PubMed]
- Zeng, W.B.; Yu, H.; Ge, F.; Yang, J.Y.; Chen, Z.H.; Wang, Y.B.; Dai, Y.D.; Adams, A. Distribution of nucleosides in populations of Cordyceps. cicadae. Molecules 2014, 19, 6123–6141. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Duan, J.A.; Tang, Y.P.; Zhu, Z.H.; Qian, Y.F.; Yang, N.Y.; Shang, E.X.; Qian, D.W. Characterization of nucleosides and nucleobases in fruits of Ziziphus jujuba by UPLC-DAD-MS. J. Agric. Food Chem. 2010, 58, 10774–10780. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Duan, J.A.; Qian, D.W.; Wang, H.Q.; Tang, Y.P.; Qian, Y.F.; Wu, D.W.; Su, S.L.; Shang, E.X. Hydrophilic interaction ultra-high performance liquid chromatography coupled with triple quadrupole mass spectrometry for determination of nucleotides, nucleosides and nucleobases in Ziziphus plants. J. Chromatogr. A 2013, 1301, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.Q.; Wang, X.; Li, H.M.; Yang, B.; Yang, H.J.; Huang, L.Q. Characterization of nucleosides and nucleobases in natural Cordyceps by HILIC-ESI/TOF/MS and HILIC–ESI/MS. Molecules 2013, 18, 9755–9769. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Bicker, W.; Wu, J.Y.; Xie, M.Y.; Lindner, W. Simultaneous determination of 16 nucleosides and nucleobases by hydrophilic interaction chromatography and its application to the quality evaluation of Ganoderma. J. Agric. Food Chem. 2012, 60, 4243–4252. [Google Scholar] [CrossRef] [PubMed]
- Zuo, H.L.; Yang, F.Q.; Xia, Z.N. Simultaneous determination of 17 nucleotides, nucleosides and nucleobases in Pinellia Ternata by high performance liquid chromatography. Chem. Rapid Commun. 2013, 1, 9–14. [Google Scholar]
- Qi, S.H.; Zhang, S.; Huang, H. Purine alkaloids from the South China Sea gorgonian Subergorgia suberosa. J. Nat. Prod. 2008, 71, 716–718. [Google Scholar] [CrossRef] [PubMed]
- Shi, Q.; Chen, J.H.; Zhao, H.Q.; Li, X.; Zheng, L.; Wang, X.R.; Zang, J.Y. Rapid simultaneous determination of 15 nucleosides and nucleobases in marine medicinal organism Anthopleura lanthogrammica Berkly by micellar electrokinetic capillary chromatography. J. Anal. Chem. 2014, 69, 169–178. [Google Scholar] [CrossRef]
- Zhao, H.Q.; Chen, J.H.; Shi, Q.; Li, X.; Zhou, W.H.; Zhang, D.L.; Zheng, L.; Cao, W.; Wang, X.R.; Lee, F.S.C. Simultaneous determination nucleosides in marine organisms using ultrasound-assisted extraction followed by hydrophilic interaction liquid chromatography-electrospray ionization time-of-flight mass spectrometry. J. Sep. Sci. 2011, 34, 2594–2601. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.Q.; Chen, J.H.; Shi, Q.; Li, X.; Cao, W.; Li, J.X.; Wang, X.R. New fingerprinting method based on the distribution of nucleoside compounds for marine animal drugs by HILIC-ESI-TOF/MS. Chem. J. Chin. Univ. 2012, 33, 44–48. [Google Scholar]
- Koshiishi, C.; Crozier, A.; Ashihara, H. Profiles of purine and pyrimidine nucleotides in fresh and manufactured tea leaves. J. Agric. Food Chem. 2001, 49, 4378–4382. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.P.; Zhao, S.Y.; Wang, J.H.; Kuang, H.C.; Liu, X. Distribution of nucleosides and nucleobases in edible fungi. J. Agric. Food. Chem. 2008, 56, 809–815. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Tian, J.M.; Xiao, J.; Shao, Q.; Gao, J.M. Bioactive metabolites isolated from Penicillium sp. YY-20, the endophytic fungus from Ginkgo biloba. Nat. Prod. Res. 2014, 28, 278–281. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Li, Q.L.; Ji, N.Y.; Liu, B.; Zhang, W.; Cao, X.P. Deoxyuridines from the Marine Sponge Associated Actinomycete Streptomyces microflavus. Mar. Drugs 2011, 9, 690–695. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Li, Y.; Xu, J.; Tang, X.L.; Li, P.L.; Li, G.Q. Studies on chemical constituents of Subergorgia reticulata. Chin. J. Mar. Drugs 2011, 3, 31–36. [Google Scholar]
- Xu, W.; Wang, C.; Zhou, G.X.; Yao, X.S.; Lin, B.R. Study on the bioactive constituents of antifungal actinomycete strain No. H75–11 from the sea bed mud at the mangrove zone of the South China Sea. Chin. J. Mar. Drugs 2011, 2, 1–6. [Google Scholar]
- Li, S.P.; Li, P.; Lai, C.M.; Gong, Y.X.; Kan, K.K.W.; Dong, T.T.X.; Tsim, K.W.K.; Wang, Y.T. Simultaneous determination of ergosterol, nucleosides and their bases from natural and cultured Cordyceps by pressurised liquid extraction and high-performance liquid chromatography. J. Chromatogr. A 2004, 1036, 239–243. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.; Yang, F.Q.; Li, S.P. Determination of purine and pyrimidine bases in natural and cultured Cordyceps using optimum acid hydrolysis followed by high performance liquid chromatography. J. Pharm. Biomed. Anal. 2007, 45, 141–144. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.L.; Chen, J.H.; Zhou, M.; Shi, Q.; Zhao, H.Q.; Cheng, H.Y.; Yang, H.H.; Wang, X.R. Simultaneous determination of 7 nucleosides in Asterias rollestoni using reversed-phase high performance liquid chromatography. Chin. J. Chromatogr. 2010, 28, 795–799. [Google Scholar] [CrossRef]
- Yang, F.Q.; Guan, J.; Li, S.P. Fast simultaneous determination of 14 nucleosides and nucleobases in cultured Cordyceps using ultra-performance liquid chromatography. Talanta 2007, 73, 269–273. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Li, W.; Li, Q.; Wang, Y.H.; Li, Z.G.; Ni, Y.F.; Koike, K. Identification and quantification of nucleosides and nucleobases in Geosaurus and Leech by hydrophilic-interaction chromatography. Talanta 2011, 85, 1634–1641. [Google Scholar] [CrossRef] [PubMed]
- Marrubini, G.; Mendoza, B.E.C.; Massolini, G. Separation of purine and pyrimidine bases and nucleosides by hydrophilic interaction chromatography. J. Sep. Sci. 2010, 33, 803–816. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.Q.; Li, D.Q.; Feng, K.; Hu, D.J.; Li, S.P. Determination of nucleotides, nucleosides and their transformation products in Cordyceps by ion-pairing reversed-phase liquid chromatography-mass spectrometry. J. Chromatogr. A 2010, 1217, 5501–5510. [Google Scholar] [CrossRef] [PubMed]
- Vinas, P.; Campillo, N.; Melgarejo, G.F.; Vasallo, M.I.; López-García, I.; Hernández-Córdoba, M. Ion-pair high-performance liquid chromatography with diode array detection coupled to dual electrospray atmospheric pressure chemical ionization time-of-flight mass spectrometry for the determination of nucleotides in baby foods. J. Chromatogr. A 2010, 1217, 5197–5203. [Google Scholar] [CrossRef] [PubMed]
- Rao, Y.K.; Chou, C.H.; Tzeng, Y.M. A simple and rapid method for identification and determination of cordycepin in Cordyceps militaris by capillary electrophoresis. Anal. Chim. Acta 2006, 566, 253–258. [Google Scholar] [CrossRef]
- Gong, Y.X.; Li, S.P.; Li, P.; Liu, J.J.; Wang, Y.T. Simultaneous determination of six main nucleosides and bases in natural and cultured Cordyceps by capillary electrophoresis. J. Chromatogr. A 2004, 1055, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Klampfl, C.W.; Himmelsbach, M.; Buchberger, W.; Klein, H. Determination of purines and pyrimidines in beer samples by capillary zone electrophoresis. Anal. Chim. Acta 2002, 454, 185–191. [Google Scholar] [CrossRef]
- Yang, F.Q.; Ge, L.Y.; Yong, J.W.; Tan, S.N.; Li, S.P. Determination of nucleosides and nucleobases in different species of Cordyceps by capillary electrophoresis–mass spectrometry. J. Pharm. Biomed. Anal. 2009, 50, 307–314. [Google Scholar] [CrossRef] [PubMed]
- Haunschmidt, M.; Buchberger, W.; Klampfl, C.W. Investigations on the migration behaviour of purines and pyrimidines in capillary electromigration techniques with UV detection and mass spectrometric detection. J. Chromatogr. A 2008, 1213, 88–92. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.F.; Zhao, X.P.; Mao, Y.; Cheng, Y.Y. Novel approach for developing urinary nucleosides profile by capillary electrophoresis-mass spectrometry. J. Chromatogr. A 2007, 1147, 254–260. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Gonzalo, E.; Hernández-Prieto, R.; García-Gómez, D.; Carabias-Martínez, R. Capillary electrophoresis–mass spectrometry for direct determination of urinary modified nucleosides. Evaluation of synthetic urine as asurrogate matrix for quantitative analysis. J. Chromatogr. B 2013, 942–943, 21–30. [Google Scholar] [CrossRef]
- Rodríguez-Gonzalo, E.; Hernández-Prieto, R.; García-Gómez, D.; Carabias-Martínez, R. Development of a procedure for the isolation and enrichment of modified nucleosides and nucleobases from urine prior to their determination by capillary electrophoresis-mass spectrometry. J. Pharm. Biomed. Anal. 2014, 88, 489–496. [Google Scholar] [CrossRef] [PubMed]
- Klampfl, C.W. CE with MS detection: A rapidly developing hyphenated technique. Electrophoresis 2009, 30, S83–S91. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Hernández, L.; Serra, N.S.; Marina, M.L.; Crego, A.L. Enantiomeric separation of free l- and d-amino acids in hydrolyzed protein fertilizers by capillary electrophoresis tandem mass spectrometry. J. Agric. Food Chem. 2013, 61, 5022–5030. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Hernández, L.; Castro-Puyana, M.; García-Ruiz, C.; Crego, A.L.; Marina, M.L. Determination of l- and d-carnitine in dietary food supplements using capillary electrophoresis-tandem mass spectrometry. Food Chem. 2010, 120, 921–928. [Google Scholar] [CrossRef]
- Sánchez-Hernández, L.; García-Ruiz, C.; Crego, A.L.; Marina, M.L. Sensitive determination of d-carnitine as enantiomeric impurity of levo-carnitine in pharmaceutical formulations by capillary electrophoresis-tandem mass spectrometry. J. Pharm. Biomed. Anal. 2010, 53, 1217–1223. [Google Scholar] [CrossRef] [PubMed]
- Eilertsen, H.C.; Huseby, S.; Degerlund, M.; Eriksen, G.K.; Ingebrigtsen, R.A.; Hansen, E. The effect of freeze/thaw cycles on reproducibility of metabolic profiling of marine microalgal extracts using direct infusion high-resolution mass spectrometry (HR-MS). Molecules 2014, 19, 16373–16380. [Google Scholar] [CrossRef] [PubMed]
- Tong, P.; Zhang, L.; He, Y.; Tang, S.R.; Cheng, J.T.; Chen, G.N. Analysis of microcystins by capillary zone electrophoresis coupling with electrospray ionization mass spectrometry. Talanta 2010, 82, 1101–1106. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.H.; Zhao, H.Q.; Li, W.L.; Wang, X.R.; Lee, F.S.C.; Yang, H.H. Analysis of alkaloids in Coptis chinensis Franch by high performance capillary electrophoresis-electrospray ionization time of flight mass spectrometry. Acta Chim. Sin. 2007, 65, 2743–2749. [Google Scholar]
- Soga, T.; Ishikawa, T.; Igarashi, S.; Sugawara, K.; Kakazu, Y.; Tomita, M. Analysis of nucleotides by pressure-assisted capillary electrophoresis-mass spectrometry using silanol mask technique. J. Chromatogr. A 2007, 1159, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Domınguez-Alvarez, J.; Rodrıguez-Gonzalo, E.; Hernandez-Meendez, J.; Carabias-Martınez, R. Programmed nebulizing gas pressure for efficient and stable capillary electrophoresis-mass spectrometry analysis of anionic compounds in positive separation mode. Anal. Chem. 2011, 83, 2834–2839. [Google Scholar] [CrossRef] [PubMed]
- Harada, K.; Ohyama, Y.; Tabushi, T.; Kobayashi, A.; Fukusaki, E. Quantitative analysis of anionic metabolites for Catharanthus roseus by capillary electrophoresis using sulfonated capillary coupled with electrospray ionization-tandem mass spectrometry. J. Biosci. Bioeng. 2008, 105, 249–260. [Google Scholar] [CrossRef] [PubMed]
- Harada, K.; Fukusaki, E.; Kobayashi, A. Pressure-assisted capillary electrophoresis mass spectrometry using combination of polarity reversion and electroosmotic flow for metabolomics anion analysis. J. Biosci. Bioeng. 2006, 101, 403–409. [Google Scholar] [CrossRef] [PubMed]
- Gómez, M.J.; Gómez-Ramos, M.M.; Malato, O.; Mezcua, M.; Férnandez-Alba, A.R. Rapid automated screening, identification and quantification of organic micro-contaminants and their main transformation products in wastewater and river waters using liquid chromatography-quadrupole-time-of-flight mass spectrometry with an accurate-mass database. J. Chromatogr. A 2010, 1217, 7038–7054. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.Y.; Chen, J.H.; Li, X.; Zhang, R.T.; Chen, C.; Shi, Q.; Wang, S.; Zheng, L.; Wang, X.R. Rapid screening and identification of paralytic shellfish poisoning toxins in red tide algae using hydrophilic interaction liquid chromatography-high resolution mass spectrometry with an accurate-mass database. Chin. J. Anal. Chem. 2013, 41, 979–985. [Google Scholar] [CrossRef]
- Sample Availability: Samples of all the 15 compounds are available from the authors.
© 2015 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, J.; Shi, Q.; Wang, Y.; Li, Z.; Wang, S. Dereplication of Known Nucleobase and Nucleoside Compounds in Natural Product Extracts by Capillary Electrophoresis-High Resolution Mass Spectrometry. Molecules 2015, 20, 5423-5437. https://doi.org/10.3390/molecules20045423
Chen J, Shi Q, Wang Y, Li Z, Wang S. Dereplication of Known Nucleobase and Nucleoside Compounds in Natural Product Extracts by Capillary Electrophoresis-High Resolution Mass Spectrometry. Molecules. 2015; 20(4):5423-5437. https://doi.org/10.3390/molecules20045423
Chicago/Turabian StyleChen, Junhui, Qian Shi, Yanlong Wang, Zhaoyong Li, and Shuai Wang. 2015. "Dereplication of Known Nucleobase and Nucleoside Compounds in Natural Product Extracts by Capillary Electrophoresis-High Resolution Mass Spectrometry" Molecules 20, no. 4: 5423-5437. https://doi.org/10.3390/molecules20045423
APA StyleChen, J., Shi, Q., Wang, Y., Li, Z., & Wang, S. (2015). Dereplication of Known Nucleobase and Nucleoside Compounds in Natural Product Extracts by Capillary Electrophoresis-High Resolution Mass Spectrometry. Molecules, 20(4), 5423-5437. https://doi.org/10.3390/molecules20045423






