Novel Cationic Carotenoid Lipids as Delivery Vectors of Antisense Oligonucleotides for Exon Skipping in Duchenne Muscular Dystrophy
Abstract
:1. Introduction
2. Results and Discussion
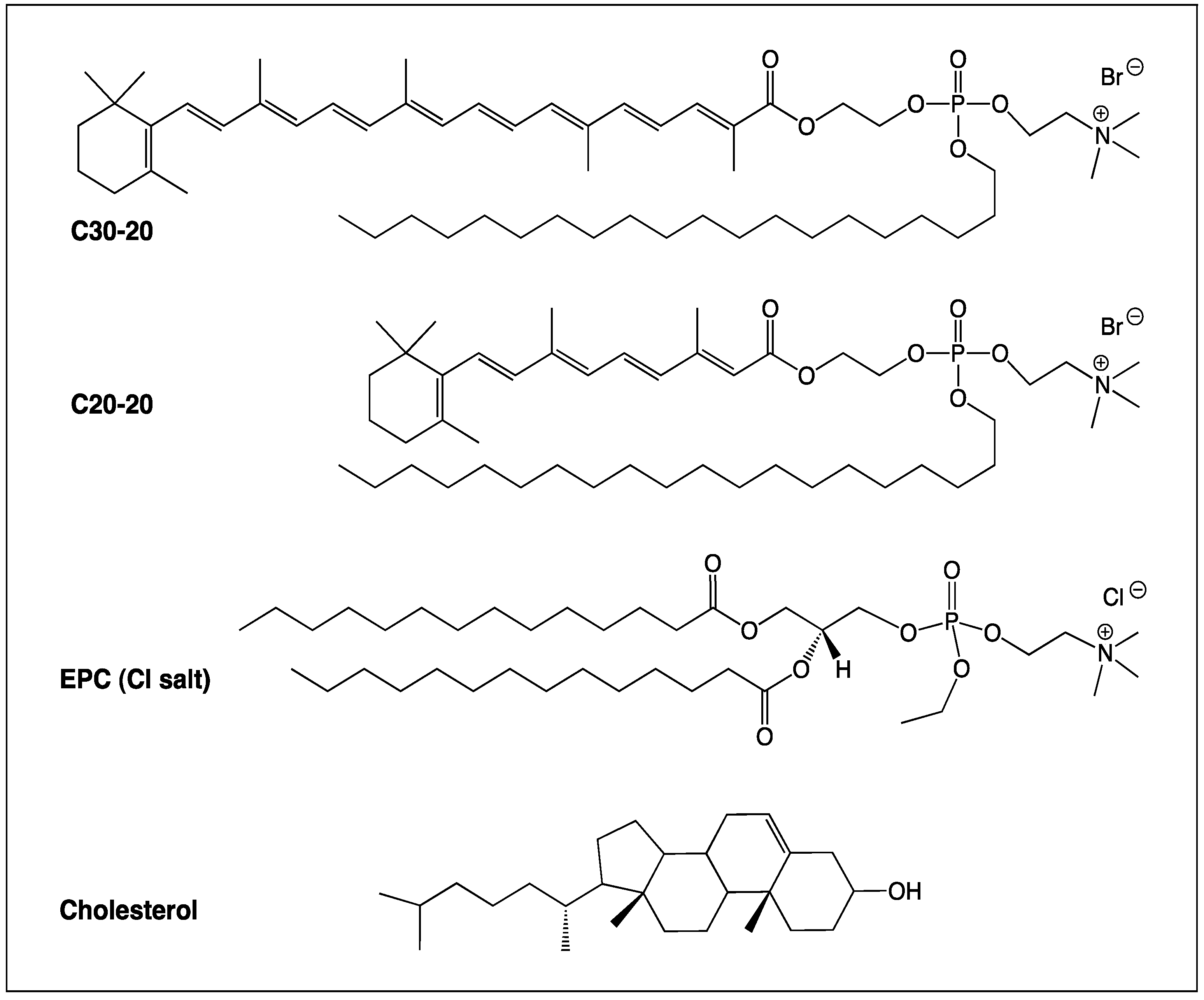
2.1. Synthesis of the Cationic Carotenoid Lipids C30-20 and C20-20
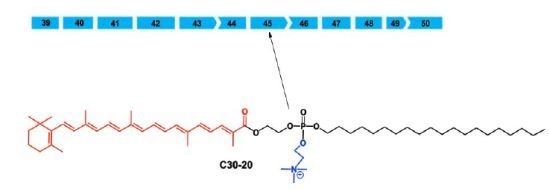
2.2. Verification of Annealing PMO to Leash
2.3. Gel Retardation Assay

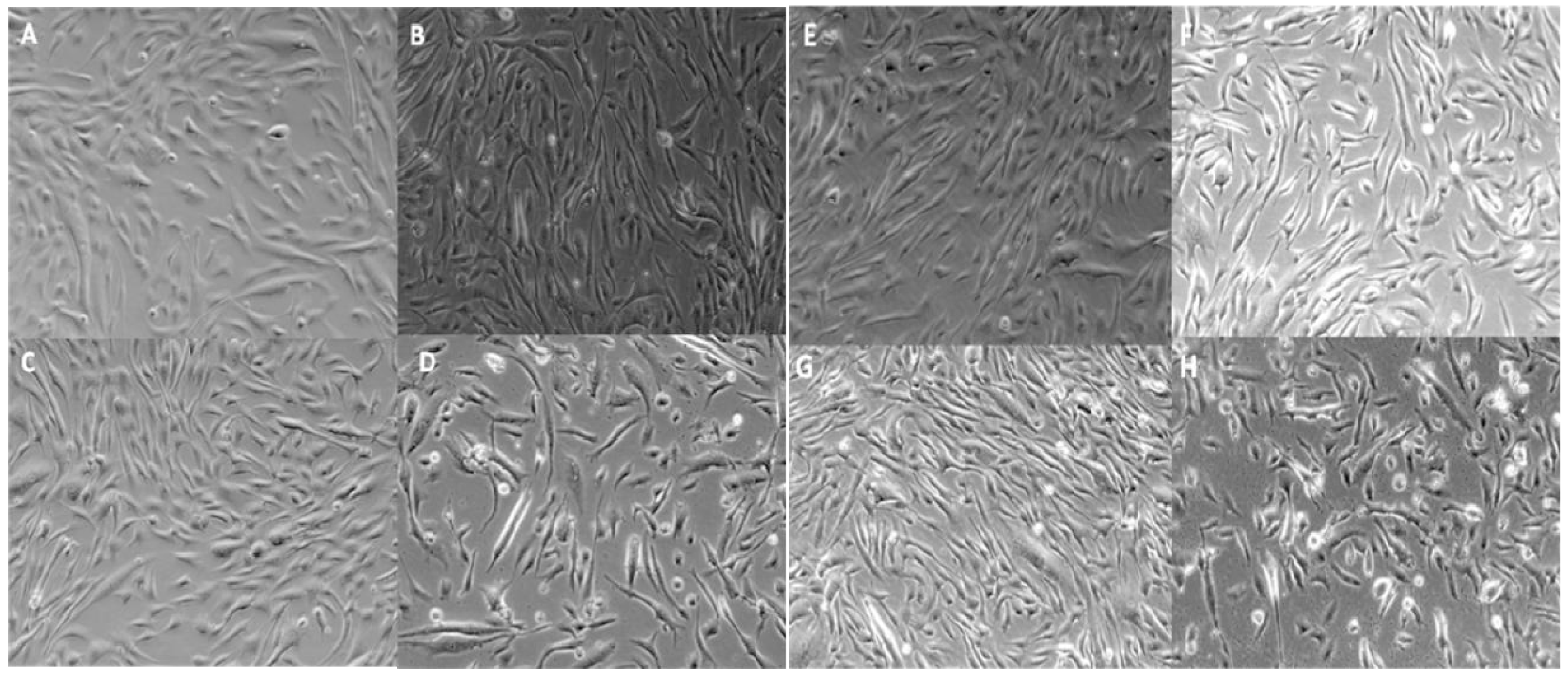
2.4. Qualitative Assessment of Cell Viability
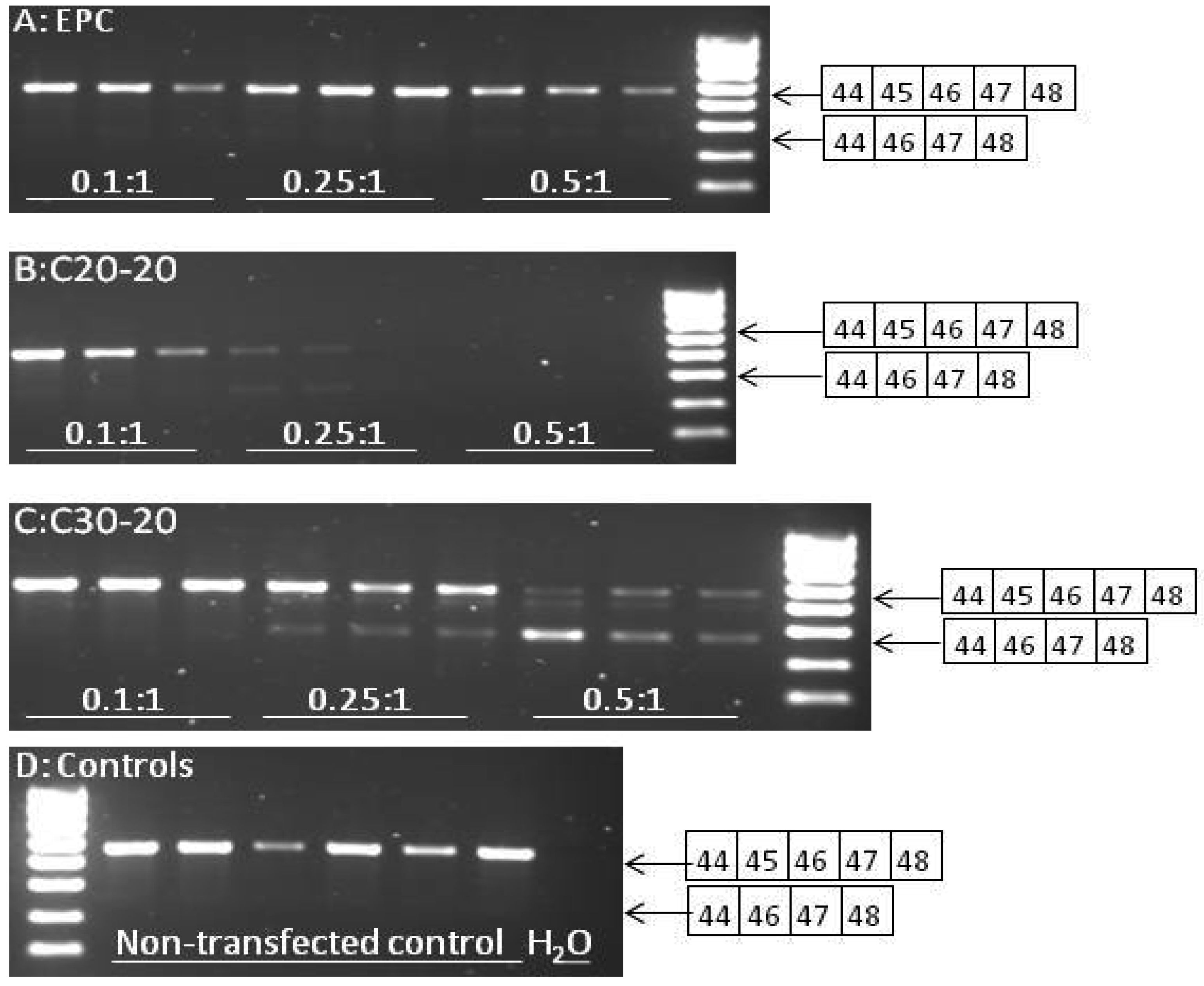
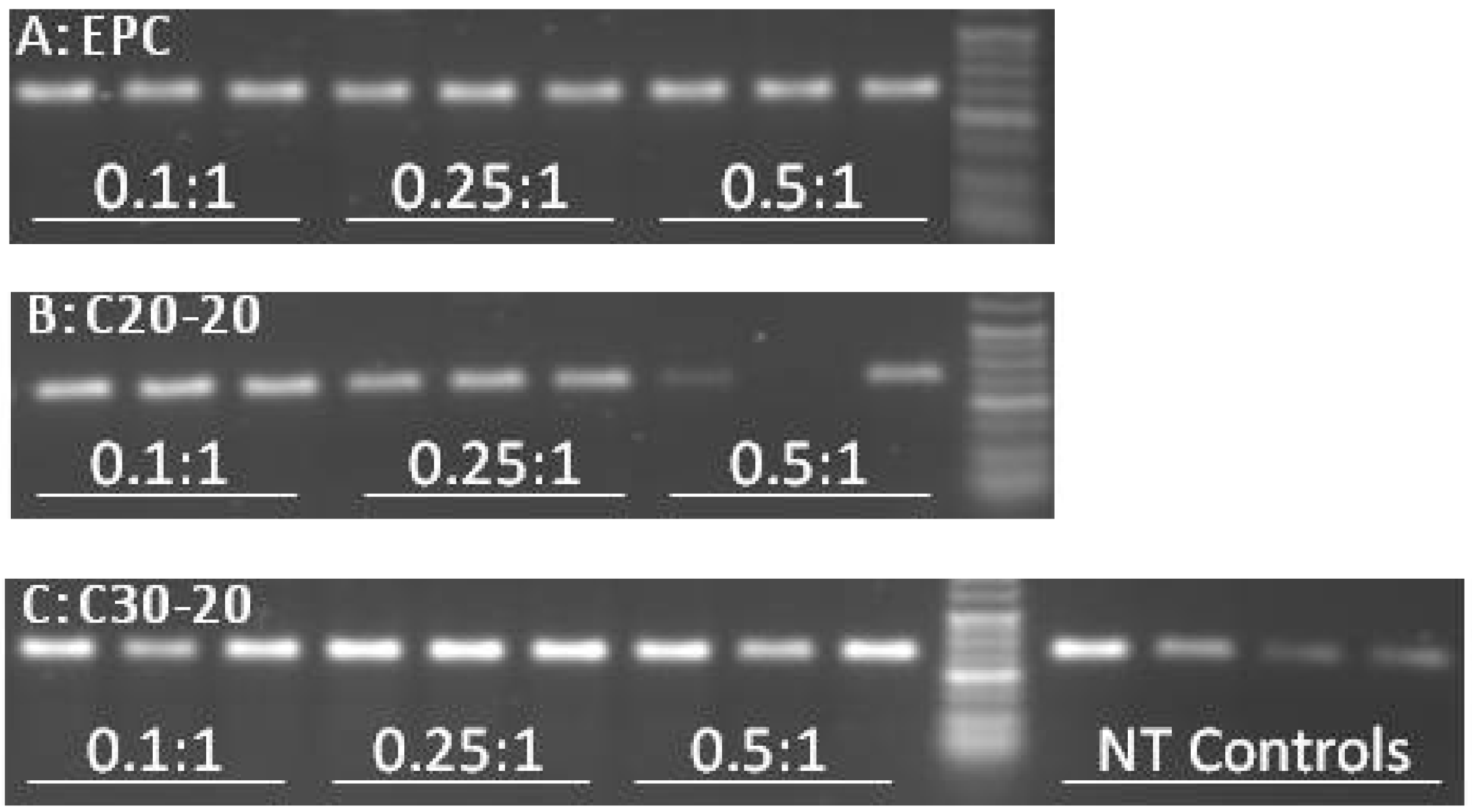
2.5. Reverse Transcriptase-PCR Results

3. Experimental
3.1. Materials and Methods
3.2. Synthesis of Cationic Carotenoid Lipids, C30-20 and C20-20
3.3. Liposome and Lipoplex Formulation
3.3.1. Ethanolic stock Solutions
3.3.2. Liposome Formulations
3.3.3. Preparation of Leashed PMO-AO
3.3.4. Verification of Annealing PMO to Leash
3.3.5. Lipid/PMO-AO Lipoplexes
3.4. Bioassays
3.4.1. Gel Retardation Assay
3.4.2. Transfection of normal human skeletal muscle primary cells (hSkMCs)
3.4.3. RNA Extraction and Purification
3.4.4. RT-PCR
4. Conclusions
Acknowledgments
Conflict of Interest
References and Notes
- Krinsky, N.I. Carotenoids as antioxidants. Nutrition 2001, 17, 815–817. [Google Scholar] [CrossRef]
- Baltschun, D.; Beutner, S.; Briviba, K.; Martin, H.-D.; Paust, J.; Peters, M.; Röver, S.; Sies, H.; Stahl, W.; Steigel, A.; et al. Singlet Oxygen Quenching Abilities of Carotenoids. Liebigs Ann. 1997, 1997, 1887–1893. [Google Scholar]
- Sharoni, Y.; Danilenko, M.; Dubi, N.; Ben-Dor, A.; Levy, J. Carotenoids and transcription. Arch. Biochem. Biophys. 2004, 430, 89–96. [Google Scholar] [CrossRef]
- Lockwood, S.F.; Gross, G.J. Disodium disuccinate astaxanthin (Cardax): Antioxidant and antiinflammatory cardioprotection. Cardiovasc. Drug Rev. 2005, 23, 199–216. [Google Scholar] [CrossRef]
- Krinsky, N.I.; Johnson, E.J. Carotenoid actions and their relation to health and disease. Mol. Aspects Med. 2005, 26, 459–516. [Google Scholar] [CrossRef]
- Goemans, N.M.; Tulinius, M.; van den Akker, J.T.; Burm, B.E.; Ekhart, P.F.; Heuvelmans, N.; Holling, T.; Janson, A.A.; Platenburg, G.J.; Sipkens, J.A.; et al. Systemic administration of PRO051 in Duchenne's muscular dystrophy. N. Engl. J. Med. 2011, 364, 1513–1522. [Google Scholar]
- Popplewell, L.J.; Adkin, C.; Arechavala-Gomeza, V.; Aartsma-Rus, A.; de Winter, C.L.; Wilton, S.D.; Morgan, J.E.; Muntoni, F.; Graham, I.R.; Dickson, G. Comparative analysis of antisense oligonucleotide sequences targeting exon 53 of the human DMD gene: Implications for future clinical trials. Neuromuscul. Disord. 2010, 20, 102–110. [Google Scholar]
- Kinali, M.; Arechavala-Gomeza, V.; Feng, L.; Cirak, S.; Hunt, D.; Adkin, C.; Guglieri, M.; Ashton, E.; Abbs, S.; Nihoyannopoulos, P.; et al. Local restoration of dystrophin expression with the morpholino oligomer AVI-4658 in Duchenne muscular dystrophy: A single-blind, placebo-controlled, dose-escalation, proof-of-concept study. Lancet Neurol. 2009, 8, 918–928. [Google Scholar] [CrossRef]
- Popplewell, L.J.; Trollet, C.; Dickson, G.; Graham, I.R. Design of phosphorodiamidate morpholino oligomers (PMOs) for the induction of exon skipping of the human DMD gene. Mol. Ther. 2009, 17, 554–561. [Google Scholar] [CrossRef]
- Arechavala-Gomeza, V.; Graham, I.R.; Popplewell, L.J.; Adams, A.M.; Aartsma-Rus, A.; Kinali, M.; Morgan, J.E.; van Deutekom, J.C.; Wilton, S.D.; Dickson, G.; et al. Comparative analysis of antisense oligonucleotide sequences for targeted skipping of exon 51 during dystrophin pre-mRNA splicing in human muscle. Hum. Gene Ther. 2007, 18, 798–810. [Google Scholar] [CrossRef]
- van Deutekom, J.C.; Janson, A.A.; Ginjaar, I.B.; Frankhuizen, W.S.; Aartsma-Rus, A.; Bremmer-Bout, M.; den Dunnen, J.T.; Koop, K.; van der Kooi, A.J.; Goemans, N.M.; et al. Local dystrophin restoration with antisense oligonucleotide PRO051. N. Engl. J. Med. 2007, 357, 2677–2686. [Google Scholar] [CrossRef]
- Aartsma-Rus, A.; Fokkema, I.; Verschuuren, J.; Ginjaar, I;, van Deutekom; van Ommen, G.J.; den Dunnen, J.T. Theoretic applicability of antisense-mediated exon skipping for Duchenne muscular dystrophy mutations. Hum. Mutat. 2009, 30, 293–299. [Google Scholar] [CrossRef]
- Gebski, B.L.; Mann, C.J.; Fletcher, S.; Wilton, S.D. Morpholino antisense oligonucleotide induced dystrophin exon 23 skipping in mdx mouse muscle. Hum. Mol. Genet. 2003, 12, 1801–1811. [Google Scholar] [CrossRef]
- Popplewell, L.J.; Graham, I.R.; Malerba, A.; Dickson, G. Bioinformatic and functional optimization of antisense phosphorodiamidate morpholino oligomers (PMOs) for therapeutic modulation of RNA splicing in muscle. Methods Mol. Biol. 2011, 709, 153–178. [Google Scholar] [CrossRef]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Popplewell, L.J.; Abu-Dayya, A.; Khanna, T.; Flinterman, M.; Khalique, N.A.; Raju, L.; Øpstad, C.L.; Sliwka, H.-R.; Partali, V.; Dickson, G.; et al. Novel Cationic Carotenoid Lipids as Delivery Vectors of Antisense Oligonucleotides for Exon Skipping in Duchenne Muscular Dystrophy. Molecules 2012, 17, 1138-1148. https://doi.org/10.3390/molecules17021138
Popplewell LJ, Abu-Dayya A, Khanna T, Flinterman M, Khalique NA, Raju L, Øpstad CL, Sliwka H-R, Partali V, Dickson G, et al. Novel Cationic Carotenoid Lipids as Delivery Vectors of Antisense Oligonucleotides for Exon Skipping in Duchenne Muscular Dystrophy. Molecules. 2012; 17(2):1138-1148. https://doi.org/10.3390/molecules17021138
Chicago/Turabian StylePopplewell, Linda J., Aseel Abu-Dayya, Tushar Khanna, Marcella Flinterman, Nada Abdul Khalique, Liji Raju, Christer L. Øpstad, Hans-Richard Sliwka, Vassilia Partali, George Dickson, and et al. 2012. "Novel Cationic Carotenoid Lipids as Delivery Vectors of Antisense Oligonucleotides for Exon Skipping in Duchenne Muscular Dystrophy" Molecules 17, no. 2: 1138-1148. https://doi.org/10.3390/molecules17021138
APA StylePopplewell, L. J., Abu-Dayya, A., Khanna, T., Flinterman, M., Khalique, N. A., Raju, L., Øpstad, C. L., Sliwka, H.-R., Partali, V., Dickson, G., & Pungente, M. D. (2012). Novel Cationic Carotenoid Lipids as Delivery Vectors of Antisense Oligonucleotides for Exon Skipping in Duchenne Muscular Dystrophy. Molecules, 17(2), 1138-1148. https://doi.org/10.3390/molecules17021138