Thymidine Analogues for Tracking DNA Synthesis
Abstract
:1. Introduction
2. A Window of Opportunity: DNA Synthesis
3. Thymidine Analogues and Their Utility
3.1. Radiolabeled Nucleoside Analogues
3.2. Halogen-Based Nucleoside Analogues
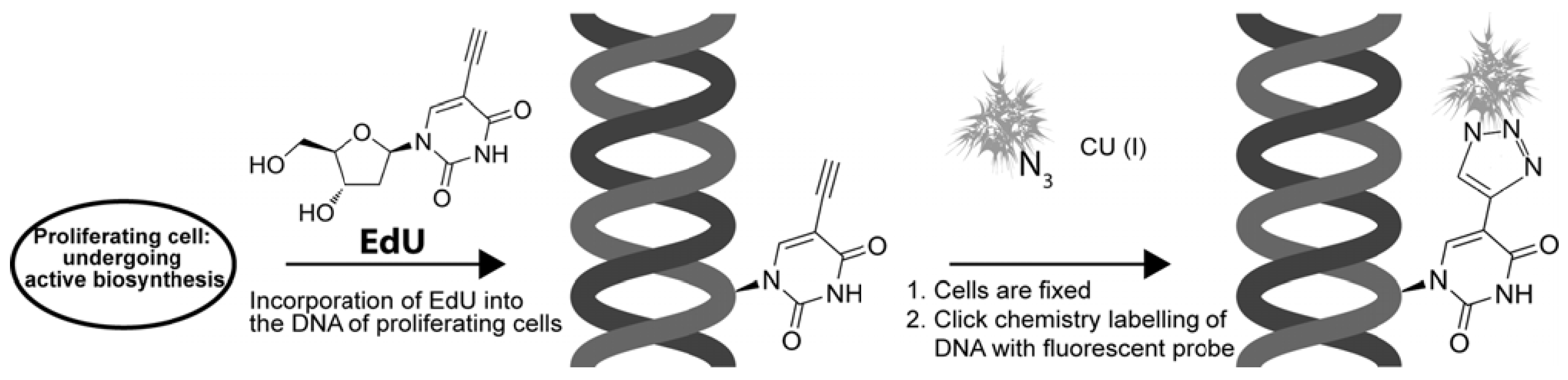
4. EdU and “Click Chemistry” Light Multiple Paths
4.1. EdU and “Click Chemistry”
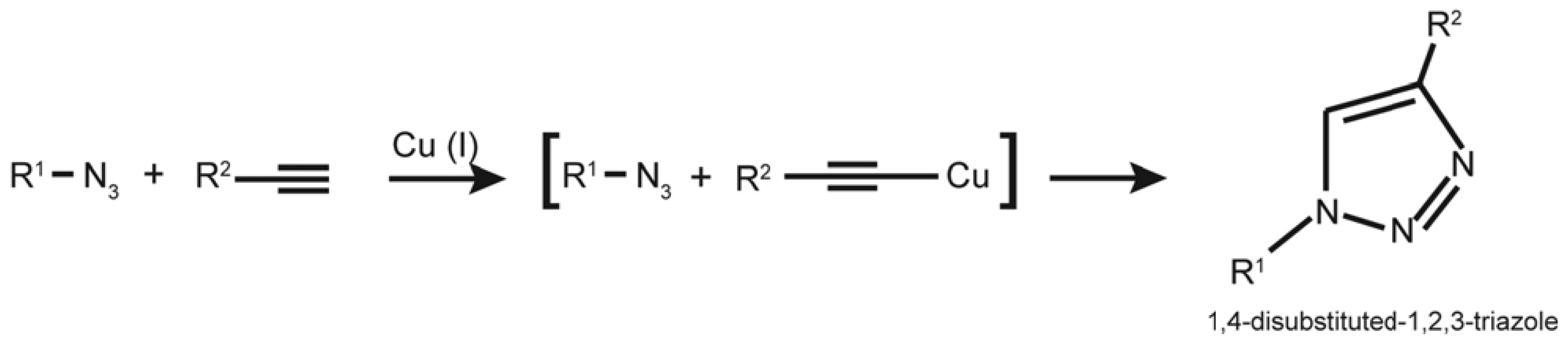
4.2. BrdU vs. EdU
4.2.1. Utility in Characterising Zones of Proliferation and the “Stem Cell Niche”
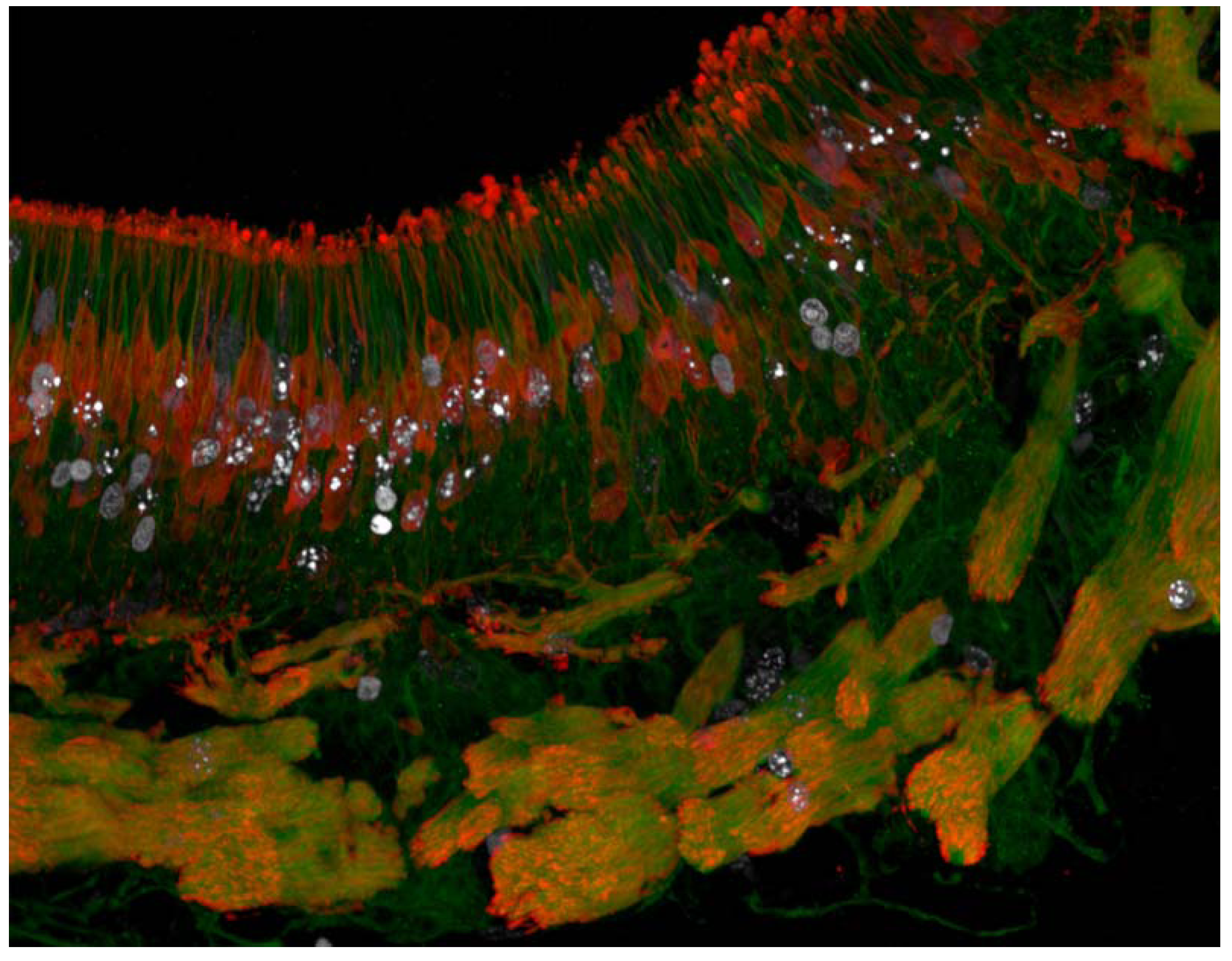
4.2.2. Isolation of Proliferative Cells for Cell Counting
4.2.3. The Next Dimension: Proliferating Cells for Molecular Assays
5. EdU for Probing Living Systems
| Catalysis-free cycloadditions | Structure Example | Application | Reference |
|---|---|---|---|
| Cyclooctyne Derivaties |  | Detection of glycan- associated azides on cell surface | [58] |
| Difluorinated Cyclooctynes |  | Visualization of cell surface glycan- associated azides / glycan trafficking | [60] |
| Hydrophilic Azacyclooctyne | 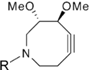 | Visualization of glycan- associated azides within cell lysates and on the surface of live cells | [61] |
| Dibenzocylooctynol Derivatives | 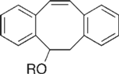 | Visualization of cell surface glycan- associated azides / glycan trafficking | [62] |
| Photo-Triggering of Cyclopropenone | 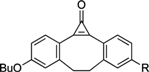 | Visualization of cell surface glycan- associated azides / glycan trafficking | [63] |
| Cyclooctyne-FLAG Conjugate |  | Visualization of cell surface glycan- associated azides in various organs | [64] |
| Electron-Deficient Alkynes |  | Introducing functional groups to DNA – in vitro | [65] |
EdU, A Unique Probe
6. Summary
References
- Klug, W.S.; Cummings, M.R. Concepts of Genetics, 8th ed; Pearson Education: Upper Saddle River, NJ, USA, 2006. [Google Scholar]
- Taupin, P. BrdU immunohistochemistry for studying adult neurogenesis: Paradigms, pitfalls, limitations, and validation. Brain Res. Rev. 2007, 53, 198–214. [Google Scholar] [CrossRef]
- Messier, B.; Leblond, C.P.; Smart, I. Presence of DNA synthesis and mitosis in the brain of young adult mice. Exp. Cell Res. 1958, 14, 224–226. [Google Scholar] [CrossRef]
- Nunez-Ramirez, R.; Klinge, S.; Sauguet, L.; Melero, R.; Recuero-Checa, M.A.; Kilkenny, M.; Perera, R.L.; Garcia-Alvarez, B.; Hall, R.J.; Nogales, E.; et al. Flexible tethering of primase and DNA Pol {alpha} in the eukaryotic primosome. Nucleic Acids Res. 2011. [Google Scholar] [CrossRef]
- Taylor, J.H.; Woods, P.S.; Hughes, W.L. The organization and duplication of chromosomes as revealed by autoradiographic studies using tritium-labeled thymidinee. Proc. Natl. Acad. Sci. USA 1957, 43, 122–128. [Google Scholar] [CrossRef]
- Altman, J. Autoradiographic and histological studies of postnatal neurogenesis. IV. Cell proliferation and migration in the anterior forebrain, with special reference to persisting neurogenesis in the olfactory bulb. J. Comp. Neurol. 1969, 137, 433–457. [Google Scholar] [CrossRef]
- Plotnik, D.A.; Emerick, L.E.; Krohn, K.A.; Unadkat, J.D.; Schwartz, J.L. Different modes of transport for 3H-thymidine, 3H-FLT, and 3H-FMAU in proliferating and nonproliferating human tumor cells. J. Nucl. Med. 2009, 51, 1464–1471. [Google Scholar]
- Huber-Ruano, I.; Pastor-Anglada, M. Transport of nucleoside analogs across the plasma membrane: A clue to understanding drug-induced cytotoxicity. Curr. Drug Metab. 2009, 10, 347–358. [Google Scholar] [CrossRef]
- Molina-Arcas, M.; Casado, F.J.; Pastor-Anglada, M. Nucleoside transporter proteins. Curr. Vasc. Pharmacol. 2009, 7, 426–434. [Google Scholar] [CrossRef]
- Molina-Arcas, M.; Trigueros-Motos, L.; Casado, F.J.; Pastor-Anglada, M. Physiological and pharmacological roles of nucleoside transporter proteins. Nucleosides Nucleotid. Nucl. 2008, 27, 769–778. [Google Scholar] [CrossRef]
- Pastor-Anglada, M.; Cano-Soldado, P.; Molina-Arcas, M.; Lostao, M.P.; Larrayoz, I.; Martinez-Picado, J.; Casado, F.J. Cell entry and export of nucleoside analogues. Virus Res. 2005, 107, 151–164. [Google Scholar] [CrossRef]
- Altman, J.; Das, G.D. Autoradiographic and histological evidence of postnatal hippocampal neurogenesis in rats. J. Comp. Neurol. 1965, 124, 319–335. [Google Scholar] [CrossRef]
- Bayer, S.A.; Yackel, J.W.; Puri, P.S. Neurons in the rat dentate gyrus granular layer substantially increase during juvenile and adult life. Science 1982, 216, 890–892. [Google Scholar]
- Gould, E.; Reeves, A.J.; Graziano, M.S.; Gross, C.G. Neurogenesis in the neocortex of adult primates. Science 1999, 286, 548–552. [Google Scholar] [CrossRef]
- Kaplan, M.S.; Hinds, J.W. Neurogenesis in the adult rat: Electron microscopic analysis of light radioautographs. Science 1977, 197, 1092–1094. [Google Scholar]
- Quinones-Hinojosa, A.; Sanai, N.; Soriano-Navarro, M.; Gonzalez-Perez, O.; Mirzadeh, Z.; Gil-Perotin, S.; Romero-Rodriguez, R.; Berger, M.S.; Garcia-Verdugo, J.M.; Alvarez-Buylla, A. Cellular composition and cytoarchitecture of the adult human subventricular zone: A niche of neural stem cells. J. Comp. Neurol. 2006, 494, 415–434. [Google Scholar] [CrossRef]
- Gratzner, H.G. Monoclonal antibody to 5-bromo- and 5-iododeoxyuridine: A new reagent for detection of DNA replication. Science 1982, 218, 474–475. [Google Scholar]
- Gratzner, H.G.; Leif, R.C.; Ingram, D.J.; Castro, A. The use of antibody specific for bromodeoxyuridine for the immunofluorescent determination of DNA replication in single cells and chromosomes. Exp. Cell Res. 1975, 95, 88–94. [Google Scholar] [CrossRef]
- Zhao, C.; Deng, W.; Gage, F.H. Mechanisms and functional implications of adult neurogenesis. Cell 2008, 132, 645–660. [Google Scholar] [CrossRef]
- Doetsch, F.; Caille, I.; Lim, D.A.; Garcia-Verdugo, J.M.; Alvarez-Buylla, A. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell 1999, 97, 703–716. [Google Scholar] [CrossRef]
- Givogri, M.I.; de Planell, M.; Galbiati, F.; Superchi, D.; Gritti, A.; Vescovi, A.; de Vellis, J.; Bongarzone, E.R. Notch signaling in astrocytes and neuroblasts of the adult subventricular zone in health and after cortical injury. Dev. Neurosci. 2006, 28, 81–91. [Google Scholar] [CrossRef]
- Nowakowski, R.S.; Lewin, S.B.; Miller, M.W. Bromodeoxyuridine immunohistochemical determination of the lengths of the cell cycle and the DNA-synthetic phase for an anatomically defined population. J. Neurocytol. 1989, 18, 311–318. [Google Scholar] [CrossRef]
- Eriksson, P.S.; Perfilieva, E.; Bjork-Eriksson, T.; Alborn, A.M.; Nordborg, C.; Peterson, D.A.; Gage, F.H. Neurogenesis in the adult human hippocampus. Nat. Med. 1998, 4, 1313–1317. [Google Scholar] [CrossRef]
- Doetsch, F.; Garcia-Verdugo, J.M.; Alvarez-Buylla, A. Cellular composition and three-dimensional organization of the subventricular germinal zone in the adult mammalian brain. J. Neurosci. 1997, 17, 5046–5061. [Google Scholar]
- Eriksson, P.S.; Perfilieva, E.; Bjork-Eriksson, T.; Alborn, A.M.; Nordborg, C.; Peterson, D.A.; Gage, F.H. Neurogenesis in the adult human hippocampus. Nat. Med. 1998, 4, 1313–1317. [Google Scholar] [CrossRef]
- Garcia-Verdugo, J.M.; Doetsch, F.; Wichterle, H.; Lim, D.A.; Alvarez-Buylla, A. Architecture and cell types of the adult subventricular zone: in search of the stem cells. J. Neurobiol. 1998, 36, 234–248. [Google Scholar] [CrossRef]
- Lois, C.; Alvarez-Buylla, A. Long-distance neuronal migration in the adult mammalian brain. Science 1994, 264, 1145–1148. [Google Scholar]
- Lois, C.; Garcia-Verdugo, J.M.; Alvarez-Buylla, A. Chain migration of neuronal precursors. Science 1996, 271, 978–981. [Google Scholar]
- Yokochi, T.; Gilbert, D.M. Replication labeling with halogenated thymidine analogs. Curr. Protoc. Cell Biol. 2007, Chapter 22. Unit 22 10. [Google Scholar]
- Tuttle, A.H.; Rankin, M.M.; Teta, M.; Sartori, D.J.; Stein, G.M.; Kim, G.J.; Virgilio, C.; Granger, A.; Zhou, D.; Long, S.H.; et al. Immunofluorescent detection of two thymidine analogues (CldU and IdU) in primary tissue. J. Vis. Exp. 2010, 46, 2166. [Google Scholar]
- Sampath, D.; Rao, V.A.; Plunkett, W. Mechanisms of apoptosis induction by nucleoside analogs. Oncogene 2003, 22, 9063–9074. [Google Scholar] [CrossRef]
- Galmarini, C.M.; Mackey, J.R.; Dumontet, C. Nucleoside analogues and nucleobases in cancer treatment. Lancet Oncol. 2002, 3, 415–424. [Google Scholar] [CrossRef]
- Heidelberger, C.; Chaudhuri, N.K.; Danneberg, P.; Mooren, D.; Griesbach, L.; Duschinsky, R.; Schnitzer, R.J.; Pleven, E.; Scheiner, J. Fluorinated pyrimidines, a new class of tumour-inhibitory compounds. Nature 1957, 179, 663–666. [Google Scholar]
- Anisimov, V.N. The sole DNA damage induced by bromodeoxyuridine is sufficient for initiation of both aging and carcinogenesis in vivo. Ann. NY Acad. Sci. 1994, 719, 494–501. [Google Scholar] [CrossRef]
- Cameron, H.A.; McKay, R.D. Adult neurogenesis produces a large pool of new granule cells in the dentate gyrus. J. Comp. Neurol. 2001, 435, 406–417. [Google Scholar] [CrossRef]
- Kee, N.; Sivalingam, S.; Boonstra, R.; Wojtowicz, J.M. The utility of Ki-67 and BrdU as proliferative markers of adult neurogenesis. J. Neurosci. Method. 2002, 115, 97–105. [Google Scholar] [CrossRef]
- Salic, A.; Mitchison, T.J. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc. Natl. Acad. Sci. USA 2008, 105, 2415–2420. [Google Scholar] [CrossRef]
- Buck, S.B.; Bradford, J.; Gee, K.R.; Agnew, B.J.; Clarke, S.T.; Salic, A. Detection of S-phase cell cycle progression using 5-ethynyl-2'-deoxyuridine incorporation with click chemistry, an alternative to using 5-bromo-2'-deoxyuridine antibodies. Biotechniques 2008, 44, 927–929. [Google Scholar] [CrossRef]
- Huisgen, R. Centenary lecture-1,3-Dipolar cycloadditions. Proc. Chem. Soc. 1961, 357–396. [Google Scholar]
- Kolb, H.C.; Finn, M.G.; Sharpless, K.B. Click chemistry: Diverse chemical function from a few good reactions. Angew. Chem. Int. Ed. Engl. 2001, 40, 2004–2021. [Google Scholar] [CrossRef]
- Diermeier-Daucher, S.; Clarke, S.T.; Hill, D.; Vollmann-Zwerenz, A.; Bradford, J.A.; Brockhoff, G. Cell type specific applicability of 5-ethynyl-2'-deoxyuridine (EdU) for dynamic proliferation assessment in flow cytometry. Cytometry A 2009, 75, 535–546. [Google Scholar]
- Hua, H.; Kearsey, S.E. Monitoring DNA replication in fission yeast by incorporation of 5-ethynyl-2'-deoxyuridine. Nucleic Acids Res. 2011, 39, e60. [Google Scholar] [CrossRef]
- Kaiser, C.L.; Kamien, A.J.; Shah, P.A.; Chapman, B.J.; Cotanche, D.A. 5-Ethynyl-2'-deoxyuridine labeling detects proliferating cells in the regenerating avian cochlea. Laryngoscope 2009, 119, 1770–1775. [Google Scholar] [CrossRef]
- Takagi, S.; McFadden, M.L.; Humphreys, R.E.; Woda, B.A.; Sairenji, T. Detection of 5-bromo-2-deoxyuridine (BrdUrd) incorporation with monoclonal anti-BrdUrd antibody after deoxyribonuclease treatment. Cytometry 1993, 14, 640–648. [Google Scholar] [CrossRef]
- Wiley, L.A.; Shui, Y.B.; Beebe, D.C. Visualizing lens epithelial cell proliferation in whole lenses. Mol. Vis. 2010, 16, 1253–1259. [Google Scholar]
- Nguyen, M.N.; Cavanagh, B.; Davenport, T.; Norazit, A.; Meedeniya, A.C.B. Tissue sectioning for epifluorescence microscopy. In Microscopy: Science, Technology, Applications and Education; Méndez-Vilas, A., Díaz, J., Eds.; Formatex: Badajoz, Spain, 2010; Volume 2, pp. 907–913. [Google Scholar]
- Zeng, C.; Pan, F.; Jones, L.A.; Lim, M.M.; Griffin, E.A.; Sheline, Y.I.; Mintun, M.A.; Holtzman, D.M.; Mach, R.H. Evaluation of 5-ethynyl-2'-deoxyuridine staining as a sensitive and reliable method for studying cell proliferation in the adult nervous system. Brain Res. 2010, 1319, 21–32. [Google Scholar] [CrossRef]
- Chehrehasa, F.; Meedeniya, A.C.; Dwyer, P.; Abrahamsen, G.; Mackay-Sim, A. EdU, a new thymidine analogue for labelling proliferating cells in the nervous system. J. Neurosci. Method. 2009, 177, 122–130. [Google Scholar] [CrossRef]
- Li, K.; Lee, L.A.; Lu, X.; Wang, Q. Fluorogenic "click" reaction for labeling and detection of DNA in proliferating cells. Biotechniques 2010, 49, 525–527. [Google Scholar]
- Sunthankar, P.; Pastuszak, I.; Rooke, A.; Elbein, A.D.; van de Rijn, I.; Canfield, W.M.; Drake, R.R. Synthesis of 5-azido-UDP-N-acetylhexosamine photoaffinity analogs and radiolabeled UDP-N-acetylhexosamines. Anal. Biochem. 1998, 258, 195–201. [Google Scholar] [CrossRef]
- Speers, A.E.; Cravatt, B.F. Profiling enzyme activities in vivo using click chemistry methods. Chem. Biol. 2004, 11, 535–546. [Google Scholar] [CrossRef]
- Yu, X.; Luo, Y.; Zhou, Y.; Zhang, Q.; Wang, J.; Wei, N.; Mi, M.; Zhu, J.; Wang, B.; Chang, H.; et al. BRCA1 overexpression sensitizes cancer cells to lovastatin via regulation of cyclin D1-CDK4-p21WAF1/CIP1 pathway: Analyses using a breast cancer cell line and tumoral xenograft model. Int. J. Oncol. 2008, 33, 555–563. [Google Scholar]
- Cavanagh, B.; Jesuadian, S.; Ng, A.; Dwyer, P.; Nguyen, M.; Bellette, B.; Karunaratne, A.; Poulsen, S.-A.; Mackay-Sim, A.; Meedeniya, A. EdU in Multiple Applications and Cell Types. In Proceedings of the Australian Neuroscience Society 31st Annual Meeting, Auckland, New Zealand,, 31 Januay-3 February 2011.
- Soares, E.V.; Hebbelinck, K.; Soares, H.M. Toxic effects caused by heavy metals in the yeast Saccharomyces cerevisiae: A comparative study. Can. J. Microbiol. 2003, 49, 336–343. [Google Scholar] [CrossRef]
- Becer, C.R.; Hoogenboom, R.; Schubert, U.S. Click chemistry beyond metal-catalyzed cycloaddition. Angew. Chem. Int. Ed. Engl. 2009, 48, 4900–4908. [Google Scholar] [CrossRef]
- Best, M.D. Click chemistry and bioorthogonal reactions: Unprecedented selectivity in the labeling of biological molecules. Biochemistry 2009, 48, 6571–6584. [Google Scholar] [CrossRef]
- Wittig, G.; Krebs, A. Zur Existenz niedergliedriger Cycloalkine, I. Chem. Ber. 1961, 94, 3290–3275. [Google Scholar]
- Agard, N.J.; Prescher, J.A.; Bertozzi, C.R. A strain-promoted [3+2] azide-alkyne cycloaddition for covalent modification of biomolecules in living systems. J. Am. Chem. Soc. 2004, 126, 15046–15047. [Google Scholar] [CrossRef]
- Lutz, J.F. Copper-free azide-alkyne cycloadditions: New insights and perspectives. Angew. Chem. Int. Ed. Engl. 2008, 47, 2182–2184. [Google Scholar] [CrossRef]
- Baskin, J.M.; Prescher, J.A.; Laughlin, S.T.; Agard, N.J.; Chang, P.V.; Miller, I.A.; Lo, A.; Codelli, J.A.; Bertozzi, C.R. Copper-free click chemistry for dynamic in vivo imaging. Proc. Natl. Acad. Sci. USA 2007, 104, 16793–16797. [Google Scholar]
- Sletten, E.M.; Bertozzi, C.R. A hydrophilic azacyclooctyne for Cu-free click chemistry. Org. Lett. 2008, 10, 3097–3099. [Google Scholar] [CrossRef]
- Ning, X.; Guo, J.; Wolfert, M.A.; Boons, G.J. Visualizing metabolically labeled glycoconjugates of living cells by copper-free and fast huisgen cycloadditions. Angew. Chem. Int. Ed. Engl. 2008, 47, 2253–2255. [Google Scholar] [CrossRef]
- Poloukhtine, A.A.; Mbua, N.E.; Wolfert, M.A.; Boons, G.J.; Popik, V.V. Selective labeling of living cells by a photo-triggered click reaction. J. Am. Chem. Soc. 2009, 131, 15769–15776. [Google Scholar]
- Chang, P.V.; Prescher, J.A.; Sletten, E.M.; Baskin, J.M.; Miller, I.A.; Agard, N.J.; Lo, A.; Bertozzi, C.R. Copper-free click chemistry in living animals. Proc. Natl. Acad. Sci. USA 2010, 107, 1821–1826. [Google Scholar]
- Li, Z.M.; Seo, T.S.; Ju, J.Y. 1,3-dipolar cycloaddition of azides with electron-deficient alkynes under mild condition in water. Tetrahedron Lett. 2004, 45, 3143–3146. [Google Scholar] [CrossRef]
- Agard, N.J.; Baskin, J.M.; Prescher, J.A.; Lo, A.; Bertozzi, C.R. A comparative study of bioorthogonal reactions with azides. ACS Chem. Biol. 2006, 1, 644–648. [Google Scholar] [CrossRef]
- Ess, D.H.; Jones, G.O.; Houk, K.N. Transition states of strain-promoted metal-free click chemistry: 1,3-Dipolar cycloadditions of phenyl azide and cyclooctynes. Org. Lett. 2008, 10, 1633–1636. [Google Scholar] [CrossRef]
- Codelli, J.A.; Baskin, J.M.; Agard, N.J.; Bertozzi, C.R. Second-generation difluorinated cyclooctynes for copper-free click chemistry. J. Am. Chem. Soc. 2008, 130, 11486–11493. [Google Scholar] [CrossRef]
- Yamakoshi, H.; Dodo, K.; Okada, M.; Ando, J.; Palonpon, A.; Fujita, K.; Kawata, S.; Sodeoka, M. Imaging of EdU, an alkyne-tagged cell proliferation probe, by Raman microscopy. J. Am. Chem. Soc. 2011, 133, 6102–6105. [Google Scholar]
© 2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Cavanagh, B.L.; Walker, T.; Norazit, A.; Meedeniya, A.C.B. Thymidine Analogues for Tracking DNA Synthesis. Molecules 2011, 16, 7980-7993. https://doi.org/10.3390/molecules16097980
Cavanagh BL, Walker T, Norazit A, Meedeniya ACB. Thymidine Analogues for Tracking DNA Synthesis. Molecules. 2011; 16(9):7980-7993. https://doi.org/10.3390/molecules16097980
Chicago/Turabian StyleCavanagh, Brenton L., Tom Walker, Anwar Norazit, and Adrian C.B. Meedeniya. 2011. "Thymidine Analogues for Tracking DNA Synthesis" Molecules 16, no. 9: 7980-7993. https://doi.org/10.3390/molecules16097980
APA StyleCavanagh, B. L., Walker, T., Norazit, A., & Meedeniya, A. C. B. (2011). Thymidine Analogues for Tracking DNA Synthesis. Molecules, 16(9), 7980-7993. https://doi.org/10.3390/molecules16097980