Overexpression of the Pdx-1 Homeodomain Transcription Factor Impairs Glucose Metabolism in Cultured Rat Hepatocytes
Abstract
:Introduction
Results and Discussion
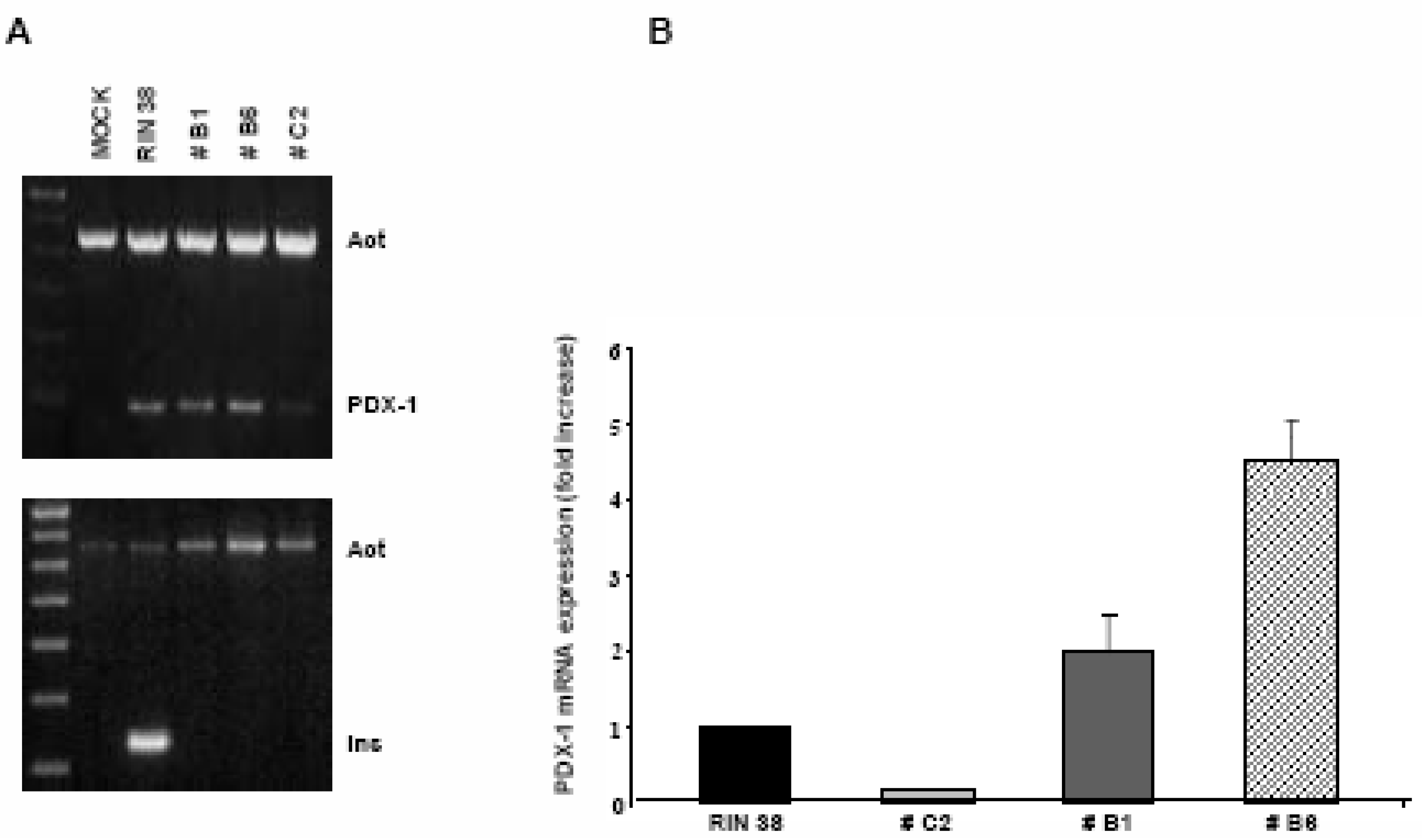
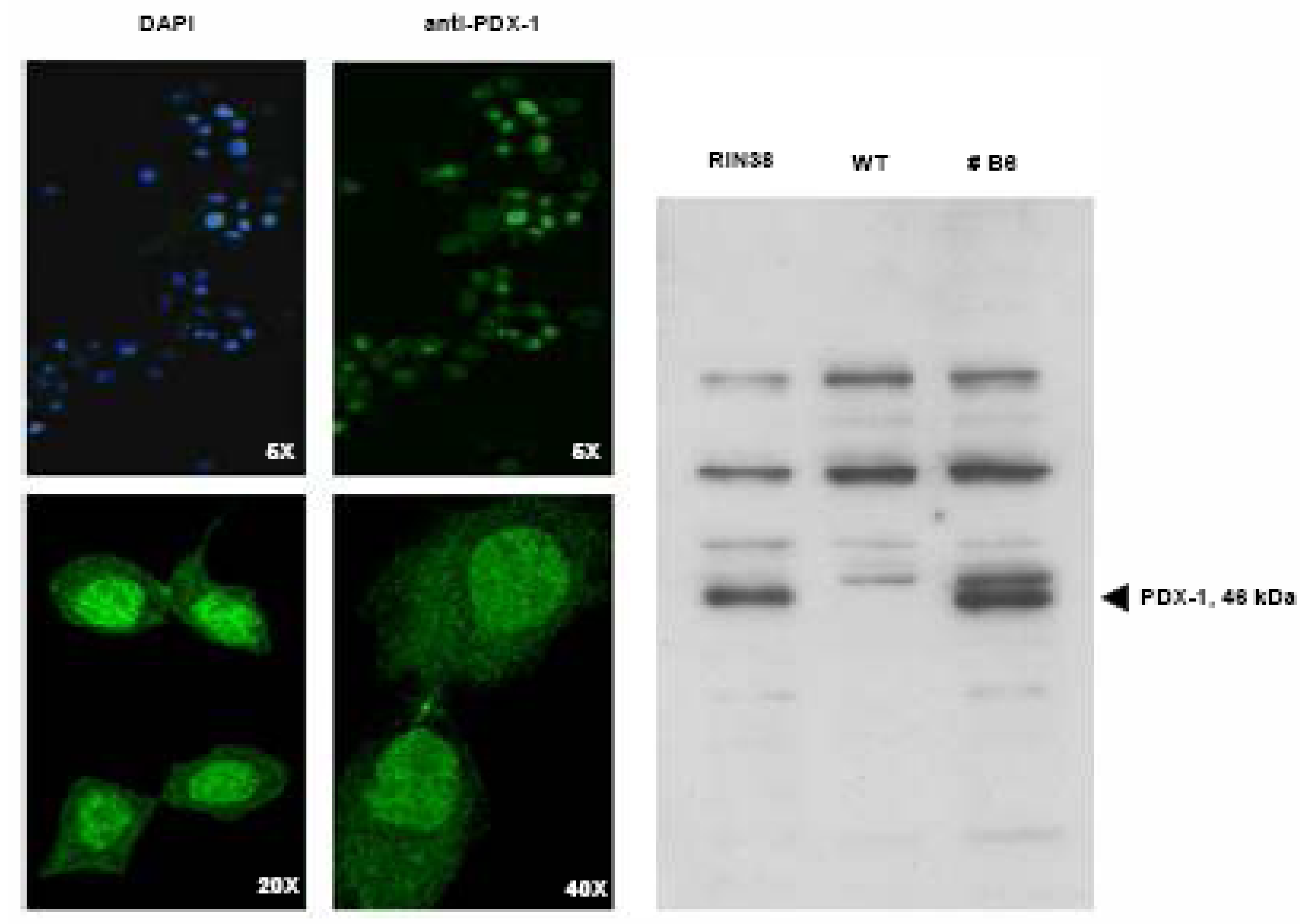
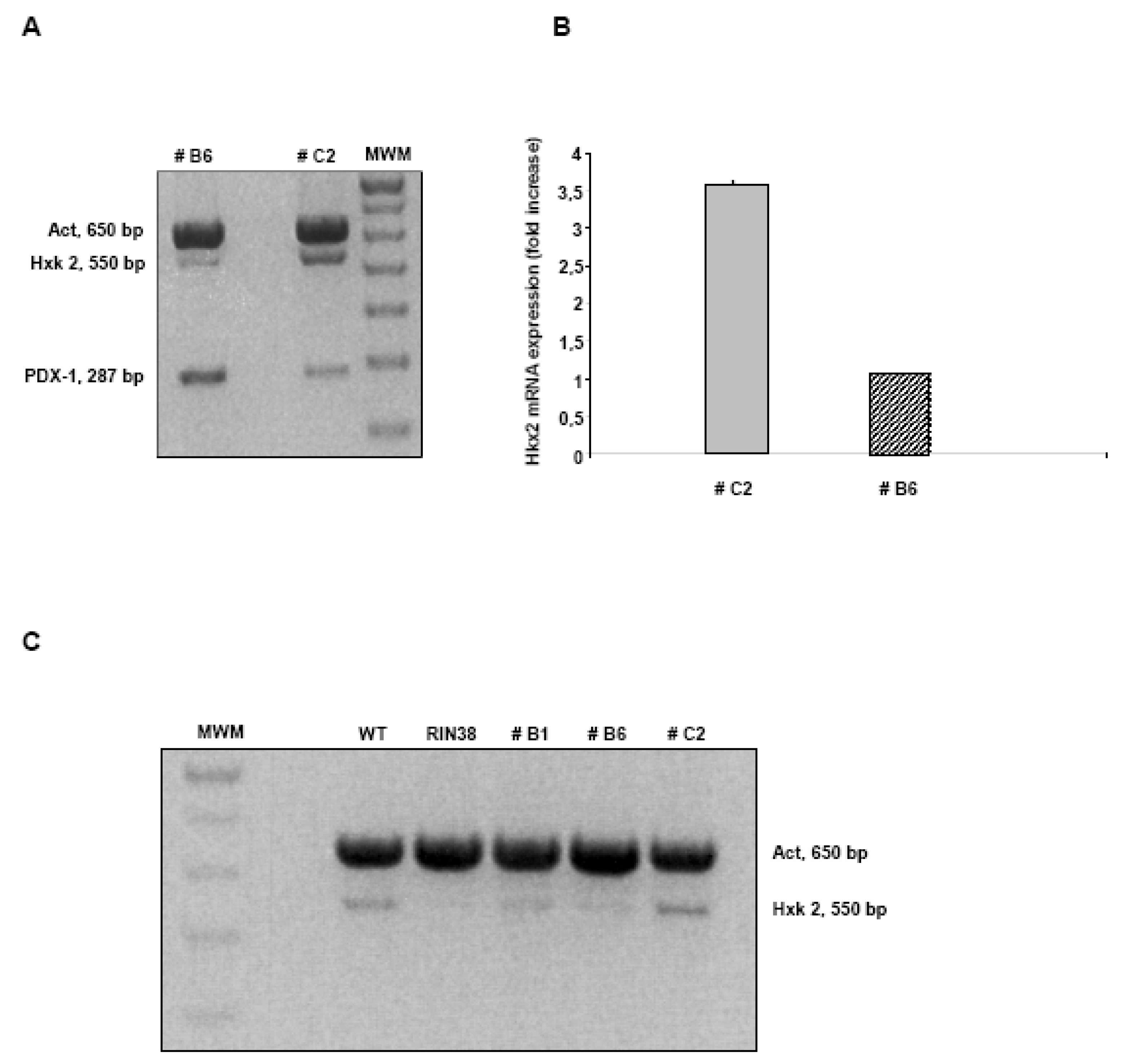
Over-expression of the Pdx-1 gene in cultured Clone 9 rat hepatocytes.
Modulation of the Hexokinase 2 (Hxk2) gene in Clone 9 hepatocytes over-expressing Pdx-1.
 |
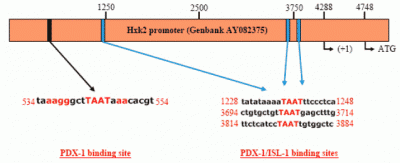
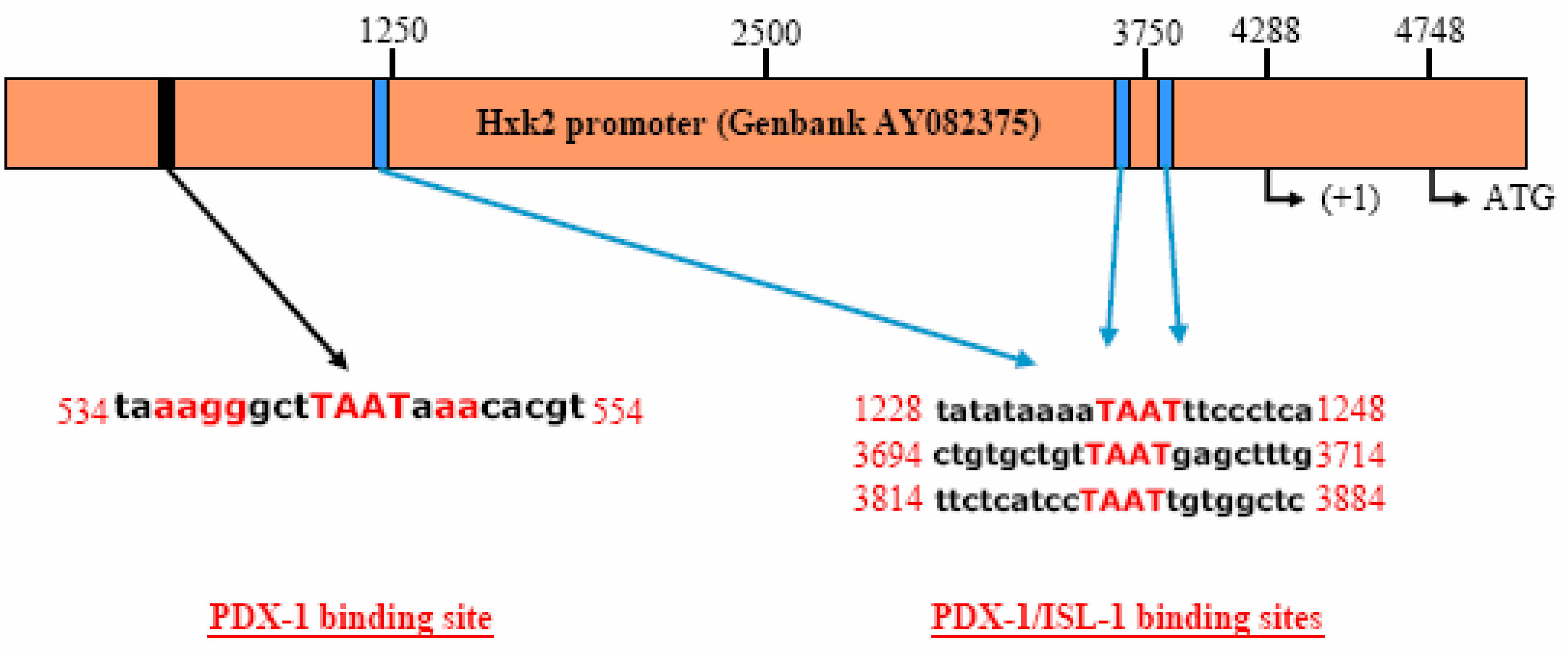
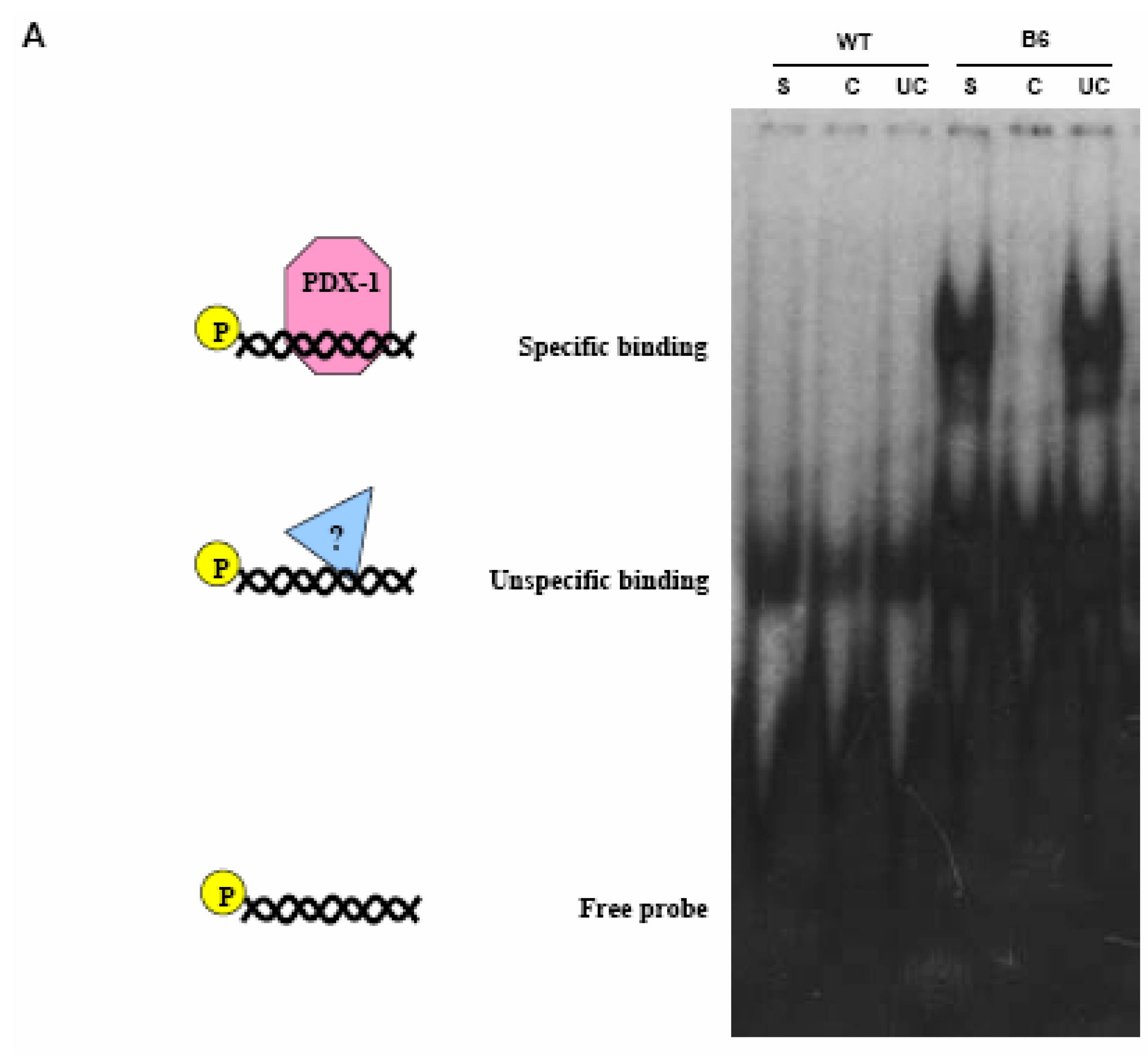
PDX-1 binds to and regulates the Hxk 2 gene promoter.

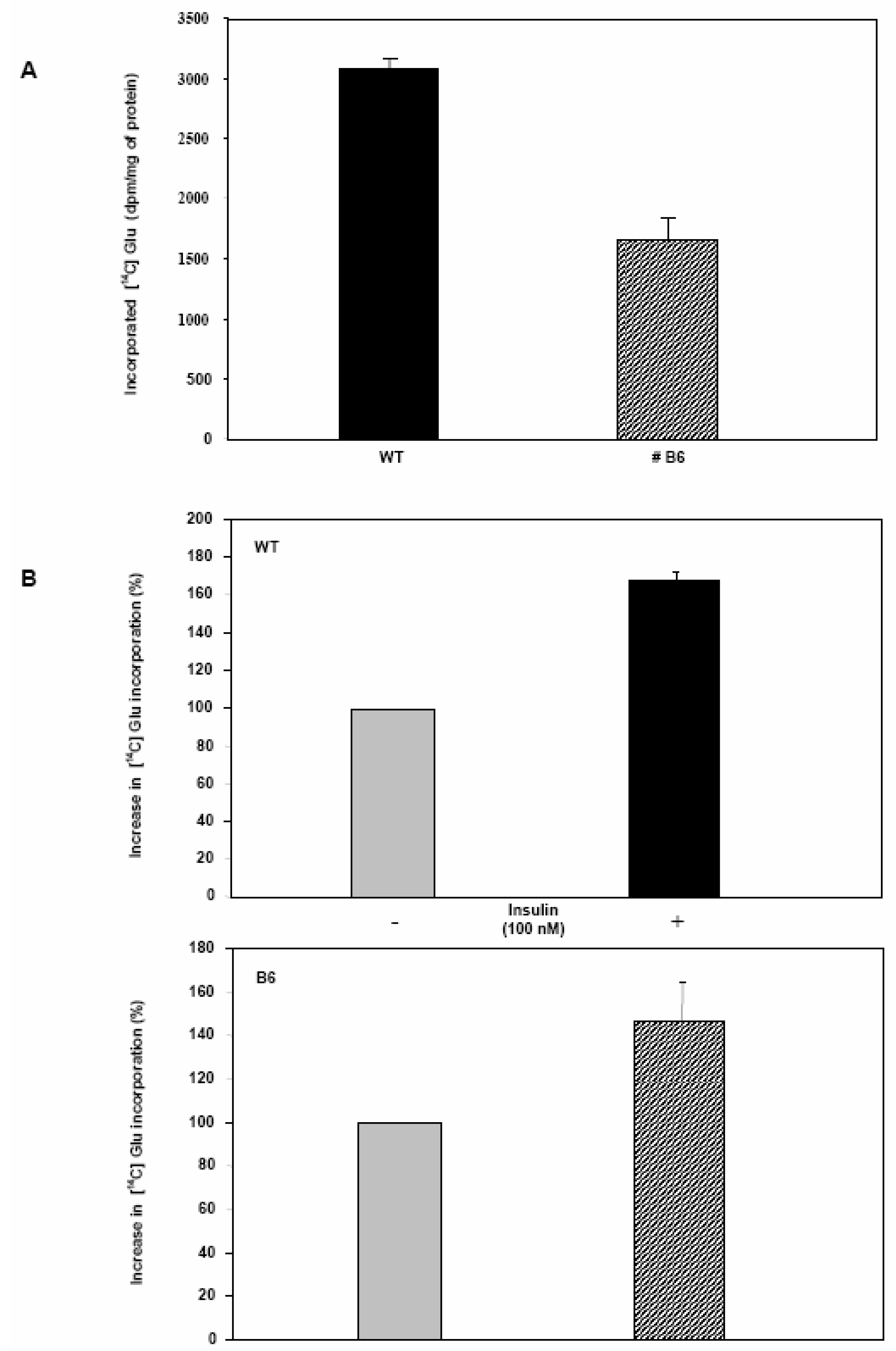
Glycogen synthesis is impaired in Clone 9 hepatocytes over-expressing Pdx-1.
Conclusions
Experimental Section
Acknowledgements
References
- Thorens, B.; Flier, J. S.; Lodish, H. F.; Kahn, B.B. Differential regulation of two glucose transporters in rat liver by fasting and refeeding and by diabetes and insulin treatment. Diabetes 1990, 39, 712–719. [Google Scholar] [CrossRef]
- Asano, T.; Katagiri, H.; Tsukuda, K.; Lin, J. L.; Ishihara, H.; Yazaki Oka, Y. Upregulation of GLUT2 mRNA by glucose, mannose, and fructose in isolated rat hepatocytes. Diabetes 1992, 41, 22–25. [Google Scholar] [CrossRef]
- Khan, A. H.; Pessin, J. E. Insulin regulation of glucose uptake: a complex interplay of intracellular signalling pathway. Diabetologia 2002, 45, 1475–1483. [Google Scholar] [CrossRef]
- Cardenas, M. L.; Cornish-Bowden, A.; Ureta, T. Evolution and regulatory role of the hexokinases. Biochim. Biophys. Acta. 1999, 1401, 242–264. [Google Scholar]
- Hofmann, S.; Pette, D. Low-frequency stimulation of rat fast-twitch muscle enhances the expression of hexokinase II and both the translocation and expression of glucose transporter 4 (GLUT-4). Eur. J. Biochem. 1994, 219, 307–315. [Google Scholar] [CrossRef]
- Ovadi, J.; Srere, P. A. Macromolecular compartmentation and channeling. Int. Rev. Cytol. 2000, 192, 255–280. [Google Scholar] [CrossRef]
- Dutta, S.; Gannon, M. B.; Peers, C.; Wright, Bonner-Weir; Montminy, S. M. PDX-PBX complexes are required for normal proliferation of pancreatic cells during development. Proc. Natl. Acad. Sci. USA. 2001, 98, 065–1070. [Google Scholar]
- Li, H. S.; Arber, S.; Jessell, T. M.; Edlund, H. Selective agenesis of the dorsal pancreas in mice lacking homeobox gene Hlxb9. Nature Genet. 1999, 23, 67–70. [Google Scholar]
- Lu, M.; Miller, C.; Habener, J. F. Functional regions of the homeodomain protein IDX-1 required for transactivation of the rat somatostatin gene. Endocrinology 1996, 137, 2959–2967. [Google Scholar]
- Peshavaria, M. E.; Henderson, E.; Sharma, A.; Wright, C.V.; Stein, R. Functional characterization of the transactivation properties of the PDX-1 homeodomain protein. Mol. Cell. Biol. 1997, 17, 3987–3996. [Google Scholar]
- Sharma, S.; Leonard, J.; Lee, S.; Chapman, H. D.; Leiter, E. H.; Montminy, M. R. Pancreatic islet expression of the homeobox factor STF-1 relies on an E-box motif that binds USF. J. Biol. Chem. 1996, 271, 2294–2299. [Google Scholar]
- Wu, K.L.; Gannon, M. M.; Peshavaria, M. F.; Offield, E.; Henderson, E.; Ray, A.; Marks, L. W.; Gamer, C.V.; Wright, Stein, R. Hepatocyte nuclear factor 3beta is involved in pancreatic beta-cell-specific transcription of the pdx-1 gene. Mol. Cell. Biol. 1997, 17, 6002–6013. [Google Scholar]
- Marshak, S.; Benshushan, E.; Shoshkes, M.; Havin, L.; Cerasi, E.; Melloul, D. Functional conservation of regulatory elements in the pdx-1 gene: PDX-1 and hepatocyte nuclear factor 3beta transcription factors mediate beta-cell-specific expression. Mol. Cell. Biol. 2000, 20, 7583–7590. [Google Scholar] [CrossRef]
- Sharma, A.; Moore, M.; Marcora, E.; Lee, J. E.; Qiu, Y.; Samaras, S.; Stein, R. The NeuroD1/BETA2 sequences essential for insulin gene transcription co-localize with those necessary for neurogenesis and p300/CREB binding protein. Mol. Cell. Biol. 1999, 19, 704–713. [Google Scholar]
- Ben-Shushan, E.; Marshak, S.; Shoshkes, M.; Cerasi, E.; Melloul, D. A pancreatic beta-cell-specific enhancer in the human PDX-1 gene is regulated by hepatocyte nuclear factor 3beta (HNF-3beta ), HNF-1alpha, and SPs transcription factors. J. Biol. Chem. 2001, 276, 17533–17540. [Google Scholar]
- Elrick, L. J.; Docherty, K. Phosphorylation-dependent nucleocytoplasmic shuttling of pancreatic duodenal homeobox-1. Diabetes 2001, 50, 2244–2252. [Google Scholar] [CrossRef]
- Boucher, M. J.; Selander, L. ; Carlsson, L.; Edlund, H. Phosphorylation marks IPF1/PDX1 protein for degradation by glycogen synthase kinase 3-dependent mechanisms. J. Biol. Chem. 2006, 281, 6395–6403. [Google Scholar]
- Kishi, A.; Nakamura, T.; Nishio, Y.; Maegawa, A. Kashiwagi, Sumoylation of Pdx1 is associated with its nuclear localization and insulin gene activation. Am. J. Physiol Endocrinol. Metab. 2003, 284, 830–40. [Google Scholar]
- Macfarlane, W .M.; Smith, S. B.; James, R. F.; Clifton, A. D.; Doza, Y. N.; Cohen, P.; Docherty, K. The p38/reactivating kinase mitogen-activated protein kinase cascade mediates the activation of the transcription factor insulin upstream factor 1 and insulin gene transcription by high glucose in pancreatic beta-cells. J. Biol. Chem. 1997, 15, 20936–20944. [Google Scholar]
- Petersen, H. V.; Peshavaria, M.; Pedersen, A. A.; Philippe, J.; Stein, R.; Madsen, O. D.; Serup, P. Glucose stimulates the activation domain potential of the PDX-1 homeodomain transcription factor. FEBS Lett. 1998, 431, 362–366. [Google Scholar] [CrossRef]
- Stoffers, D. A.; Kieffer, T. J.; Hussain, M. A.; Drucker, D. J.; Bonner-Weir, S.; Habener, J. F.; Egan, J. M. Insulinotropic glucagon-like peptide 1 agonists stimulate expression of homeodomain protein IDX-1 and increase islet size in mouse pancreas. Diabetes 2000, 49, 741–748. [Google Scholar] [CrossRef]
- Sayo, Y.; Hosokawa, H.; Imachi, H.; Murao, K.; Sato, M.; Wong, N. C.; Ishida, T.; Takahara, J. Transforming growth factor beta induction of insulin gene expression is mediated by pancreatic and duodenal homeobox gene-1 in rat insulinoma cells. Eur. J. Biochem. 2000, 267, 971–978. [Google Scholar] [CrossRef]
- Sapir, T.; Shternhall, K.; Meivar-Levy, I.; Blumenfeld, H. T.; Cohen, S.; Skutelsky, S.; Eventov-Friedman, I.; Barshack, I.; Goldberg, S.; Pri-Chen, L.; Ben-Dor, S.; Polak-Charcon, A.; Karasik, I.; Shimon, E.; Mor, S.; Ferber, S. Cell-replacement therapy for diabetes: Generating functional insulin-producing tissue from adult human liver cells. Proc. Natl. Acad. Sci. USA. 2005, 102, 7964–7969. [Google Scholar] [CrossRef]
- Shternhall-Ron, K.; Quintana, F. J.; Perl, S.; Meivar-Levy, I.; Barshack, I.; Cohen, S.; Ferber, S. Ectopic PDX-1 expression in liver ameliorates type 1 diabetes. J. Autoimmun. 2007, 28, 134–142. [Google Scholar] [CrossRef]
- Meivar-Levy, I.; Ferber, S. New organs from our own tissues: liver-to-pancreas trans- differentiation. Trends Endocrinol Metab. 2003, 14, 460–466. [Google Scholar] [CrossRef]
- Reddy, D.; Pollock, A.S.; Clark, S. A.; Sooy, K.; Vasavada, R. C.; Stewart, A. F.; Honeyman, T.; Christakos, S. Transfection and overexpression of the calcium binding protein calbindin-D28k results in a stimulatory effect on insulin synthesis in a rat beta cell line (RIN 1046-38). Proc. Natl. Acad. Sci. USA 1997, 94, 1961–1966. [Google Scholar] [CrossRef]
- Lee, M. G.; Pedersen, P. L. Glucose metabolism in cancer: importance of transcription factor- DNA interactions within a short segment of the proximal region of the type II hexokinase promoter. J. Biol. Chem. 2003, 278, 41047–41058. [Google Scholar] [CrossRef]
- Wilson, J. E. Isozymes of mammalian hexokinase: structure, subcellular localization and metabolic function. J Exp Biol. 2003, 206, 2049–2057. [Google Scholar] [CrossRef]
- Kojima, H.; Fujimiya, M.; Matsumura, K.; Younan, P.; Imaeda, H.; Maeda, M.; Chan, L. NeuroD-betacellulin gene therapy induces islet neogenesis in the liver and reverses diabetes in mice. Nat Med. 2003, 9, 596–603. [Google Scholar] [CrossRef]
- Sample Availability: Cell lines used in this work are available from the authors.
© 2008 by the authors. Licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Pillich, R.T.; Scarsella, G.; Risuleo, G. Overexpression of the Pdx-1 Homeodomain Transcription Factor Impairs Glucose Metabolism in Cultured Rat Hepatocytes. Molecules 2008, 13, 2659-2673. https://doi.org/10.3390/molecules13102659
Pillich RT, Scarsella G, Risuleo G. Overexpression of the Pdx-1 Homeodomain Transcription Factor Impairs Glucose Metabolism in Cultured Rat Hepatocytes. Molecules. 2008; 13(10):2659-2673. https://doi.org/10.3390/molecules13102659
Chicago/Turabian StylePillich, Rudolf Tito, Gianfranco Scarsella, and Gianfranco Risuleo. 2008. "Overexpression of the Pdx-1 Homeodomain Transcription Factor Impairs Glucose Metabolism in Cultured Rat Hepatocytes" Molecules 13, no. 10: 2659-2673. https://doi.org/10.3390/molecules13102659
APA StylePillich, R. T., Scarsella, G., & Risuleo, G. (2008). Overexpression of the Pdx-1 Homeodomain Transcription Factor Impairs Glucose Metabolism in Cultured Rat Hepatocytes. Molecules, 13(10), 2659-2673. https://doi.org/10.3390/molecules13102659