Abstract
A compact Modular Chemical Descriptor Language (MCDL) chemical structure editor (Java applet) is described. The small size (approximately 200 KB) of the applet allows its use to display and edit chemical structures in various Internet applications. The editor supports the MCDL format, in which structures are presented in compact canonical form and is capable of restoring bond orders as well as of managing atom and bond drawing overlap. A small database of cage and large cyclic fragment is used for optimal representation of difficult-to-draw molecules. The improved algorithm of the structure diagram generation can be used for other chemical notations that lack atomic coordinates (SMILES, InChI).
Introduction
Linear molecular descriptors are frequently used for storing, retrieval, and presentation of chemical information. Their role has increased significantly with the advent of the Internet. Although chemical structures on the Web can be presented as bitmap objects (in GIF, JPEG, or PNG formats), this method of representation is not optimal. An alternative method entails use of special chemical applets or plug-ins capable of rendering chemical structure information encoded in chemical descriptors. This method of coding reduces net traffic, allows work with a chemical structure as an object, for example, to rotate three-dimensional (3D) structure in space [1,2], and allows the structures to be edited in Web interfaces of chemical databases. Two approaches exist for representation of molecular structures as linear descriptors—chemical names or computer-readable codes.
CAS [3] and IUPAC [4] nomenclature are examples of the chemical name approach. IUPAC and CAS names are the most understandable notation for a chemist, but their use in databases or as Internet descriptors is a complicated task. Chemical names are relatively long and have sophisticated formats, thus impeding computer structure recognition. In many cases the existing name-generating programs [5,6] cannot process some classes of compounds, such as cage structures or molecules with abnormal valence elements. In addition, chemical names generated by various computer programs are often not unique. Ambiguity in IUPAC naming complicates searching and chemical identity comparison. The reverse task—chemical structure graph generation from the chemical name—has it set of problems too. As with naming, drawing of polycyclic and abnormal valence element molecules is the most difficult task, even for the latest versions of the software [7]. NameExpertTM, a product of ChemInnovation [8], converts IUPAC names to chemical structures, but the program cannot handle the names of certain specific classes of molecules, such as names of element-organic compounds. The list of rules restricting the names of compounds that are supported by this program is published on the home page of the software vendor.
Other types of linear molecular descriptors include computer-readable formats, such as Wiswesser Line Notation (WLN) [9], SYBYL® [10], SMILES [11] (Daylight Chemical Information Systems), and InChI [12] codes. The SMILES code is the de facto standard for computer-readable linear notation, although it has some disadvantages. First of all, canonical numbering requires use of a proprietary SMILES2 algorithm [13,14]. The second disadvantage of SMILES is lack of adequate representation of fragment-specific information, a useful search feature when a chemical structure descriptor is embedded in an HTML document. The information is also of value for computation of certain physical-chemical properties that can be attributed to a particular structure fragment, for example, NMR chemical shifts [15]. InChI was recommended by IUPAC as the standard for computer-readable chemical structure notations [16,17]. It is now widely used in NIST and NIH public databases. Extending InChI to accommodate 3D chemical structures was described in [18]. However, the absence of descriptors for fragment-centered properties can be considered as disadvantage.
These shortcomings were taken into consideration during the development of the Modular Chemical Descriptor Language (MCDL), which is designed for linear representation of chemical structures and compound properties [19]. Composition and connectivity modules of the MCDL string are coded in canonical form and therefore can be used directly for structure comparison. Supplementary modules are designed to store various compound properties (e.g., chemical names, elemental analysis data, MS-spectra, common physical-chemical properties, etc.). These features make MCDL coding convenient and attractive for presenting chemical structures in chemical databases and HTML documents. The MCDL and InChI formats are structurally similar. For example, both use separate modules to describe composition, connectivity table and charges. MCDL provides direct placement of hydrogen atoms, whereas InChI uses a separate block.
Special chemical browser plug-ins, Java applets, or Microsoft.NET technology can be used to draw chemical structures embedded as text codes in HTML documents. Plug-ins are specific to a particular Internet browser and operating system. The new Microsoft.NET method is not distributed widely because of its incompatibility with the popular Java technology. Java applets are widely used to process chemical structures [20,21,22,23,24,25]. Peter Ertl’s popular JME applet [22] can be used to create and to edit chemical structures, as well as to embed chemical structure information into HTML documents. It can also generate structure codes in SMILES and in JME (similar to MDL molfile [26]) formats. However, only the JME format, which stores Cartesian coordinates, is suitable for chemical structure rendering using the JME applet.
An open source JchemPaint Java applet, integrated into the Chemistry Development Kit (CDK) [23,24] allows generation of structural diagrams from coordinateless structure formats. This is considered to be one of the most important features of the CDK software package. As a result, this applet allows rendering of chemical structures encoded in SMILES format. In addition, the CDK software suite contains various supplementary software libraries: NMR spectra prediction, 3D structure visualization, and calculation of some topological indexes. JChemPaint supports various chemical structure formats (MOL, PDB, CML, XYZ, XML, SMILES) using a structure generator, verification of graph connectivity, and HOSE code [27] (to predict atom-centered properties). All these features make the applet relatively “heavy”—the size of the current DEMO version of JChemPaint *.jar file is above 1.5 MB [28]. A simple Java applet that creates/draws/edits chemical structure as MCDL strings was created and is described in the present paper. Software development was performed with particular attention to MCDL-specific problems, such as bond order reconstruction, as well as more general ones, such as optimal rendering and polycyclic compounds, and visual fragment overlapping.
Results and Discussion
Java 1.1 was used to create the applet (since the main function of the applet is to view structures within an Internet browser). This avoids the “heavy” Java Run-Time Environment plug-in [29], which is necessary to execute Java 1.3 applications in Microsoft Internet Explorer (currently the most popular Web browser). The applet architecture is relatively simple. There is a class SimpleMolecule, in which vectors atoms and bonds are defined. Vector atoms contains the group of Atom class, vector bonds —the group of Bond class. Atom class consists of nA—periodic table position, nC—charge, nV—valence (nonzero if not standard), rL—radical sign, nB—the number of attached atoms, array aC[]—the numbers of attached atoms in atoms collection. The nB and aC[] fields are calculated from a connectivity matrix, which is stored in class Bonds in compact form. The Bond class contains following fields: aT1, aT2—numbers of atoms in atoms collection, tB—bond type (single, double, or triple), dB—cyclic structure indicator. The DrawMolecule class is the assessor of the SimpleMolecule class and can draw two-dimensional (2-D) chemical structures. Finally, EditedMolecule class contains methods required for chemical structure modifications: to append or to remove a bond or a fragment. The class also contains methods employed to search for a structure fragment (subgraph isomorphism).
All these classes are stored in the Molecule package. The package contains four other classes—MCDL, BondAlternate, TemplateRedraw, and ChainRotate. The MCDL class keeps the collection of methods for generation of MCDL code from connectivity matrix and reverse procedure—for generation of connectivity matrix from the MCDL string. The algorithms of the direct transformation were described previously [19]. The BondAlternate class is used to reconstruct bond order from the number of hydrogen atoms, attached to forming bond atoms. The algorithm of bond reconstruction is described below.
The TemplateRedraw class contains the database of fragments (primary polycyclic) with coordinates of atoms for the structure-rendering purposes. The database can be easily appended by addition of a string with atomic coordinates of a fragment. This string contains the number of atoms and bonds in a fragment. The X and Y coordinates for each atom are defined as 2-byte variables and a 1-byte flag (whether the new bonds can be appended or not). Each bond descriptor contains the numbers of bonded atoms. All atoms and bonds in a fragment can be compared with any atoms and bonds in a rendered structure. Other attributes (e.g., charge, atomic number, valence, bond order) are not stored, so the fragments database is very compact. For example, cubane (C8H8) is coded by a 50 bytes-long string, which is important for fast applet loading on a client computer. The same class is used to generate structure diagrams of predefined fragments in a chemical structure.
The static class ChainRotate contains the CorrectOverlapped method. It determines whether or not atoms (bonds) are visually overlapped and tries to resolve overlapping by fragment rotation around acyclic bonds.
A simple architecture allows for a small-sized applet – about only 200K. The small applet can be loaded quickly and does not require special download optimization methods, such as Obfuscation [30] (removal of unused classes or methods), or Extension Mechanism [31] (download packages “on demand”).
Reconstruction of bond order from MCDL string
Bond order (single, double, or triple) is important chemical structure information, but it is a supplementary module (which may or may not be presented) in a MCDL string. In many cases, bond order can be unambiguously restored from the number of protons attached to each atom. To recalculate bond order, all bonds are assigned to be single, and an array nHCalc[nAtoms] is formed. nHCalc[nAtoms] is the number of calculated hydrogens (assuming that the valences of all elements are standard). When an MCDL string is analyzed, the nH[nAtoms] array is formed, where nH[nAtoms] is real (the number of hydrogens attached to a particular atom). If all bonds are single, then the following Eq. (1) is true for each atom:
nHCalc[i] >= nH[i] .
Then the bond order is being increased (using double or triple bonds instead of single bonds) until the Eq. (2) is true:
nHCalc[i] = nH[i] .
The process starts with identification of bonds with orders that can be determined unambiguously. If any atom i has n attached atoms, and Eq. (2) is correct for all of them except for a single j neighbor, then the bond order between i and j atoms can be calculated according to Eq. (3):
Border = 1 + (nHCalc[i] - nH[i]) .
One bond is added in Eq. (3) to reflect the presence of at least a single bond between this pair of atoms. Accordingly, the values of nHCalc for i and j atoms are changed. If any bond order was changed as described above, then all atoms are rechecked again. Changes in nHCalc allow for possible determination of other bond orders. The iteration process is terminated when no more bond orders can be defined unambiguously. This iteration procedure is capable of restoring bond order in cumulenes and enynes. Bond order in these compounds can only be determined by taking into consideration the nature of terminal atoms. Absence of this information could lead to ambiguity of bond order identification and mismatching of these two classes of compounds.
The algorithm described above does not work for cyclic compounds with alternating bonds (e.g., for aromatic compounds). Unambiguous identification of bond orders in cyclic fragments is not possible for these molecules; therefore, Eq. (3) cannot be used. A similar situation exists for antiaromatic compounds, such as cyclooctatetraene (C8H8). Aromatic bonds can be used to demonstrate cyclic aromatic fragments, but Kekule structures are generally more appealing for chemists. In addition, Kekule structures can be incorporated into any chemical database without any restrictions. Because a similar situation exists for importing the results of quantum mechanical calculations or X-ray structure analysis into a database supporting 2D structures, the problem can be solved using any previously developed algorithm [32,33,34,35] for generation of Kekule structures for aromatic compounds.
Unfortunately, fast algorithms for generation of Kekule structures can be used only for even-numbered rings. The following steps need to be taken for odd five-member rings:
- Two cyclic bonds attached to a chalcogen (oxygen, sulphur) or a three-coordinated nitrogen of unknown order are replaced by single bonds. The same procedure is executed for a nitrogen atom linked with neighboring atoms with three aromatic bonds—junction of fused aromatic rings.
- The valences of positively charged heteroatoms are incremented by 1 relative to standard values: N+: 4; O+: 3.
- The order of an arbitrary selected bond is assigned to 1. The order of an adjacent bond is considered to be 2, next—1 and so on. For fusion atoms (any atom of a fused-ring system which is common to two or more rings), one bond is temporarily assigned as a double and the other two as a single. This temporary assignment is stored to allow future modification in case of an incorrect assignment.
- If the calculated number of hydrogens does not correspond to the MCDL string number, then the reconstruction process is considered to be unsuccessful. In this case, the algorithm returns to the last fused atom assignment (point 3) to reassign single and double bonds.
- If after all attempts, there is no acceptable bond assignment, the program returns to point 1, and the order of the first arbitrary selected bond is set to be 2.
- Finally, Kekule structure representation is considered to be impossible if no bond order assignment that corresponds to an MCDL string can be found.
This algorithm might look ineffective because of an exhaustive check of all possible assignments of single and double bonds in cycles. In practice, however, it is relatively fast due to elimination of impossible combinations (the order of a subsequent bond is defined by the order of a preceding one). A Kekule structure is generated from the first attempt if only six-member cycles are present in a compound. To speed up the process, the above procedure is performed separately for each non-fused cyclic fragment. It should be noted that in some rare cases the restoration of bond orders using only the MCDL connectivity module and the number of attached protons is not possible. These cases were examined in [19].
Structure diagram generation of polycyclic compounds
The coordinates of atoms in an MCDL string can be stored in a Cartesian coordinates supplementary module, but this module is not obligatory. Therefore, in general it is necessary to generate 2D Cartesian coordinates to draw an adequate structure diagram from the connectivity module. Although this task has an unlimited number of solutions, only a few of them can be considered as attractive (publication quality). The 2D chemical structures look neat when all bond lengths are equivalent, and all angles between the bonds are close to 2π/3. This perfect arrangement is not always possible, but it represents the highest reference point for the task.
Several structure diagrams–generation algorithms were developed in the past [36,37]. The majority of chemical structure editors have a “Clean Structure” command [7,38,39]. When executed, this command generated a structure diagram in which all bonds are equal, and the angles are as optimal as possible. Presentation of polycyclic compounds is the most difficult task for commercial chemical structure drawing programs—ISIS/Draw [38], ChemDraw [39], ChemSketch [7] as well as open source JChemPaint [24]. The cubane (C8H8) diagram, in which 2D coordinates are defined randomly, is shown in Figure 1. These structure drawings were generated from the distorted diagram by execution of “Clean structure” command. ChemSketch [7] (ACD Labs) and partially JChemPaint [23] generate an adequate, publication-quality 2D picture.
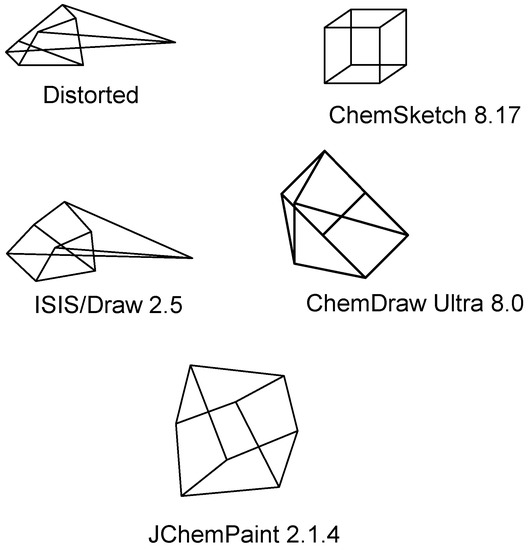
Figure 1.
The result of Clean Structure command execution in popular structure editors: ChemSketch 8.17, ISIS/DRAW 2.5, ChemDraw Ultra 8.0, and JChemPaint 2.1.4.
ChemSketch uses a proprietary, undisclosed algorithm. It is likely that templates with pre-defined atomic coordinates are being used to generate of 2D atomic coordinates in polycyclic compounds. If a molecule contains several polycyclic groups (polycyclic fragments separated from other fragments by acyclic bonds), ChemSketch uses atomic coordinates from templates for each fragment. Exact matching between a group and a fragment template is required for this method. If fused rings are added to a group, a poor structure diagram is generated (tripticene-C20H14, 1,2-trimethylenecubane- C11H12).
A simplified template algorithm is used in JChemPaint. Atomic coordinates are taken from a template for a single fragment in a chemical structure [40]. If a chemical structure contains several polycyclic fragments, a poor structure diagram is generated (1,1’-biscubane, C16H14).
There is another “cleaning” mechanism that can display polycyclic compounds as images of reasonable quality. It generates 3D chemical structures and then projects these 3D objects in 2D mode [25,39]. The drawback of this method is the absence of a universal algorithm generating fine 2D structure diagrams from 3D atomic coordinates. Sometimes these 2D projection images have distorted (non-optimal) bond lengths and angles of structural fragments that could otherwise be drawn without overlap using optimal angles and identical bond lengths. ChemDraw Ultra offers an optional interactive user interface to improve 2D structure diagram generation [39], but this interactive interface is not suitable for automatic batch conversion of large sets of chemical structures.
To resolve the problems of optimal presentation of common polycyclic and large cycle structures, modifications of the existing template algorithm are required. The content of the database is searched to find a matching fragment, and, if successful, the coordinates of atoms and the scaling factor are used together with appropriate shifting and rotating subroutines to achieve optimal drawing. The search is repeated until no more fragments from the template database are found. To avoid bond overlapping, these fragments must contain only database-defined chemical bonds. This restriction allows for accuracy in drawing polycyclic compounds (example: 1,2-tetramethylenecubane, which contains a 12-membered ring; provided that this 12-membered ring is recorded in the fragment database). Because only coordinates of vertices are being used for drawing, these templates are compatible with any atoms (bonds) in a structure. In addition, it is possible to create a very compact depository of fragments, which is critical for development of a compact MCDL applet. From our experience, the current size of the template database (105 fragments) is adequate to draw the preponderance of polycyclic compounds.
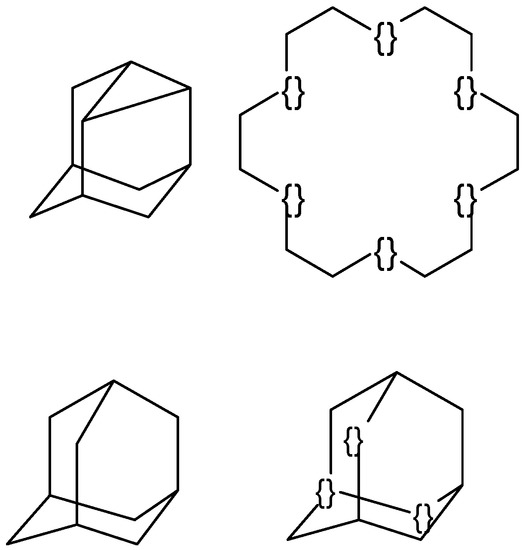
Figure 2.
Several examples of fragments used in the database of poly(cyclic) fragments. The atoms that cannot be used for bonding with other atoms are marked as {}.
Several examples of structures from the database are shown in Figure 2. The database contains polycyclic structures (adamantane, noradamantane) as well as large cycles (cyclooctadecane). Large rings are difficult to draw because the angles between the bonds tend to be small, and the size of the ring is difficult to estimate visually. Symmetrical (poly)cyclic fragments might have several possibilities for bonding with other molecular fragments in the final structure, and it is possible that a randomly selected connection point might not be the optimal one due to atom/bond overlap in visually congested areas. Examples of such poorly generated structure diagrams are shown in Figure 3. To solve the problem, some fragment atoms in the database are marked-out as “unavailable for bonding” (other fragments of a molecule should not be attached to these places). Examples of the marked-out fragments are shown in Figure 2—adamantane with three marked-out atoms and cyclooctadecane with six marked-out atoms. The unmarked adamantane structure is also stored in the database to generate the structure diagrams of the parent adamantane and its simple derivatives.
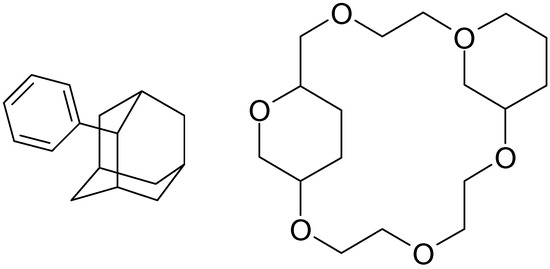
Figure 3.
Computer-generated drawings of polycyclic molecules: 2‑phenyladamantane and dicyclohexyl-18-crown-6 (some atoms are overlapped).
Fragments in the database are arranged according to their size (number of atoms). The search begins with the fragment having the maximal number of atoms or maximum number of bonds when the number of atoms is the same (otherwise the largest matching fragment may not be found among smaller ones). If there are marked fragments and unmarked fragments, the search begins with the unmarked ones.
The next problem is associated with multiple re-use of small subfragments. Without it a large database of all possible fragments would be required, and the applet would be too large. For example, the complete 1,1’-diadamantane template would be required to display its structure. Alternatively, the molecule can be successfully rendered using two smaller adamantane templates. To employ this strategy, repeated search of molecular fragments in a molecule should be performed. Consequently, if coordinates of some atoms are restored, then they are excluded from the next iteration. The algorithm of drawing structure using fragments can be summarized as follows:
- The set of the minimum number of cycles in the compound is calculated using an algorithm [41] and stored in the LIST.
- The search in the fragment database is executed, and coordinates of relevant atoms are assigned when a fragment is found. If a fragment is not found, then the maximum size cycle from the LIST is drawn. If there is no cycle, then two linked atoms with maximum substitution numbers are used as the initial fragment to generate a structure diagram.
- The fragment in the database is searched for atoms with not-yet-determined coordinates. The fragment should be linked to an atom with known 2D coordinates.
- If the fragment is found, then coordinates of corresponding atoms in the structure are considered to be assigned. Then the algorithm returns to point 3, and the next fragment is searched. If there are no more qualified fragments, then the algorithm moves to point 5.
- All cycles from the list with at least one assigned coordinate atom are added to the structure. If coordinates of only one atom are known, then a spiro-cycle is added with standard bond lengths, and angles are calculated from the size of the cycle. If coordinates of two bonded atoms are known, then a fused cycle is added with the bonds’ length equal to a known bond and with the angles calculated as 2π/N, where N is the size of the cycle. If the coordinates of three and more atoms are known (polycyclic structure), then the chain is locked. The positions of new atoms are assigned using a special subroutine to avoid bond intersection (if possible). If the position of at least one more atom is determined here, then the algorithm returns to point 3, otherwise it goes to point 6.
- Coordinates of acyclic atoms (connected to cyclic atoms with known coordinates) are calculated using the standard bond length and the optimal bond angle 2π/3. In the case of long chains, coordinates of only the first (connection) atom are calculated. If the position of at least one more atom is determined here then the algorithm returns to point 3, otherwise it goes to point 7.
- A determination is made whether coordinates of all atoms are defined. “Yes” means the process is finished; “No” means that compound is a disconnected graph with two or more substructures. The process is repeated beginning from point 1 for the next fragment until full completion.
Structure diagrams of some polycyclic compounds generated from MCDL strings are shown in Figure 4. It is quite possible that there are other polycyclic structures that cannot be presented optimally with this algorithm. They can be added to the database in the future versions of the software.
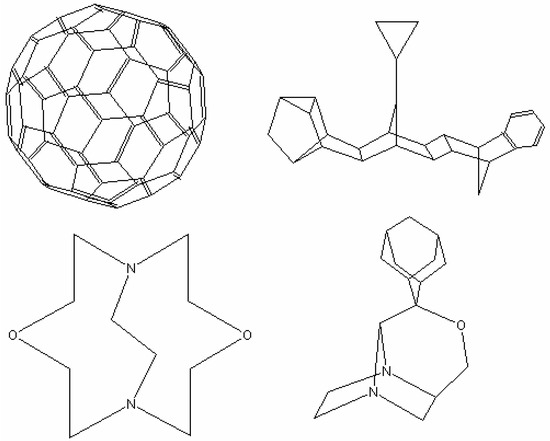
Figure 4.
Examples of structure diagrams generated from MCDL strings.
Overlapped fragments
Atom and bonds in computer-generated drawings of molecules with bulky fragments are often overlapped. In some cases, the overlap can be avoided by rotation of aromatic fragments around acyclic bonds by 180°, but not always. For example, atom overlap in 1,5-diisopropyl-1’,5’-dichlorobiphenyl (Figure 5-A) cannot be fixed by any rotation. In this case, bond lengths or bond angles should be changed, which leads to poor visual quality of the structure drawings. In another example, atom overlap in the 1,5-diethyl-1’,5’-dichlorobiphenyl (Figure 5-B) structure can be avoided using rotation around the C-C bond (Figure 5-C).
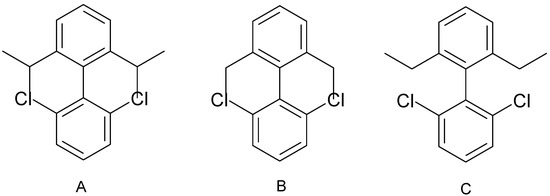
Figure 5.
Structures containing overlapped fragments (A and B) and the lack of overlapping (C).
To solve this problem, a slightly simplified algorithm [37] is employed. The algorithm uses rotation around acyclic bonds. Initially, new chain atoms are added in the direction of their “growth,” leading to formation of chains with maximum length. In many cases overlap of fragments can be avoided by using rotation around acyclic bonds. This can be accomplished by calculating the number of nonequivalent substituents for all acyclic bonds followed by identification of “spherical fragment” for each atom in the molecule (similar to HOSE [27] code). The radius of this “spherical fragment” is equal to the topological lengths of the molecule. After that, atom-centered indexes [15] are calculated. If these atom-centered indexes for two acyclic bond-neighboring atoms are equal, then they are equivalent. This is an indication that rotation across this bond cannot produce an optimal picture. The list of these bonds is stored, and all possible combinations of 180° rotations are performed to find the optimal drawing. The total number of these combinations is equal to 2N, where N is the number of qualified acyclic bonds. To minimize computation time, the maximal number of these qualified bonds is limited to 12.
Conclusions
The MCDL chemical structure applet editor source codes and executables will be available for public domain at the MCDL SourceForge development area [42,43]. The overall look of the applet (Figure 6) is made similar to a popular JME editor [22]. The new chemical drawing approaches developed for the MCDL applet have wider applications in the area of computer structure generation. For example, virtual combinatorial libraries generation [44] and virtual screening [45] are based on computer design of new molecule structures and evaluation of their properties. These chemical structures should be visualized, and improvement in drawing quality makes the software more attractive and easier to use. Even in complicated cases (such as of virtual Diels-Alder cyclization reactions), the use of atomic coordinate templates simplifies structure diagram generation.
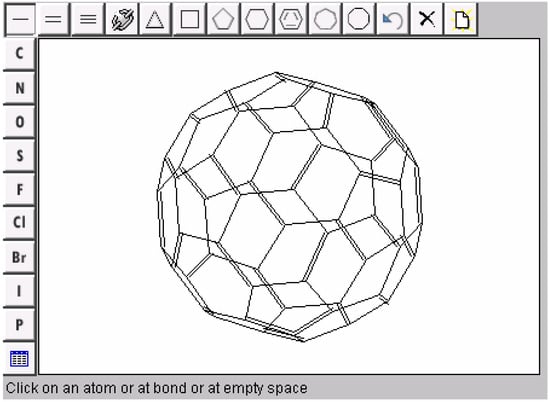
Figure 6.
Computer-generated drawing of [60] fullerene molecule in the MCDL applet window.
The proposed approach does not solve the structure generation problem entirely, but it does make it more efficient. The existing database has only 105 unique fragments, which is not adequate for structure generation of uncommon polycyclic and spiro-compounds. Further improvements can be achieved by adding more template coordinates in the database, but the growth of the database increases applet loading time and performance on the client side. The database resources can be searched more effectively using the known coordinates of atoms and atom pairs (bonds). The approach is useful for generation of the structures of spiro- and polycyclic compounds and will be included in the next version of the software package.
Supplementary materials
Source codes of MCDL structure editor are available at [43].
Acknowledgments
This research was sponsored by the IPP program. Oak Ridge National Laboratory is managed by UT-Battelle, LLC, under contract DE-AC05-00OR22725 for the U.S. Department of Energy. Contribution 18 from the Discovery Chemistry Project.
References and Notes
- Rzepa, H.S.; Cahser, O.; Leach, C. Recent Applications of Hyperactive Chemistry and the World-Wide-Web: Towards an Integrated Chemistry Information Environment. New Initiatives in Chemical Education: An On-line Symposium, June 3 - July 19, 1996. available at http://www.ch.ic.ac.uk/rzepa/cc96/cc96_intro.html accessed December 2005.
- Applets and visualization. http://www.morechemistry.com/links/Applets_and_Visualizations.html accessed December 2005.
- CAS. http://www.cas.org accessed December 2005.
- International Union of Pure and Applied Chemistry (IUPAC). Nomenclature of Organic Chemistry; Rigaudy, J., Klesney, S.P., Eds.; Pergamon Press: Oxford, U.K., 1979. [Google Scholar]
- ACD/Name. http://www.acdlabs.com/products/name_lab/name/ accessed December 2005.
- MDL/CrossFire. http://www.mimas.ac.uk/crossfire/autonom.html accessed December 2005.
- ACD/ChemSketch 8.17. http://www.acdlabs.com/products/chem_dsn_lab/chemsketch/ accessed December 2005.
- http://www.cheminnovation.com/products/nameexpert.asp accessed March 2006.
- Smith, E.G. Wisswesser-Line Formula Chemical Notation; McGraw-Hill: New York, 1968. [Google Scholar]
- Ash, S.; Cline, M.A.; Homer, R.W.; Hurst, T.; Smith, G.B. SYBYL line notation (SLN): A versatile language for chemical structure representation. J. Chem. Inf. Comput. Sci. 1997, 37, 71–79. [Google Scholar] [CrossRef]
- Weininger, D. SMILES a Chemical Language and Information System. 1. Introduction to Methodology and Encoding Rules. J. Chem. Inf. Comput. Sci. 1988, 28, 31–36. [Google Scholar] [CrossRef]
- Stein, S.E.; Heller, S.R.; Tchekhovskoi, D. An Open Standard for Chemical Structure Representation: The IUPAC Chemical Identifier. In Proceedings of the 2003 International Chemical Information Conference (Nimes), Infonortics, 2003; pp. 131–143.
- http://cactus.nci.nih.gov/services/translate/trans_info.html accessed December 2005.
- Weininger, D.; Weininger, A.; Weininger, J.L. SMILES 2. Algorithm for Generation of Unique SMILES Notation. J. Chem. Inf. Comput. Sci. 1989, 29, 97–101. [Google Scholar] [CrossRef]
- Trepalin, S.V.; Yarkov, A.V.; Dolmatova, L.M.; Zefirov, N.S.; Finch, S.A.E. WinDat: An NMR Database Compilation Tool, User Interface and Spectrum Libraries for Personal Computers. J. Chem. Inf. Comput. Sci. 1995, 35, 405–411. [Google Scholar] [CrossRef]
- Rovner, S.L.; Washington, C. Chemical ‘Naming’ method unveiled. Chem. Eng. News 2005, 83, 39–40. [Google Scholar]
- http://www.iupac.org/inchi accessed December 2005.
- Prasanna, M. D.; Vondrasek, J.; Wlodawer, A.; Bhat, T.N. Application of InChI to Curate, Index, and Query 3-D Structures. Proteins 2005, 60, 1–4. [Google Scholar]
- Gakh, A.A.; Burnett, M.N. Modular Chemical Descriptor Language (MCDL): Composition, Connectivity, and Supplementary Modules. J. Chem. Inf. Comput. Sci. 2001, 41, 1494–1499. [Google Scholar] [CrossRef] [PubMed]
- Rzepa, H.; Tonge, A. VchemLab: A Virtual Chemistry Laboratory. The Storage, Retrieval, and a Display of Chemical Information Using Standard Internet Tools. J. Chem. Inf. Comput. Sci. 1998, 38, 1048–1053. [Google Scholar]
- Csizmadia, F.J. Chem: Java Applets and Modules Supporting Chemical Database Handling from Web Browsers. J. Chem. Inf. Comput. Sci. 2000, 40, 323–324. [Google Scholar] [CrossRef] [PubMed]
- Ertl, P.; Jacob, O. WWW-based chemical information system. THEOCHEM 1997, 419, 113–120. [Google Scholar] [CrossRef]
- Krause, S.; Willighagen, E.; Steinbeck, C. JChemPaint-Using the Collaborative Forces of the Internet to Develop a Free Editor for 2D Chemical Structures. Molecules 2000, 5, 93–98. [Google Scholar]
- Steinbeck, C.; Han, Y.; Kuhn, S.; Horlacher, O.; Luttman, E.; Willighagen, E. The Chemistry Development Kit (CDK): An Open-Source Java Library for Chemo- and Bioinformatics. J. Chem. Inf. Comput. Sci. 2003, 43, 493–500. [Google Scholar]
- http://www.chemaxon.com/products.html accessed March 2006.
- Dalby, A.; Hourse, J.G.; Hounshell, W.D.; Gurchurst, A.K.I.; Grier, D.L.; Leland, B.A.; Laufer, J. Description of several chemical structure file formats used by computer program developed at Molecular Design Limited. J. Chem. Inf. Comput. Sci. 1992, 32, 244–255. [Google Scholar] [CrossRef]
- Bremser, W. HOSE—a novel substructure code. Anal. Chim. Acta 1978, 103, 355–365. [Google Scholar] [CrossRef]
- JChemPaint applet demo. http://jchempaint.sourceforge.net/applet accessed December 2005.
- Sun Java Run Time Enviroment. http://java.sun.com/j2se/downloads.html accessed December 2005.
- http://directory.google.com/Top/Computers/Programming/Languages/Java/Development_Tools/Obfuscators/ accessed December 2005.
- http://java.sun.com/j2se/1.3/docs/guide/extensions/spec.html accessed December 2005.
- Mayer, I. Charge, bond order and valence in the abinitio SCF theory. Chem. Phys. Lett. 1983, 97, 270–274. [Google Scholar] [CrossRef]
- Mayer, I. Comments on the quantum-theory of valence and bonding - choosing between alternative definitions. Chem. Phys. Lett. 1984, 110, 440–444. [Google Scholar] [CrossRef]
- Cioslowski, J.; Mixon, S.T. Covalent Bond Orders in the Topological Theory of Atoms in Molecules. J. Am. Chem. Soc. 1991, 113, 4142–4145. [Google Scholar] [CrossRef]
- Baber, J.C.; Hodgkin, E.E. Automatic assignment of chemical connectivity to organic molecules in the Cambridge Structural Database. J. Chem. Inf. Comput. Sci. 1992, 32, 401–406. [Google Scholar] [CrossRef]
- Helson, H.E. Structure Diagram Generation. Rev. Comput. Chem. 1999, 13, 313–398. [Google Scholar]
- Fricker, P.C.; Gastreich, M.; Rarey, M. Automated Drawing of Structural Molecular Formulas under Constraints. J. Chem. Inf. Comput. Sci. 2004, 44, 1065–1078. [Google Scholar] [CrossRef]
- MDL™ ISIS Draw 2.5. http://www.mdl.com/products/framework/isis_draw/index.jsp accessed: December 2005.
- Cambridge ChemDraw Ultra 8.0. http://www.cambridgesoft.com/products/family.cfm?FID=2 accessed December 2005.
- http://cdk.sf.net/api/org/openscience/cdk/layout/TemplateHandler.html and http://cdk.sf.net/api/org/openscience/cdk/layout/StructureDiagramGenerator.html accessed December 2005.
- Figueras, J. Ring Perception Using Breadth-First Search. J. Chem. Inf. Comput. Sci. 1996, 36, 986–991. [Google Scholar] [CrossRef]
- MCDL applet. http://www.zelinsky.ru/mcdl/mcdl.html.
- Source codes. https://sourceforge.net/projects/mcdl.
- Schnur, D. Design and Diversity Analysis of Large Combinatorial Libraries Using Cell-Based Methods. J. Chem. Inf. Comput. Sci. 1999, 39, 36–45. [Google Scholar]
- Bajorath, J. Selected Concepts and Investigations in Compound Classification, Molecular Descriptor Analysis, and Virtual Screening. J. Chem. Inf. Comput. Sci. 2001, 41, 233–245. [Google Scholar]
© 2006 by MDPI (http://www.mdpi.org). Reproduction is permitted for noncommercial purposes.