An Unsupervised Learning-Based Multi-Organ Registration Method for 3D Abdominal CT Images
Abstract
:1. Introduction
2. Methods
2.1. Optimization Problem Formulation
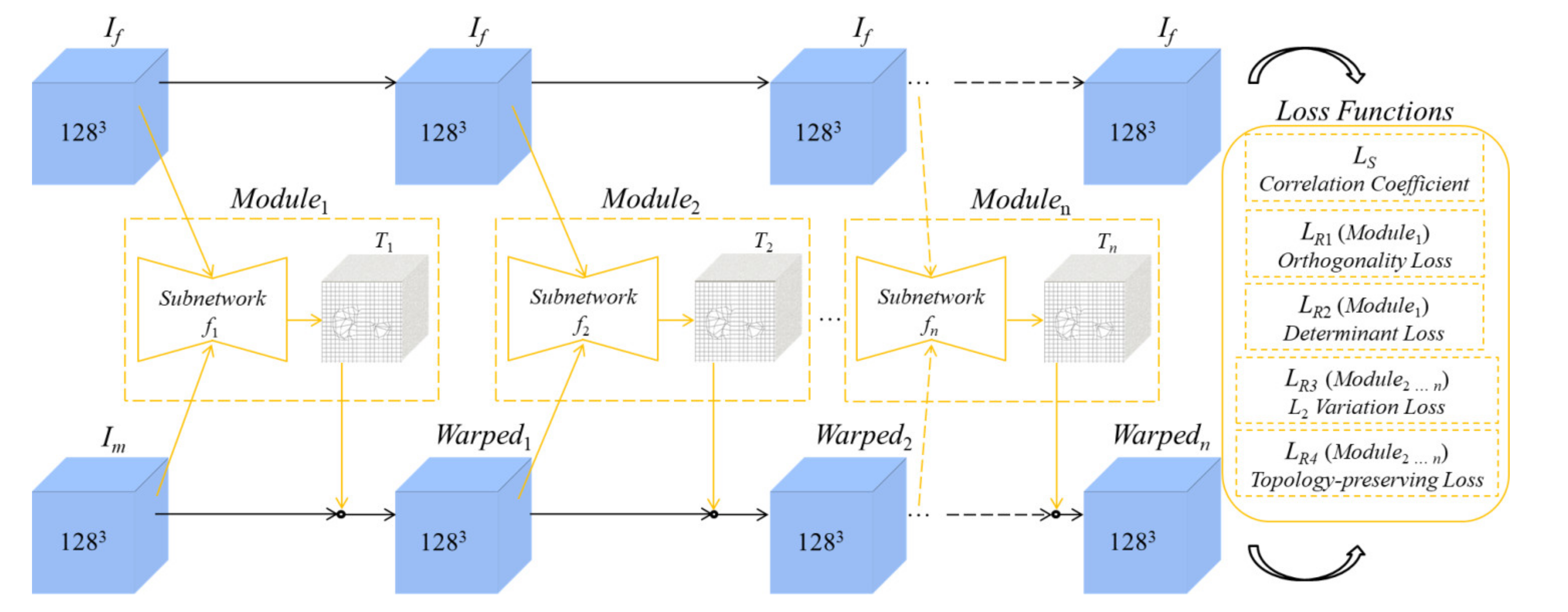
2.2. Architecture of the Unsupervised Learning-Based Networks
2.2.1. Coarse Registration
2.2.2. Fine Registration
2.3. Loss Functions
2.3.1. Similarity Loss
2.3.2. Regularization Terms for the Coarse-Subnetwork
2.3.3. Regularization Terms for the Fine-Subnetworks
3. Results and Discussion
3.1. Database
3.2. Evaluation Indexes
3.3. Experimental Analysis
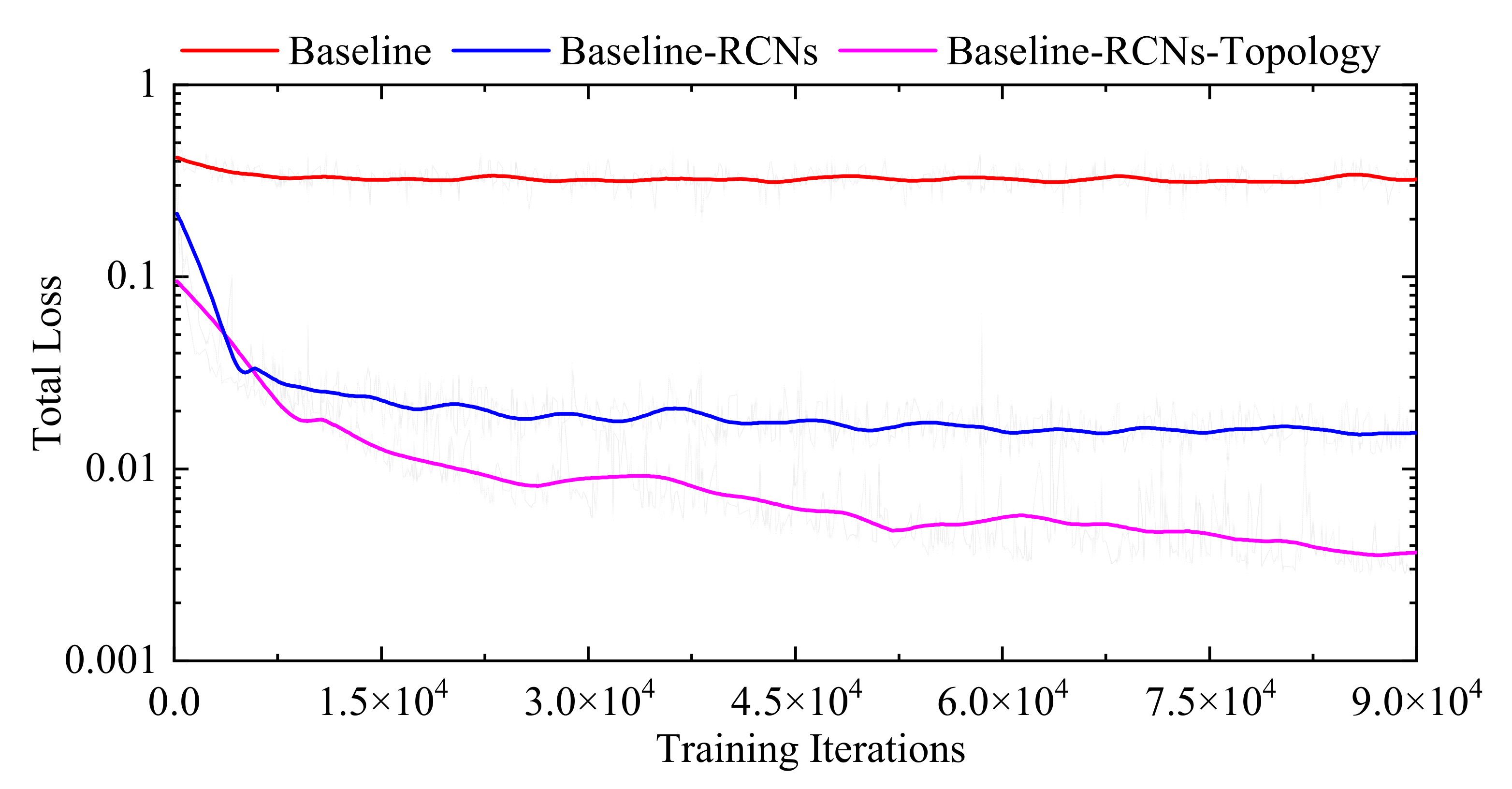
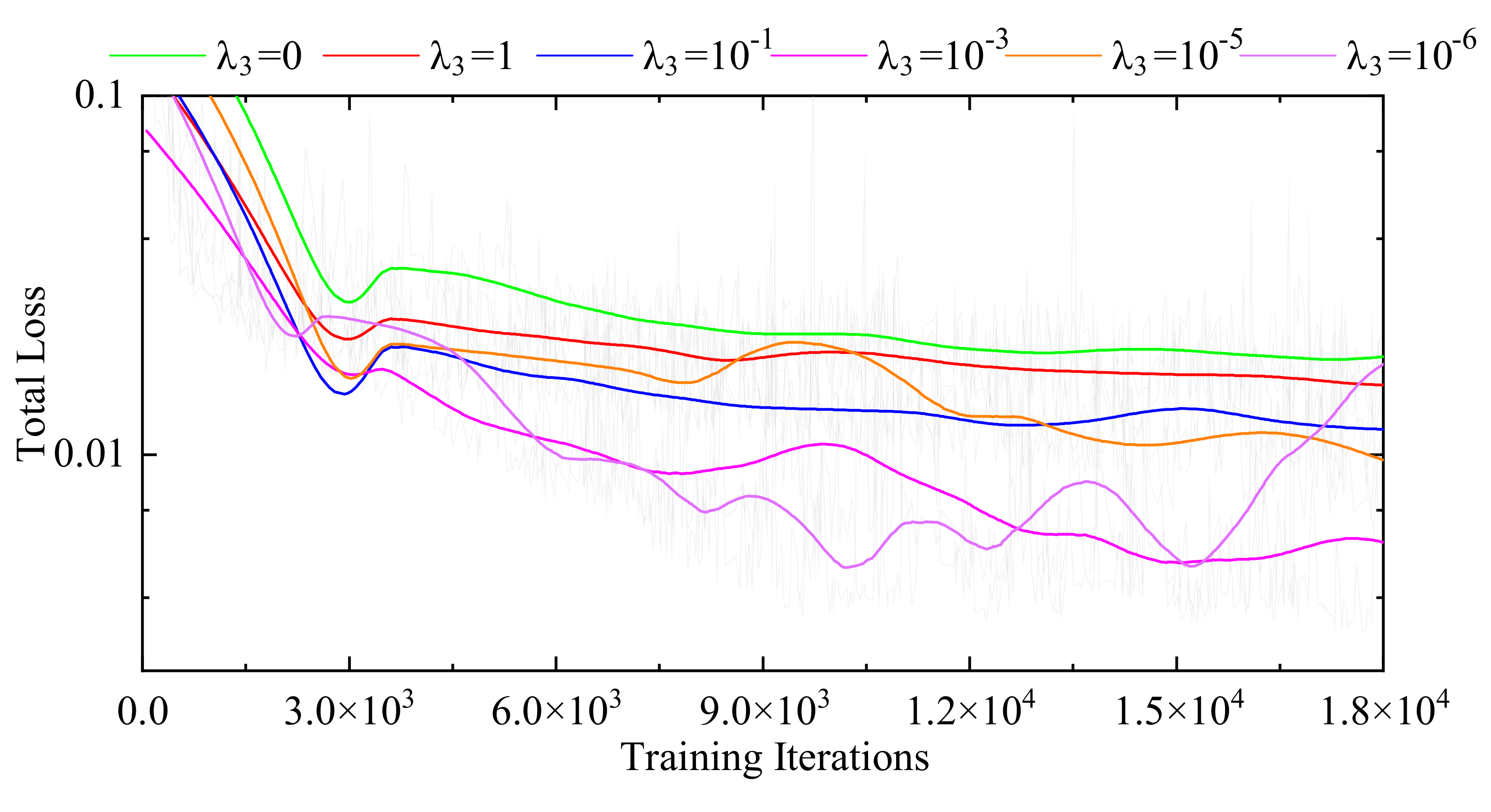
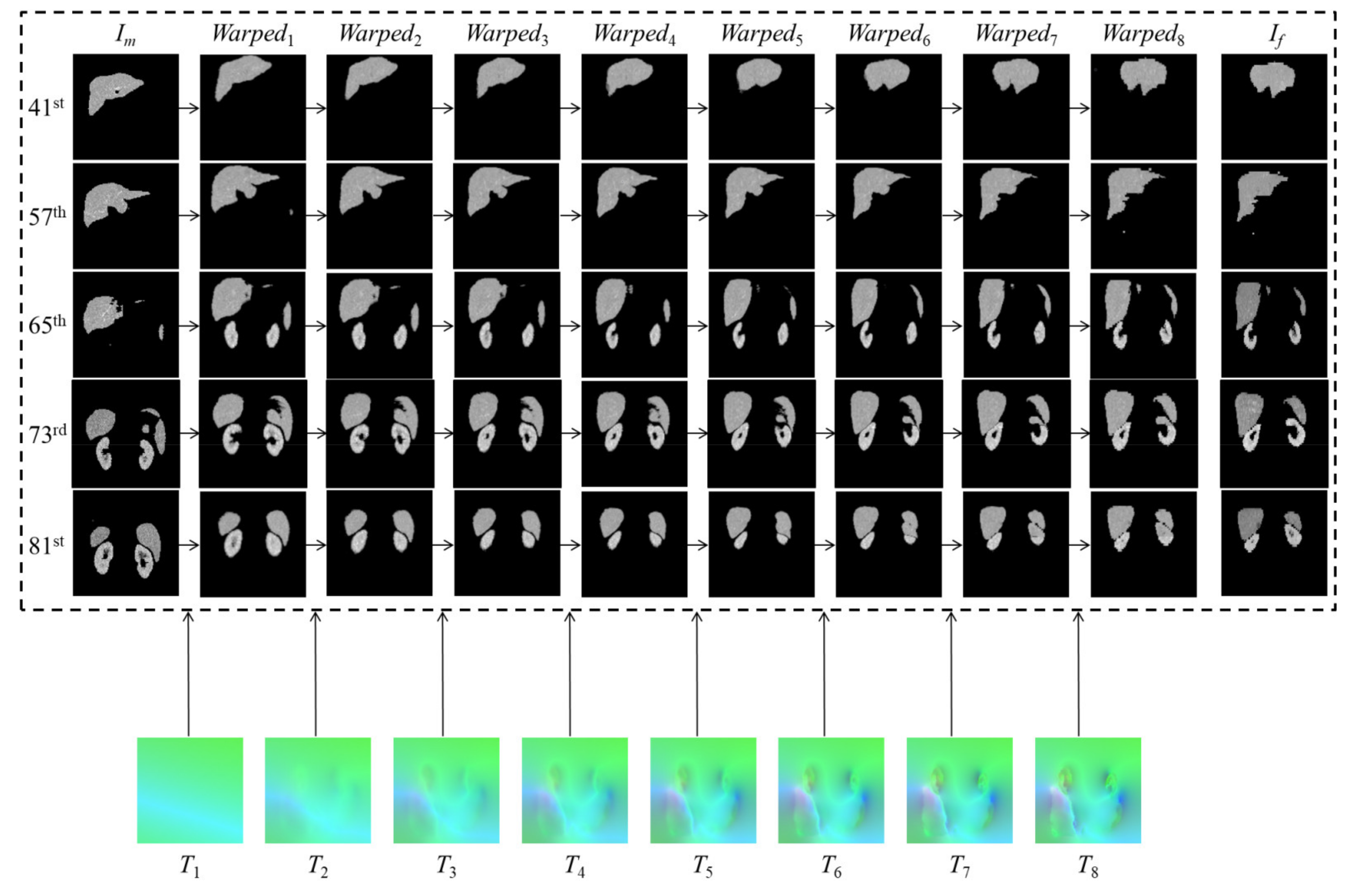
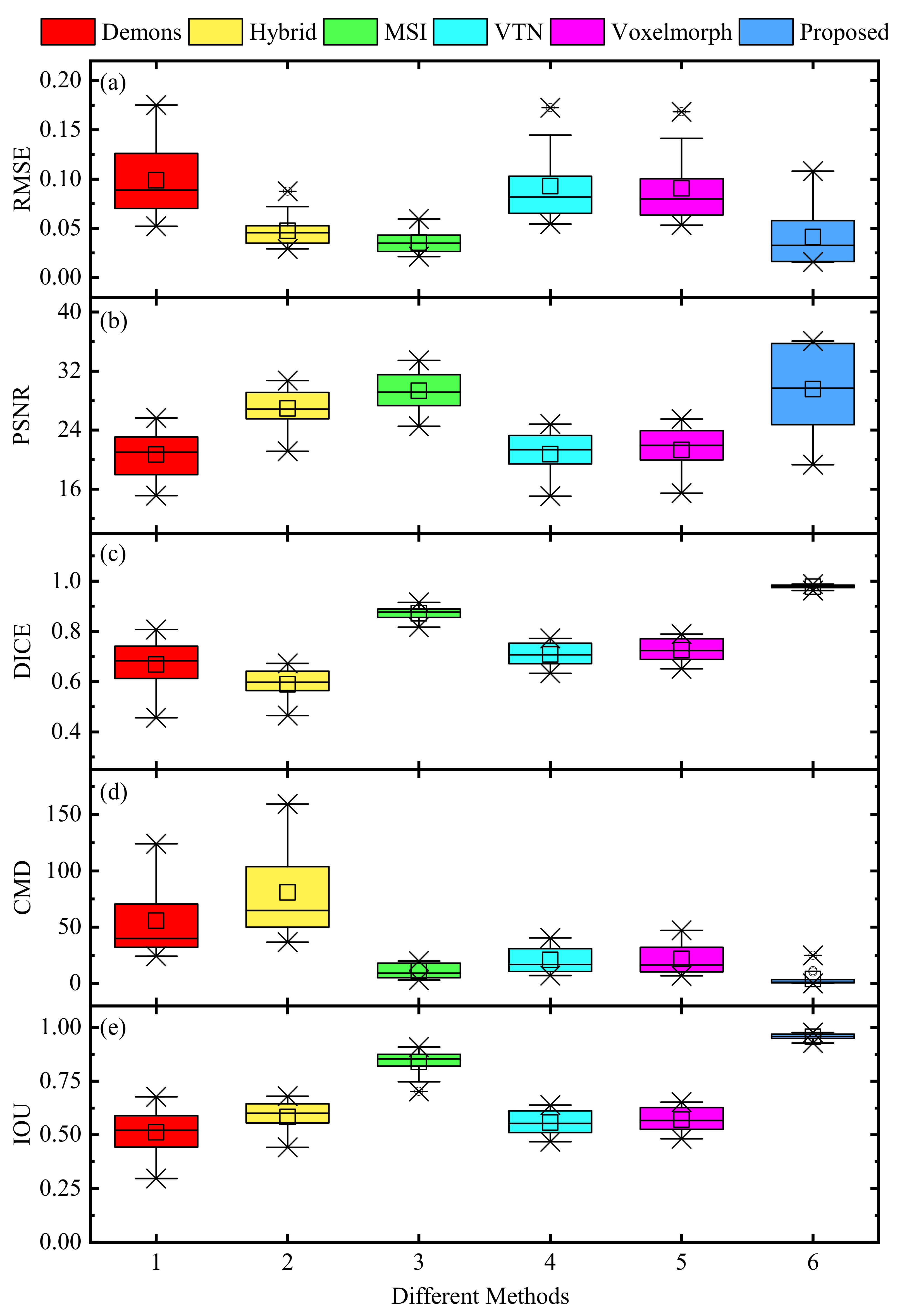
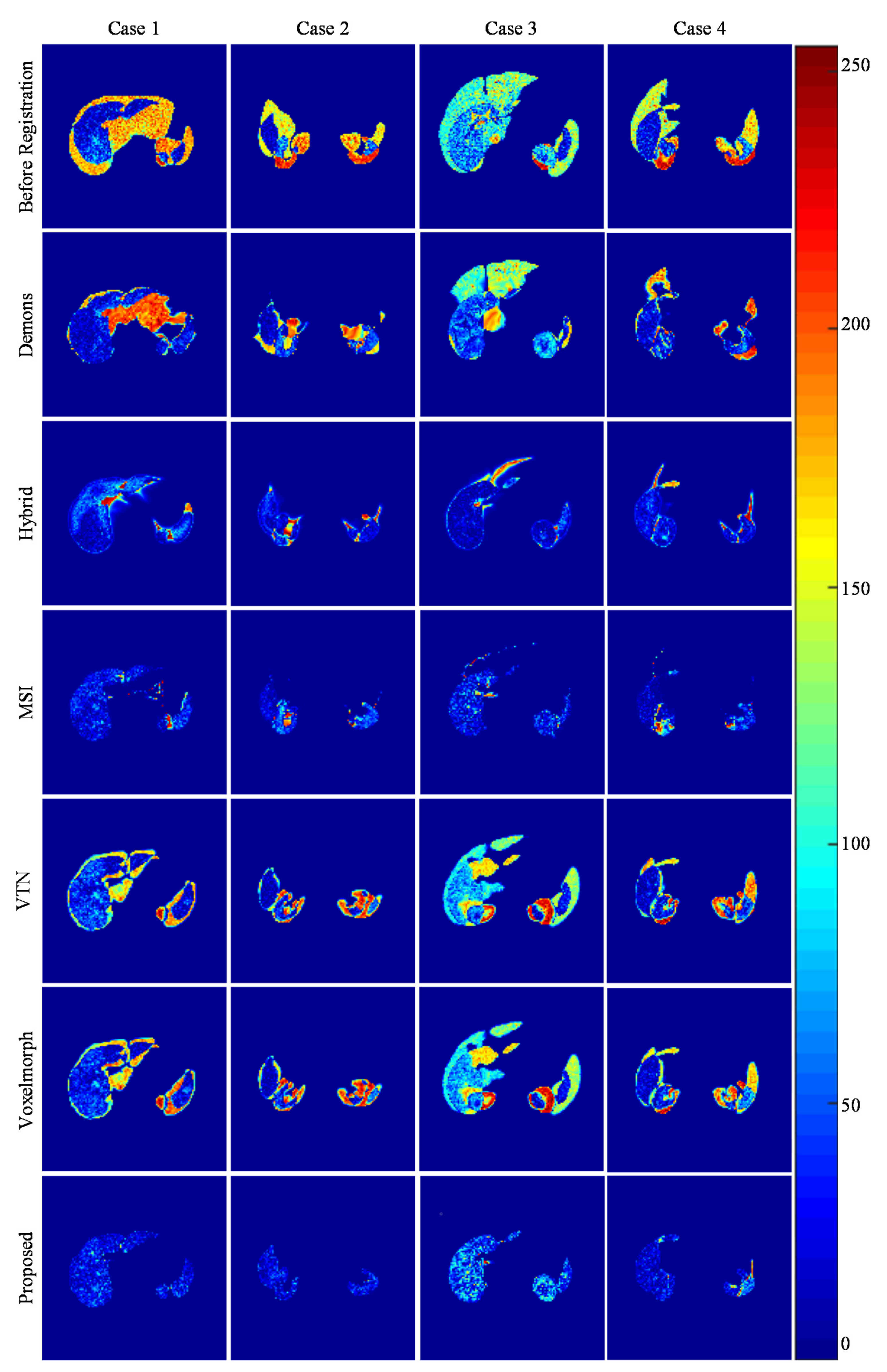
3.3.1. Internal Comparisons
3.3.2. External Comparisons
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bielecki, Z.; Stacewicz, T.; Wojtas, J.; Mikołajczyk, J.; Szabra, D.; Prokopiuk, A. Selected optoelectronic sensors in medical applications. Opto-Electron. Rev. 2018, 26, 122–133. [Google Scholar] [CrossRef]
- David, D.D.S.; Parthiban, R.; Jayakumar, D.; Usharani, S.; RaghuRaman, D.; Saravanan, D.; Palani, U. Medical Wireless Sensor Network Coverage and Clinical Application of Mri Liver Disease Diagnosis. Eur. J. Mol. Clin. Med. 2021, 7, 2559–2571. [Google Scholar]
- Gao, L.; Zhang, G.; Yu, B.; Qiao, Z.; Wang, J. Wearable human motion posture capture and medical health monitoring based on wireless sensor networks. Measurement 2020, 166, 108252. [Google Scholar] [CrossRef]
- Luo, X.; He, X.; Shi, C.; Zeng, H.-Q.; Ewurum, H.C.; Wan, Y.; Guo, Y.; Pagnha, S.; Zhang, X.-B.; Du, Y.-P. Evolutionarily Optimized Electromagnetic Sensor Measurements for Robust Surgical Navigation. IEEE Sens. J. 2019, 19, 10859–10868. [Google Scholar] [CrossRef]
- Kok, E.N.D.; Eppenga, R.; Kuhlmann, K.F.D.; Groen, H.C.; Van Veen, R.; Van Dieren, J.M.; De Wijkerslooth, T.R.; Van Leerdam, M.; Lambregts, D.M.J.; Heerink, W.J.; et al. Accurate surgical navigation with real-time tumor tracking in cancer surgery. NPJ Precis. Oncol. 2020, 4, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Ahn, S.J.; Lee, J.M.; Lee, D.H.; Lee, S.M.; Yoon, J.-H.; Kim, Y.J.; Yu, S.J.; Han, J.K. Real-time US-CT/MR fusion imaging for percutaneous radiofrequency ablation of hepatocellular carcinoma. J. Hepatol. 2017, 66, 347–354. [Google Scholar] [CrossRef]
- Li, K.; Su, Z.; Xu, E.; Huang, Q.; Zeng, Q.; Zheng, R. Evaluation of the ablation margin of hepatocellular carcinoma using CEUS-CT/MR image fusion in a phantom model and in patients. BMC Cancer 2017, 17, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Radu, C.; Fisher, P.; Mitrea, D.; Birlescu, I.; Marita, T.; Vancea, F.; Florian, V.; Tefas, C.; Badea, R.; Ștefănescu, H.; et al. Integration of Real-Time Image Fusion in the Robotic-Assisted Treatment of Hepatocellular Carcinoma. Biology 2020, 9, 397. [Google Scholar] [CrossRef]
- Li, D.; Zhong, W.; Deh, K.M.; Nguyen, T.D.; Prince, M.R.; Wang, Y.; Spincemaille, P. Discontinuity Preserving Liver MR Registration with Three-Dimensional Active Contour Motion Segmentation. IEEE Trans. Biomed. Eng. 2018, 66, 1884–1897. [Google Scholar] [CrossRef]
- Xie, Y.; Chao, M.; Xing, L. Tissue Feature-Based and Segmented Deformable Image Registration for Improved Modeling of Shear Movement of Lungs. Int. J. Radiat. Oncol. Biol. Phys. 2009, 74, 1256–1265. [Google Scholar] [CrossRef] [Green Version]
- Fu, Y.; Lei, Y.; Wang, T.; Curran, W.J.; Liu, T.; Yang, X. Deep learning in medical image registration: A review. Phys. Med. Biol. 2020, 65, 20TR01. [Google Scholar] [CrossRef] [Green Version]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [Green Version]
- Nazib, A.; Fookes, C.; Perrin, D. A comparative analysis of registration tools: Traditional vs deep learning approach on high resolution tissue cleared data. arXiv 2018, arXiv:1810.08315. preprint. [Google Scholar]
- Villena-Martinez, V.; Oprea, S.; Saval-Calvo, M.; Azorin-Lopez, J.; Fuster-Guillo, A.; Fisher, R.B. When Deep Learning Meets Data Alignment: A Review on Deep Registration Networks (DRNs). Appl. Sci. 2020, 10, 7524. [Google Scholar] [CrossRef]
- Thirion, J.P. Image matching as diffusion process: An analogy with Maxwell’s demons. Med. Image Anal. 1998, 2, 243–260. [Google Scholar] [CrossRef] [Green Version]
- Klein, S.; Staring, M.; Murphy, K.; Viergever, M.A.; Pluim, J.P.W. elastix: A Toolbox for Intensity-Based Medical Image Registration. IEEE Trans. Med. Imaging 2009, 29, 196–205. [Google Scholar] [CrossRef] [PubMed]
- Modat, M.; Ridgway, G.; Taylor, Z.; Lehmann, M.; Barnes, J.; Hawkes, D.J.; Fox, N.; Ourselin, S. Fast free-form deformation using graphics processing units. Comput. Methods Programs Biomed. 2010, 98, 278–284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, X.; Yang, J.; Zhang, J.; Nie, D.; Kim, M.-J.; Wang, Q.; Shen, D. Deformable image registration based on similarity-steered CNN regression. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Quebec City, QC, Canada, 10–14 September 2017; Springer: Cham, Switzerland, 2017; pp. 300–308. [Google Scholar]
- Ferrante, E.; Oktay, O.; Glocker, B.; Milone, D.H. On the adaptability of unsupervised CNN-based deformable image registration to unseen image domains. In Proceedings of the International Workshop on Machine Learning in Medical Imaging, Granada, Spain, 16 September 2018; Springer: Cham, Switzerland, 2018; pp. 294–302. [Google Scholar]
- Blendowski, M.; Hansen, L.; Heinrich, M.P. Weakly-supervised learning of multi-modal features for regularised iterative descent in 3D image registration. Med. Image Anal. 2021, 67, 101822. [Google Scholar] [CrossRef]
- Xu, Z.; Niethammer, M. DeepAtlas: Joint semi-supervised learning of image registration and segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Shenzhen, China, 13–17 October 2019. [Google Scholar]
- Lei, Y.; Fu, Y.; Wang, T.; Liu, Y.; Patel, P.; Curran, W.J.; Liu, T.; Yang, X. 4D-CT deformable image registration using multiscale unsupervised deep learning. Phys. Med. Biol. 2020, 65, 085003. [Google Scholar] [CrossRef]
- Heinrich, M.P.; Hansen, L. Highly Accurate and Memory Efficient Unsupervised Learning-Based Discrete CT Registration Using 2.5D Displacement Search. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Lima, Peru, 4–8 October 2020; Springer: Cham, Switzerland, 2020; pp. 190–200. [Google Scholar]
- Balakrishnan, G.; Zhao, A.; Sabuncu, M.R.; Guttag, J.; Dalca, A.V. VoxelMorph: A Learning Framework for Deformable Medical Image Registration. IEEE Trans. Med. Imaging 2019, 38, 1788–1800. [Google Scholar] [CrossRef] [Green Version]
- Zhao, S.; Lau, T.; Luo, J.; Chang, E.I.-C.; Xu, Y. Unsupervised 3D End-to-End Medical Image Registration with Volume Tweening Network. IEEE J. Biomed. Health Inform. 2020, 24, 1394–1404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, S.; Dong, Y.; Chang, E.; Xu, Y. Recursive cascaded networks for unsupervised medical image registration. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Seoul, Korea, 27 October–2 November 2019; pp. 10600–10610. [Google Scholar]
- Kuang, D.; Schmah, T. Faim—A convnet method for unsupervised 3d medical image registration. In Proceedings of the International Workshop on Machine Learning in Medical Imaging, Shenzhen, China, 13 October 2019; Springer: Cham, Switzerland, 2019; pp. 646–654. [Google Scholar]
- Mok, T.C.W.; Chung, A. Fast symmetric diffeomorphic image registration with convolutional neural networks. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 4644–4653. [Google Scholar]
- Ferrante, E.; Paragios, N. Slice-to-volume medical image registration: A survey. Med. Image Anal. 2017, 39, 101–123. [Google Scholar] [CrossRef] [Green Version]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; Springer: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Rudin, L.I.; Osher, S.; Fatemi, E. Nonlinear total variation based noise removal algorithms. Phys. D Nonlinear Phenom. 1992, 60, 259–268. [Google Scholar] [CrossRef]
- Xu, Z.; Lee, C.P.; Heinrich, M.P.; Modat, M.; Rueckert, D.; Ourselin, S.; Abramson, R.G.; Landman, B.A. Evaluation of Six Registration Methods for the Human Abdomen on Clinically Acquired CT. IEEE Trans. Biomed. Eng. 2016, 63, 1563–1572. Available online: https://competitions.codalab.org/competitions/17094 (accessed on 1 November 2018). [CrossRef] [PubMed] [Green Version]
- Bilic, P.; Christ, P.F.; Vorontsov, E.; Chlebus, G.; Chen, H.; Dou, Q.; Fu, C.-W.; Han, X.; Heng, P.-A.; Hesser, J.; et al. The Liver Tumor Segmentation Benchmark (LiTS). arXiv 2019, arXiv:1901.04056. preprint. [Google Scholar]
- Heimann, T.; Ginneken, B.V.; Styner, M.A. Segmentation of the Liver 2007(SLIVER07). Available online: http://sliver07.isi.uu.nl/ (accessed on 12 December 2018).
- Soler, L.; Hosttettle, A.; Charnoz, A.; Fasquel, J.; Moreau, J. 3D Image Reconstruction for Comparison of Algorithm Database: A Patient Specific Anatomical and Medical Image Database. Available online: https://www.ircad.fr/research/3dircadb/ (accessed on 16 April 2018).
- Sara, U.; Akter, M.; Uddin, M.S. Image quality assessment through FSIM, SSIM, MSE and PSNR—A comparative study. J. Comput. Commun. 2019, 7, 8–18. [Google Scholar] [CrossRef] [Green Version]
- Pei, H.-Y.; Yang, D.; Liu, G.-R.; Lu, T. MPS-Net: Multi-Point Supervised Network for CT Image Segmentation of COVID-19. IEEE Access 2021, 9, 47144–47153. [Google Scholar] [CrossRef]
- Lombaert, H.; Grady, L.; Pennec, X.; Ayache, N.; Cheriet, F. Spectral Log-Demons: Diffeomorphic Image Registration with Very Large Deformations. Int. J. Comput. Vis. 2014, 107, 254–271. [Google Scholar] [CrossRef] [Green Version]
- Chan, C.L.; Anitescu, C.; Zhang, Y.; Rabczuk, T. Two and Three Dimensional Image Registration Based on B-Spline Composition and Level Sets. Commun. Comput. Phys. 2017, 21, 600–622. [Google Scholar] [CrossRef]
- Aganj, I.; Iglesias, J.E.; Reuter, M.; Sabuncu, M.R.; Fischl, B. Mid-space-independent deformable image registration. NeuroImage 2017, 152, 158–170. [Google Scholar] [CrossRef] [Green Version]

| Metric | Base-Net | Base-Net-1 | Base-Net-3 | Base-Net-5 | Base-Net-7 |
|---|---|---|---|---|---|
| RMSE | 0.0898 | 0.0516 | 0.0430 | 0.0419 | 0.0413 |
| PSNR | 21.3938 | 26.8639 | 28.9574 | 29.3259 | 29.5584 |
| SSIM | 0.9018 | 0.9572 | 0.9701 | 0.9720 | 0.9732 |
| DICE | 0.7305 | 0.9155 | 0.9621 | 0.9724 | 0.9775 |
| IOU | 0.5772 | 0.8451 | 0.9272 | 0.9445 | 0.9562 |
| SS | 0.8494 | 0.9757 | 0.9944 | 0.9974 | 0.9982 |
| SC | 0.6459 | 0.8631 | 0.9320 | 0.9468 | 0.9578 |
| HD (mm) | 17.8924 | 15.0090 | 13.1231 | 12.6245 | 12.2646 |
| CMD (mm) | 21.8383 | 7.0733 | 4.7972 | 4.3341 | 3.9296 |
| Metrics | Demons | Hybrid | MSI | VTN | Voxelmorph | Proposed |
|---|---|---|---|---|---|---|
| RMSE | 0.0987 | 0.0474 | 0.0355 | 0.0928 | 0.0906 | 0.0413 |
| PSNR | 20.6969 | 26.9121 | 29.3418 | 20.7456 | 21.3212 | 29.5584 |
| SSIM | 0.8925 | 0.9412 | 0.9592 | 0.8769 | 0.9009 | 0.9732 |
| DICE | 0.6678 | 0.5891 | 0.8715 | 0.7078 | 0.7253 | 0.9775 |
| IOU | 0.5113 | 0.5833 | 0.8386 | 0.5569 | 0.5709 | 0.9562 |
| SS | 0.8077 | 0.9189 | 0.9268 | 0.8256 | 0.8450 | 0.9982 |
| SC | 0.5836 | 0.6137 | 0.8997 | 0.6268 | 0.6407 | 0.9578 |
| HD (mm) | 27.7843 | 30.7378 | 25.7123 | 18.1536 | 17.7319 | 12.2646 |
| CMD (mm) | 55.6456 | 80.7641 | 10.7022 | 20.8790 | 21.8432 | 3.9296 |
| CPU (s) | 381.0294 | 510.1189 | - | 7.5612 | 8.6247 | 9.8922 |
| GPU (s) | - | - | 536.4378 | 1.3998 | 1.5212 | 1.6371 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, S.; Zhao, Y.; Liao, M.; Zhang, F. An Unsupervised Learning-Based Multi-Organ Registration Method for 3D Abdominal CT Images. Sensors 2021, 21, 6254. https://doi.org/10.3390/s21186254
Yang S, Zhao Y, Liao M, Zhang F. An Unsupervised Learning-Based Multi-Organ Registration Method for 3D Abdominal CT Images. Sensors. 2021; 21(18):6254. https://doi.org/10.3390/s21186254
Chicago/Turabian StyleYang, Shaodi, Yuqian Zhao, Miao Liao, and Fan Zhang. 2021. "An Unsupervised Learning-Based Multi-Organ Registration Method for 3D Abdominal CT Images" Sensors 21, no. 18: 6254. https://doi.org/10.3390/s21186254
APA StyleYang, S., Zhao, Y., Liao, M., & Zhang, F. (2021). An Unsupervised Learning-Based Multi-Organ Registration Method for 3D Abdominal CT Images. Sensors, 21(18), 6254. https://doi.org/10.3390/s21186254






