Characteristics of Food Protein-Derived Antidiabetic Bioactive Peptides: A Literature Update
Abstract
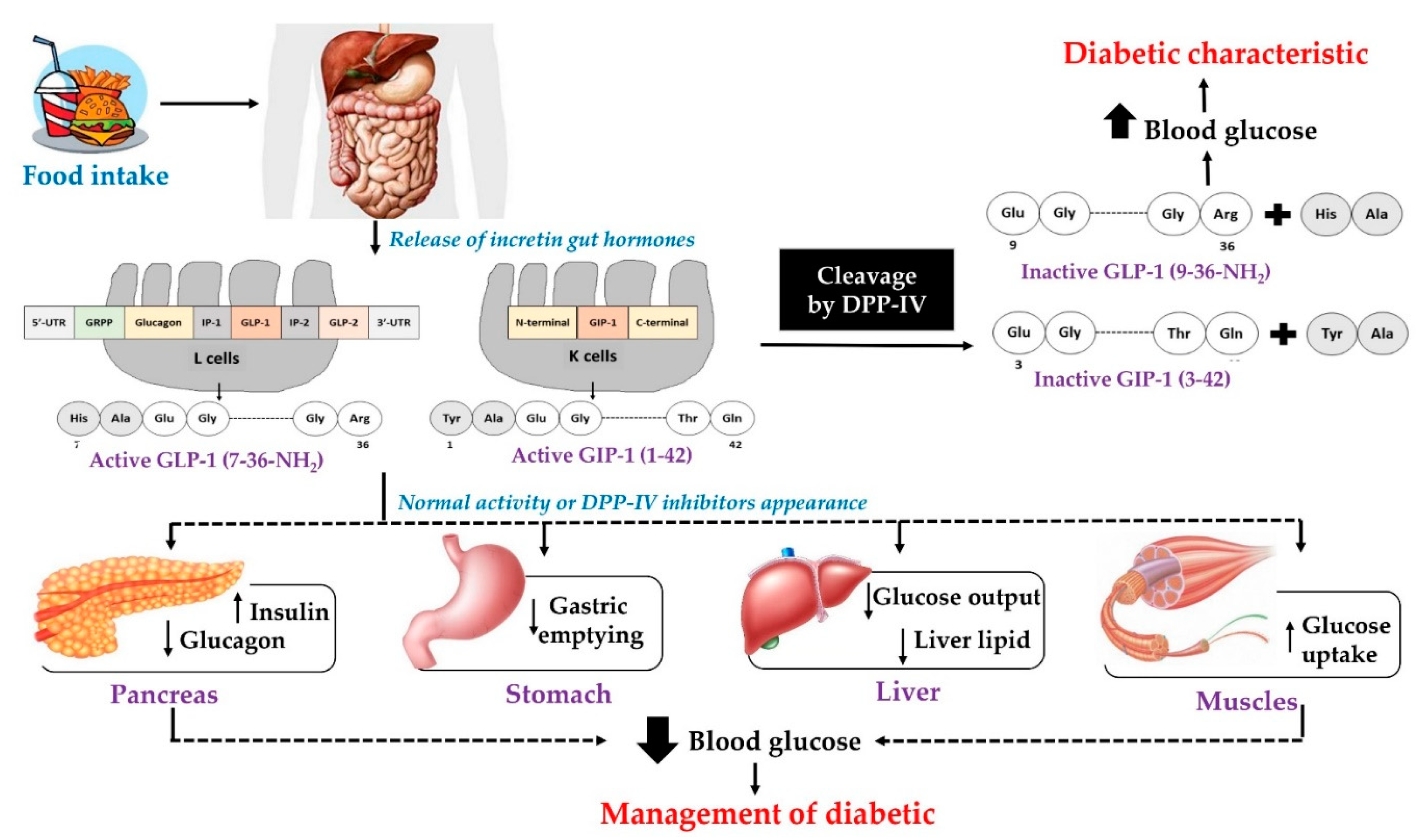
:1. Introduction
2. Dietary Proteins as Precursors of Anti-Diabetes Peptides
2.1. Dipeptidyl Peptidase—IV Inhibitors
2.2. PTP-1B Inhibitors
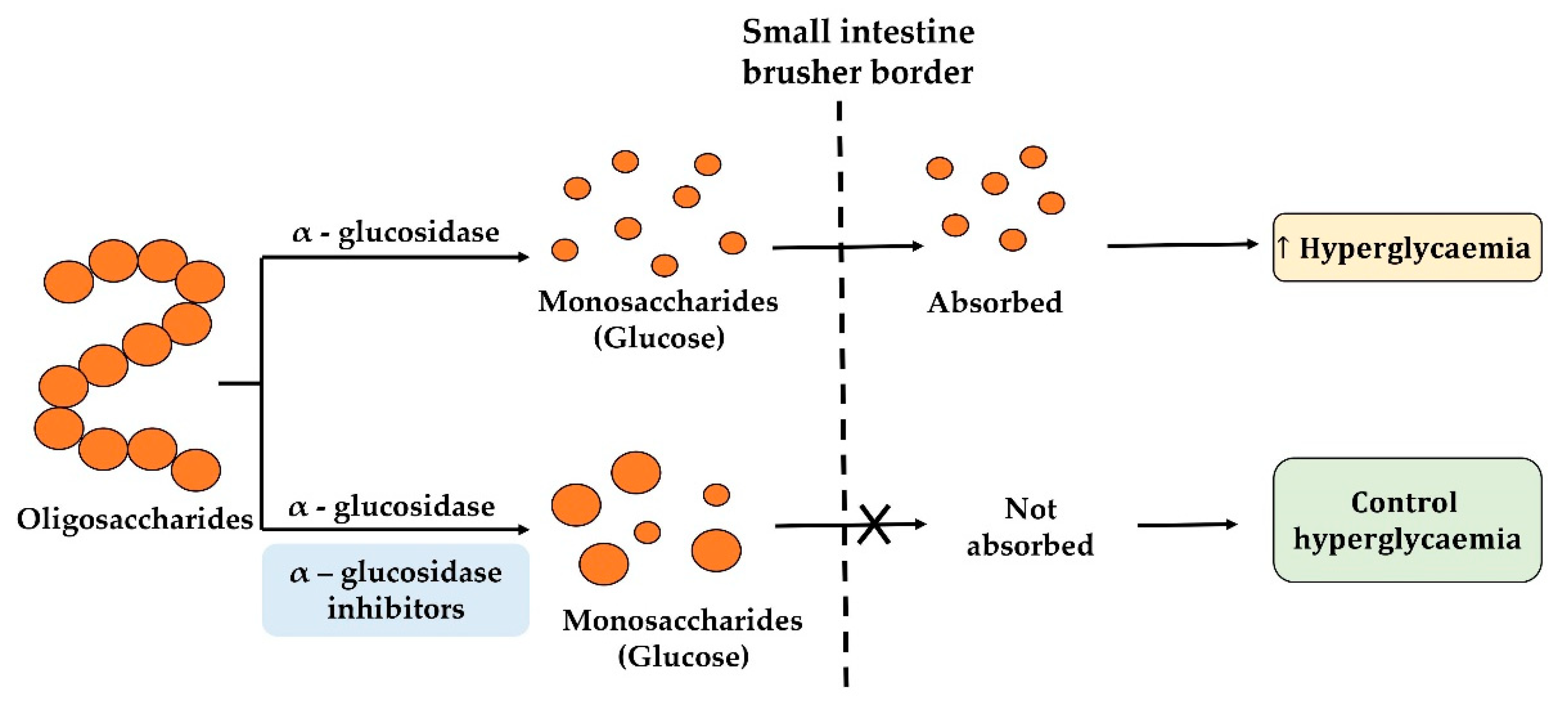
2.3. α-Glucosidase Inhibitory Peptides
2.4. Food Protein-Derived α-glucosidase Inhibition Peptides
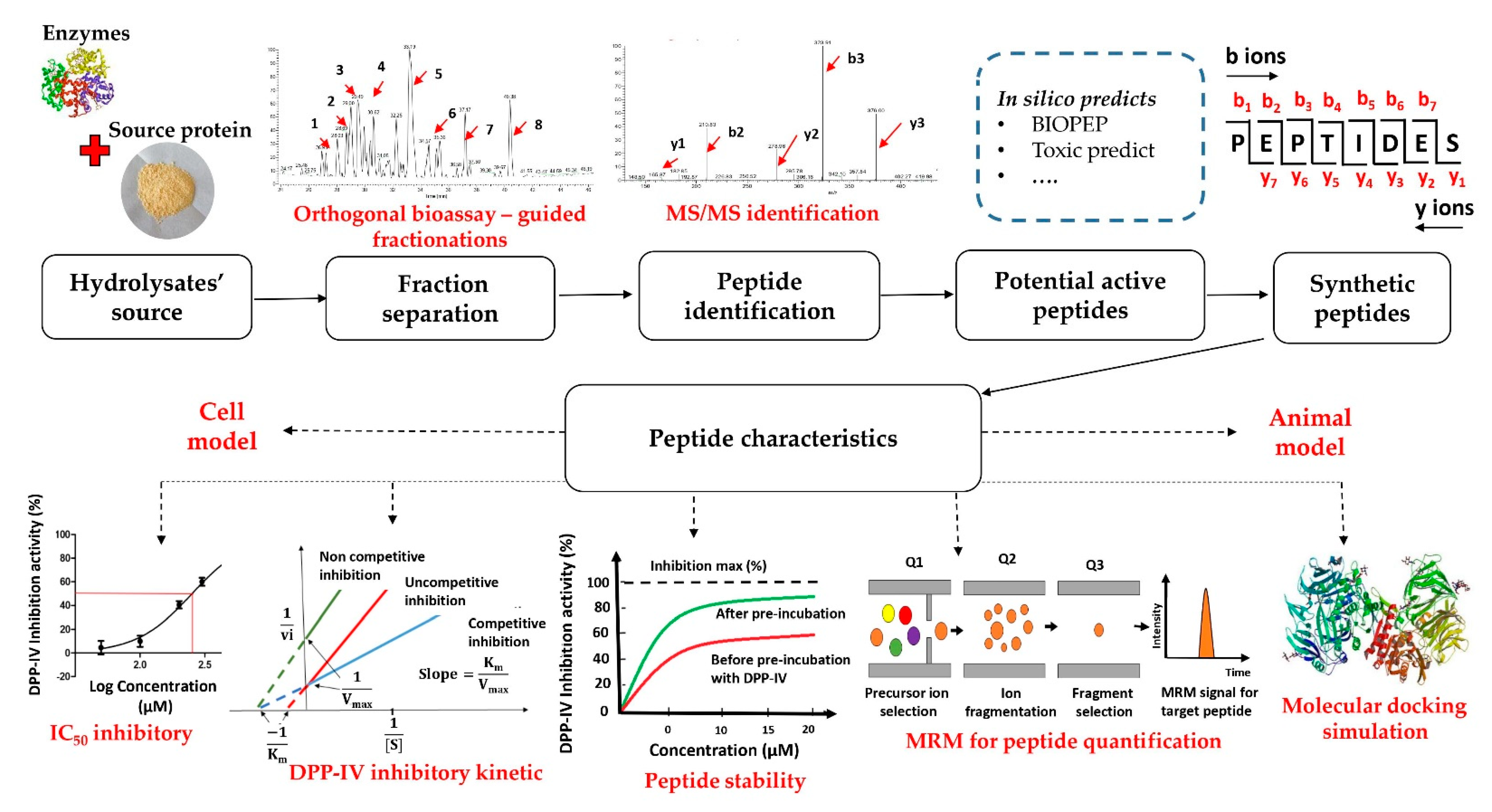
3. Common Acquisition Procedures for DPP-IV Inhibitory Peptides
3.1. The Strategy of the DPP-IV Inhibitory Peptide Discovery
3.2. Enzymatic Hydrolysates of Food Proteins
3.3. Bioassay-Guided Fractionation Methods
3.4. Peptide Sequence Identification Using Tandem Mass Spectrometry (MS/MS) Analysis Coupled with Database-Assisted Sequence Matching or de Novo Sequencing
3.5. In Silico Prediction of Potential DPP-IV Inhibitory Peptides
3.6. Characteristics of Inhibitory Peptides
3.6.1. Features and Structure–Function Activity of DPP-IV Inhibitory Peptides
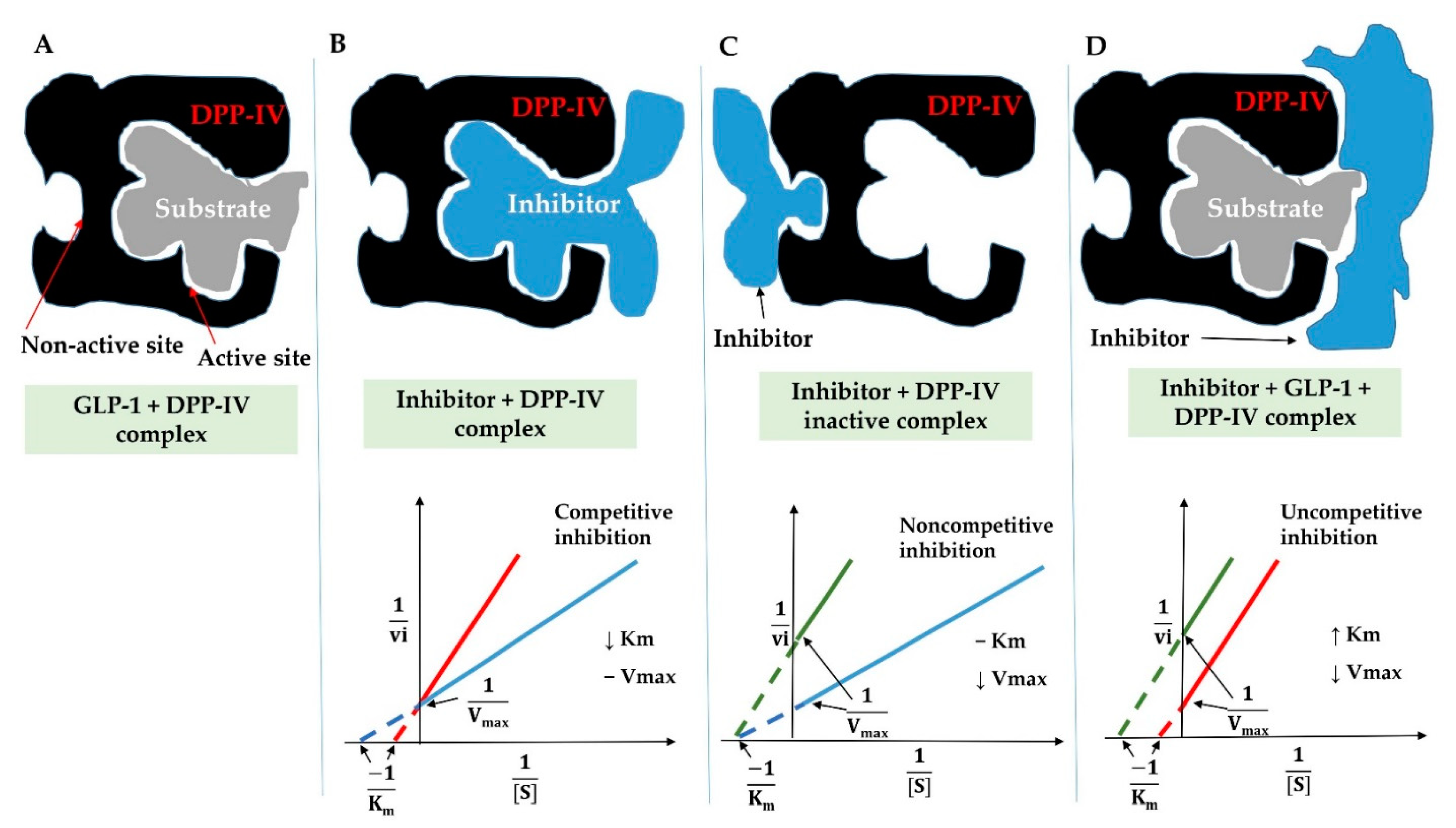
3.6.2. Inhibition Modes of DPP-IV Inhibitory Peptides
3.6.3. The Stability of the DPP-IV Inhibition Peptides
3.6.4. Quantification of Peptides
3.6.5. Molecular Docking
3.7. The Specific In Vitro Models of GLP-1 Secretion Studies
3.8. In Vivo Effect of Food-Derived DPP-IV Inhibitory Peptides
3.8.1. In Vivo Animal Studies
3.8.2. Clinical Studies
4. Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Source | Enzyme | Sequence | IC50 | Reference |
|---|---|---|---|---|
| Quinoa hydrolysate | Gastrointestinal enzyme | IQAEGGLT | 109.48 µM | [53] |
| Almond oil manufacture residue | Prote Ax and protease M | WH | 17.03 µM (17.03 μmol/L) | [37] |
| WS | 2.47 µM (24.71 μmol/L) | |||
| Rational in silico design | SVPA | 4.56 mM | [76] | |
| SEPA | 0.79 mM | |||
| Sprouted quinoa yoghurt | Lactobacillus casei | VAHPVF | 13.47 µM (9 ppm) | [77] |
| LAHMIVAGA | 12.37 µM (10.9 ppm) | |||
| KSFGSSNI | 22.07 µM (18.5 ppm) | |||
| KDLQL | 26.01 µM (16 ppm) | |||
| MIKLRSTAKN | 12.15 µM (14.1 ppm) | |||
| Soy protein | Alkaline proteinase | LLPLPVLK | 237.43 µM (237.43 μmol/L) | [75] |
| SWLRL | 182.05 µM (182.05 μmol/L) | |||
| WLRL | 162.29 µM (162.29 μmol/L) | |||
| Silkworm pupae | In silico digestion | QPGR | 65.6 µM (65.8 µmol/L) | [28] |
| SQSPA | 20 µM (20 µmol/L) | |||
| QPPT | 560 µM (560 µmol/L) | |||
| NSPR | 205 µM (205 µmol/L) | |||
| Dark tea protein | Trypsin | TAELLPR | 0.53 mM (0.43 mg/mL) | [31] |
| CGKKFVR | 0.62 mM (0.52 mg/mL) | |||
| AVPANLVDLNVPALLK | 0.62 mM (1.03 mg/mL) | |||
| VVDLVFFAAAK | 33.95 µM (0.04 mg/mL) | |||
| Dibble insects | Simulated intestinal digestion (pancreatin and bile extract) | IIAPPER | 28.79 µM (22.86 µg/mL) | [13] |
| LAPSTIK | 62.63 µM (45.60 µg/mL) | |||
| KVEGDLK | 23.34 µM (18.37 µg/mL) | |||
| NYVADGLG | 25.24 µM (20.37 µg/mL) | |||
| AAAPVAVAK | 13.71 µM (10.92 µg/mL) | |||
| AGDDAPR | 27.81 µM (19.47 µg/mL) | |||
| GKDAVIV | 22.77 µM (15.94 µg/mL) | |||
| AIGVGAIER | 14.75 µM (13.04 µg/mL) | |||
| FDPFPK | 7.94 µM (5.95 µg/mL) | |||
| Spanish dry-cured ham | AEEEYPDL | 5.58 mM | [34] | |
| LGVGG | 6.36 mM | |||
| GGLGP | 8.71 mM | |||
| EA | 17 mM | |||
| PP | 18.3 mM | |||
| VE | 22.17 mM | |||
| PE | 25.05 mM | |||
| AD | 25.66 mM |
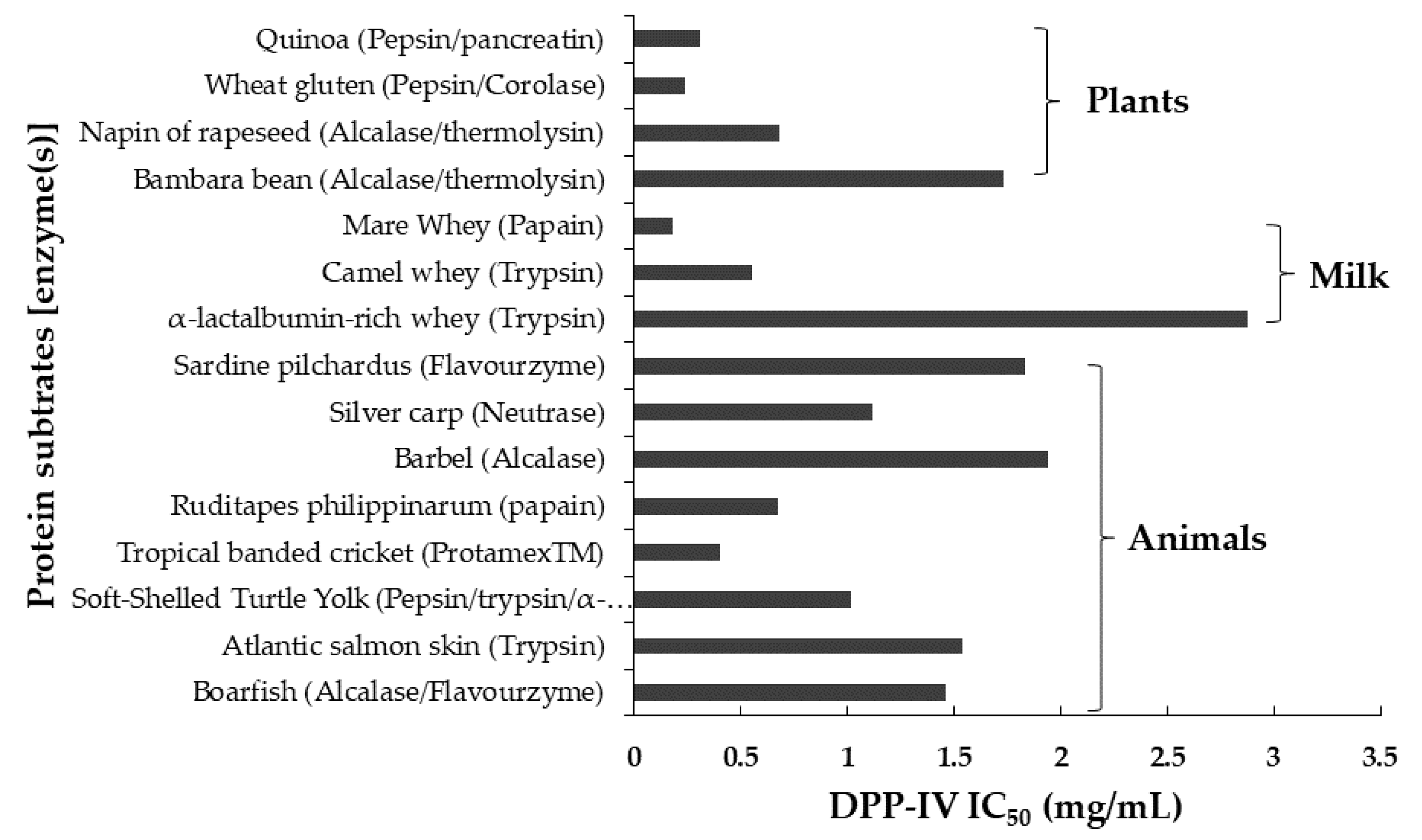
| Protein Source | Enzymatic Treatment | IC50 DPP-IV of Hydrolysate | Fraction Method | Materials | Reference |
|---|---|---|---|---|---|
| Bovine α-Lactalbumin hydrolysates | Alcalase | 1. Gel filtration chromatography 2. Reversed-phase high-performance liquid chromatography (RP-HPLC) | 1. Sephadex G-25 column (2.5 cm × 70 cm) 2. C18 RP-HPLC (4.6 mm × 250 mm, 5 µm) | [4] | |
| Millet protein hydrolysates | α-amylase and cellulase | 1. Gel filtration chromatography 2. RP-HPLC | 1. Sephadex G-25 column (2.5 cm × 70 cm) 2. C18 RP-HPLC (250 × 4.6 mm, 5 µm) | [38] | |
| Boarfish protein hydrolysate | Alcalase 2.4L and flavourzyme 500L | 1.46 ± 0.06 mg/mL | Semi-preparative reverse phase-high performance liquid chromatography (SP-RP-HPLC) | C18 semi-preparative column (250 × 15 mm I.D., 10 µM) | [36] |
| α-lactalbumin-rich whey protein | Trypsin | 2.88 ± 0.150 mg/mL | Gel filtration chromatography | Superdex peptide 10/300 GL | [35] |
| Atlantic salmon (Salmo salar) skin | Trypsin | 1.54 ± 0.06 mg/mL | 1. Gel filtration chromatography 2. SP-RP-HPLC | 1. Sephadex G–25 gel column (1.6 cm × 60 cm) 2. C18 column (250 × 4.6 mm, 5 µm) | [3] |
| A Porphyra dioica protein | Alcalase and flavourzyme | SP-RP-HPLC | C18 semi-preparative column (250 × 15 mm I.D., 10 mm) | [97] | |
| Soft-shelled turtle yolk hydrolysate | Pepsin, trypsin, and α-chymotrypsin | 1.017 ± 0.0356 mg/mL | 1. Ultrafiltration 2. RP-HPLC 3. Strong cation exchange (SCX) chromatography | 1. <3 kDa MWCO 2. C18 column (10 mm × 250 mm; 5 µm) | [2] |
| Dark tea protein | Trypsin | Ultrafiltration | [31] | ||
| Brewers’ spent grain | Pepsin | 1. Gel permeation-high performance liquid chromatography (GP-HPLC) 2. Reversed phase (RP-)-ultra-high performance liquid chromatography (UPLC) | 1. TSK G2000 SW separating column (600 × 7.5 mm ID) connected to a TSKGEL SW guard column (75 × 7.5 mm) 2. C18 column (2.1 mm × 50 mm × 1.7 µm) | [52] | |
| Camel whey protein | Trypsin | Between 0.55 ± 0.05 and 1.52 ± 0.16mg/L | 1. UPLC 2. GP-HPLC | 1. C18 column (2.1 mm × 50 mm × 1.7 µm) 2. TSK G2000 SW separating column (600 × 7.5 mm) connected to a TSKGEL SW guard column (75 × 7.5 mm) | [8] |
| Bambara bean protein hydrolysates | Alcalase and thermolysin | 1.73 mg/mL | 1. RP-HPLC 2. Gel permeation chromatography with UV/fluorescence detection | [91] | |
| Camel milk protein hydrolysate | Alcalase, bromelain, and papain | RP-HPLC | C4 column (3.4 μm HPLC Column) | [49] | |
| Silver carp (Hypophthalmichthys molitrix) muscle hydrolysate | Neutrase | 1. Ultrafiltration 2. Gel filtration chromatography 3. SP-RP-HPLC | 1. <3 kDa MWCO 2. Sephadex column (0.6 × 100 cm) 3. C18 column (4.6 × 250 mm) | [96] | |
| Egg white | Simulated digestion (pepsin and trypsin) | RP-HPLC | C18 column (250 × 4.6 mm, 5 µm) | [41] | |
| Napin of rapeseed (Brassica napus) | Alcalase and trypsin | 0.68 mg/mL | 1. Ultrafiltration 2. High-performance size-exclusion chromatography (SEC-HPLC) | 1. <1 kDa MWCO 2. TSK gel 2000 SWXL column (300 × 7.8 mm) | [80] |
| Tropical banded cricket (Gryllodes sigillatus) proteins | ProtamexTM | Between 0.40 ± 0.03 and 1.01 ± 0.07 mg/mL | 1. Ultrafiltration 2. Gel permeation high performance liquid chromatography (GP-HPLC) | 1. <1 kDa MWCO 2. TSK G2000 SW separating column (600 × 7.5 mm) connected to a TSKGEL SW guard column (75 × 7.5 mm) | [9] |
| Water-soluble extracts from natto | 5.35 ± 0.27 mg/mL | 1. Ultrafiltration 2. Gel filtration high-performance liquid chromatography (HPLC) | 1. <3 kDa MWCO 2. Superdex peptide 10/300 GL | [94] | |
| Wheat gluten hydrolysate | Pepsin and corolase PP | Between 0.24 ± 0.02 and 0.66 ± 0.06 mg/mL | 1. Ultrafiltration 2. GP-HPLC 3. RP_UHPLC | 1. <1 kDa MWCO 2. TSK G2000 SW separating column (600 × 7.5 mm) connected to a TSKGEL SW guard column (75 × 7.5 mm) 3. C18 column (2.1 mm × 50 mm × 1.7 µm) | [81] |
| Bighead carp (hypophthalmichthys nobilis) muscle hydrolysate | Pepsin | 1. Ultrafiltration 2. Gel filtration chromatography 3. SP-RP-HPLC | 1. <3 kDa MWCO 2. Sephadex G-15 column (0.6 × 100 cm) 3. C18 column (4.6 × 250 mm) | [90] | |
| Ruditapes philippinarum hydrolysate | Papain | 672.12 µg/mL | 1. Ethanol precipitation 2. RP-HPLC | 1. 2. C18 column (5 µm, 19 × 150 mm) | [89] |
| Barbel protein hydrolysate | Alcalase® | 1.94 mg/mL | 1. Size exclusion chromatography (SEC) 2. RP-HPLC 3. RP-HPLC | 1. Superdex peptide HR 10/30 2. TRACER-Excel 1200 3. C18 column (2.1 × 150 mm, 2.7 m) | [92] |
| Silver carp protein (SCP) hydrolysates | Neutrase | 1.12 mg/mL | 1. Ultrafiltration 2. Thin-layer chromatography (TLC) fractionation 3. RP-HPLC | 1. <3 kDa MWCO 2. Silica gel 60 TLC plate 3. C18 column (250 × 4.6 mm, 5 µm) | [54] |
| Sardine pilchardus protein | Flavourzyme | 1.83 ± 0.05 mg/mL | Size exclusion chromatography (SEC) | Superdex peptide 10/300GL column | [93] |
| Mare Whey Proteins | Papain | 0.18 mg/mL | 1. Gel filtration chromatography 2. RP-HPLC | 1. Sephadex G-25 column (2.5 cm × 70 cm) 2. C18 column (250 × 4.6 mm, 5 µm) | [79] |
| Quinoa (Chenopodium quinoa Willd.) | Pepsin and pancreatin | 0.31 ± 0.01 mg/mL | 1. Ultrafiltration 2. RP-HPLC | 1. <5 kDa MWCO 2. Hi-pore reversed phase RP-318 (250 21.5 mm) column | [53] |
| Protein Data Bank (PDB) ID | Software | Peptide | Binding Sites | Reference |
|---|---|---|---|---|
| 1WCY | Discovery studio 4.0 | FAGDDAPR, | Arg125, Arg358, Lys554, Phe357 | [89] |
| LAPSTM | Ser630, Arg125, Ser209, Try662, Tyr666, Arg125 | |||
| FAGDDAPRA | Ser209, Tyr547, Tyr585 | |||
| FLMESH | Arg125, Arg358 | |||
| 1WCY | Discovery studio 4.0 software | VPW | Arg358, Tyr662, Ser630 | [146] |
| IPR | Glu205, Glu206, Tyr662 | |||
| 3Q8W, 3F8S, and 5T4B | Discovery studio (DS) 2017 client software | IRDLLER | Glu205, Val207, Arg125, Arg429, Val546, Tyr585, Trp627, and Tyr752, Glu206, Arg125, Arg356 Arg429, and Asp545, Trp629, Phe357 | [41] |
| YAEERYP | Glu206, Arg356, Arg358, Glu361, Tyr456, Gln553, Asp556, Arg358, Arg429, Asp556, Arg356, Lys554 and Arg560 | |||
| IRNVLQPS | Arg125, Phe357, Arg560, Ser630, and Tyr666 | |||
| 5J3J | Discovery studio (DS) 2017 client software | ADF | Glu205, Glu206, Arg125, Arg358, Phe357 | [30] |
| MIR | Phe357, Arg358, Glu205, Glu206, Arg125 | |||
| FGR | Tyr547, Tyr631, Val656, Phe357, Trp659, Arg669, Tyr666, Tyr662, Glu205, Glu206, Asp663, Arg125, His126 | |||
| 5Y7H | Auto dock tools | YYGYTGAFR | Phe461, Ile407, Glu361, Gln469, Leu60, Ser59, Arg61, Asp104 | [3] |
| LDKVFR | Glu408, Arg356, Tyr585, Glu205, Glu206, Ser209, Ile470, Gly406, Arg358, Glu361, Ile405, Cys551, Phe357, and Arg669 | |||
| VLATSGPG | Val459, Ser59, Ser460, Arg61 | |||
| 4PNZ | Maestrov software | ELKDLKGY | Arg125, Ser630, Asn710, Tyr547, Ser209, Val207, Glu205, Glu361, Glu408, Arg429, Tyr456, and Asp556, Arg125, Glu361, Glu408, and Arg429, Tyr666 | [4] |
| ILDKVGINY | Arg356, Arg357, Arg358, Glu408, Arg429, Arg560, Arg358, Arg125 | |||
| 4PNZ | Maestrov software | NDWHTGPLS | Tyr666, Ser630, Ser209, Arg125, Glu205, Glu206, Tyr662, Tyr456, Arg560, Arg429, Tyr547 | [38] |
| TYPHQQPPILT | Glu206, Ser630, Arg125, Arg358, Lys554, Tyr547, Ser662, His126, Ser209, Glu408 | |||
| 4PNZ | Discovery studio 4.0 | PAGPF | Asn740, Ser630 | [80] |
| IPQVS | Asn740, Arg125, Glu206 | |||
| ELHQEEPL | Arg125, Glu206, Arg358 | |||
| KTMPGP | Tyr547, Arg358 |
| Number | Source | Substrate | Cell Line | Benefits | Disadvantage | Reference |
|---|---|---|---|---|---|---|
| 1 | Egg white | - Fractionation hydrolysate - Peptide: PFL, RVASMASEKM | STC-1 | Peptides but not free amino acids, showed a potent GLP-1 and CCK secretagogue effect | Proteins only had a modest GLP-1 secretagogue effect, no CCK release from proteins | [33] |
| 2 | Casein | - Whey hydrolysate - GPVRGPFPIIV | GLUTag | Could stimulate GLP-1 release | [153] | |
| 3 | Bovine hemoglobin | VVYPW, VVYPWQRF, and YPWQRF | STC-1 | Potent inhibitors of the GLP-1 inactivating DPP-IV | Did not show significant modulatory effects on the corresponding prohormones mRNA levels | [165] |
| 4 | Skin secretions of the crowned bullfrog | Family of tigerinin peptides | GLUTag and BRIN-BD11 | - Released GLP-1 from GLUTag -Released Insulin from BRIN-BD11 | [167] | |
| 5 | Frog skin | Hymenochirin-1b (Hym-1B; IKLSPETKDNLKKV- LKGAIKGAIAVAKMV.NH 2) | GLUTag and BRIN-BD11 | - Stimulated GLP-1 secretion from GLUTag cells - Increase in the rate of insulin release from BRIN-BD11 cells | [168] | |
| 6 | Bacterial peptide | (MAADIISTIGDLVK WIIDTVNKFKK), (MAQDIISTIGDLVK WIIDTVNKFTKK), (MAADIISTIGDLVK WIIDTVNKFTKK), and (MAQDIISTIGDLVK WIIDTVNKFKK) | NCI H716 cells | [162] | ||
| 7 | Bovine hemoglobin | ANVST, TKAVEH, and KAAVT | STC-1 | - Secretion of CCK and GLP-1 levels - Expression of DPP-IV inhibition action | [164] |
| Food Component | Animal Model | Method Research | Main Findings | Reference |
|---|---|---|---|---|
| Extract of H. rosa-sinensis leaves | Male Long-Evans rats, streptozotocin (STZ)—induced diabetic | Dose: 250 and 500 mg/kg; oral gavage Duration: 28 days | - Decreased serum glucose, cholesterol, and triglycerides - Increased circulating insulin, hdl cholesterol, and hepatic glycogen without increasing body weight | [50] |
| Rhazya stricta root extracts | Male BALB/c albino mice, Alloxan-induced diabetic | Dose: 20 mg/kg b.wt/day; oral Duration: 28 days | - Inhibiting enzymes DPP-IV - Increase in the GLP1 secretion - Reduction in blood glucose | [176] |
| Terminalia arjuna extract | Male Wistar rats, streptozotocin (STZ)—induced diabetic | Dose: 500 mg/kg, oral Duration: 4 weeks | - Superior DPP-IV inhibitory activity | [51] |
| The extract of Pueraria tuberosa | Male Charles Foster rats, streptozotocin (STZ)—induced diabetic | Dose: a dose of 500 mg/kg bw Duration: 35 days | - Increase in secretion of GLP-1 and GIP - Decrease in DPP-IV specific activities | [178] |
| Amaranth grain | Male Wistar rats, streptozotocin (STZ)—induced diabetic | Dose: 30 g/day, oral Duration: 12 weeks | - Reduced DPP-IV activity | [179] |
| Urchin gonad tissue extracts | Male rats, streptozotocin (STZ)—induced diabetic | Dose: 1.8 mg/kg/day, intragastric Duration: 10 days | - Decreased fasting serum glucose levels - Reduced blood malondiadehyde | [56] |
| Grape seed procyanidin extract | Female Zucker rats | Dose: 25–35 mg/kg/day, oral Duration: 19–60 days | - Decreased DPP-IV activity | [180] |
| Porcine skin gelatin hydrolysate (<1kDa fraction) released from Flavourzyme | Sprague Dawley male rats, streptozotocin (STZ)—induced diabetic | Dose: 300 mg/day; oral Duration: 42 days | - Improvement of glucose tolerance - Inhibition of DPP-IV activity - Increase in insulin levels - Increase in active GLP-1 levels - Reduction of glucagon levels | [181] |
| Atlantic salmon skin gelatin hydrolysate released from Flavourzyme | Sprague Dawley male rats, streptozotocin (STZ)—induced diabetic | Dose: 300 mg/day; oral administration Duration: 5 weeks | - Improvement of glucose tolerance - Increased active GLP-1 concentrations - Reduction of glucagon levels - Enhancement of insulin secretion - Inhibition in plasma DPP-IV activity - Improvement of glycemic control | [182] |
| Rice endosperm and rice bran protein hydrolysate released from pepsin or papain | Male Sprague Dawley rats | Dose: 2 g/kg body weight once (oral); and 500 mg once (ileal) | - No significant difference in plasma glucose during OGTT - Reduced glycemic response - Increased GLP-1 concentrations - Reduced plasma DPP-IV activity - Increased active GLP-1 | [183] |
| Halibut (HSGH) and tilapia (TSGH) fish skin gelatin hydrolysates (<1.5 kDa fraction) released from Flavourzyme | Sprague Dawley male rats, streptozotocin (STZ)—induced diabetic | Dose: 750 mg/kg bw/day; oral Duration: 30 days | - Decreased plasma glucose - Reduced plasma DPP-IV activity - Increased total and active GLP-1 concentrations - Increased plasma insulin levels - TSGH is more potent as an antihyperglycemic agent than HSGH | [55] |
| Corn zein hydrolysate released from papain | Male sprague Dawley rats | Dose: 500 mg/rat; Ileal, once Duration: acute | - Enhancement of insulin secretion - Increase in total GLP-1 concentration - Decrease in plasma DPP-IV activity - Prevention of hyperglycemia | [184] |
| Sodium caseinate hydrolysate (<1 kDa fraction) released from bromelain | Sprague Dawley male streptozotocin (STZ)—induced diabetic rats | Dose: 250 and 500 mg/kg/day; administration Duration: 6 weeks | - Improvement of glucose tolerance - Inhibition of plasma DPP-IV activity - Increase of active GLP-1 levels - Enhancement of insulin secretion | [185] |
| ß-casein-derived peptide LPQNIPPL from gouda-type cheese | Female Sprague Dawley rats | Dose: 300 mg/kg, once, oral administration Duration: acute | - Decreased plasma glucose - No change in insulin plasma concentrations | [187] |
| Peptide AGFAGĐAPR from Chinese black tea | Male ICR mice, streptozotocin (STZ)—induced diabetic | Dose: 400 mg/day; oral administration Duration: 57 days | - Increase of GLP-1 levels - Increase of insulin levels | [188] |
References
- Salim, M.A.S.M.; Gan, C.-Y. Dual-function peptides derived from egg white ovalbumin: Bioinformatics identification with validation using in vitro assay. J. Funct. Foods 2020, 64, 103618. [Google Scholar] [CrossRef]
- Nong, N.T.P.; Chen, Y.-K.; Shih, W.-L.; Hsu, J.-L. Characterization of Novel Dipeptidyl Peptidase-IV Inhibitory Peptides from Soft-Shelled Turtle Yolk Hydrolysate Using Orthogonal Bioassay-Guided Fractionations Coupled with In Vitro and In Silico Study. Pharmaceuticals 2020, 13, 308. [Google Scholar] [CrossRef]
- Jin, R.; Teng, X.; Shang, J.; Wang, D.; Liu, N. Identification of novel DPP–IV inhibitory peptides from Atlantic salmon (Salmo salar) skin. Food Res. Int. 2020, 133, 109161. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Gong, H.; Mao, X. Dipeptidyl peptidase-IV inhibitory activity and related molecular mechanism of bovine α-lactalbumin-derived peptides. Molecules 2020, 25, 3009. [Google Scholar] [CrossRef]
- Fosgerau, K.; Hoffmann, T. Peptide therapeutics: Current status and future directions. Drug Discov. Today 2015, 20, 122–128. [Google Scholar] [CrossRef] [Green Version]
- de la Torre, B.G.; Albericio, F. Peptide Therapeutics 2.0. Molecules 2020, 25, 2293. [Google Scholar] [CrossRef] [PubMed]
- Lacroix, I.M.; Li-Chan, E.C. Food-derived dipeptidyl-peptidase IV inhibitors as a potential approach for glycemic regulation–Current knowledge and future research considerations. Trends Food Sci. Technol. 2016, 54, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Nongonierma, A.B.; Cadamuro, C.; Le Gouic, A.; Mudgil, P.; Maqsood, S.; FitzGerald, R.J. Dipeptidyl peptidase IV (DPP-IV) inhibitory properties of a camel whey protein enriched hydrolysate preparation. Food Chem. 2019, 279, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Nongonierma, A.B.; Lamoureux, C.; FitzGerald, R.J. Generation of dipeptidyl peptidase IV (DPP-IV) inhibitory peptides during the enzymatic hydrolysis of tropical banded cricket (Gryllodes sigillatus) proteins. Food Funct. 2018, 9, 407–416. [Google Scholar] [CrossRef] [Green Version]
- Hartmann, R.; Meisel, H. Food-derived peptides with biological activity: From research to food applications. Curr. Opin. Biotechnol. 2007, 18, 163–169. [Google Scholar] [CrossRef]
- Li-Chan, E.C. Bioactive peptides and protein hydrolysates: Research trends and challenges for application as nutraceuticals and functional food ingredients. Curr. Opin. Food. Sci. 2015, 1, 28–37. [Google Scholar] [CrossRef] [Green Version]
- Nongonierma, A.B.; FitzGerald, R.J. Bioactive properties of milk proteins in humans: A review. Peptides 2015, 73, 20–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zielinska, E.; Karas, M.; Baroniak, B.; Jakubczyk, A. Evaluation of ACE, α-glucosidase, and lipase inhibitory activities of peptides obtained by in vitro digestion of selected species of edible insects. Eur. Food Res. Technol. 2020, 246, 1361–1369. [Google Scholar] [CrossRef] [Green Version]
- FitzGerald, R.J.; Cermeño, M.; Khalesi, M.; Kleekayai, T.; Amigo-Benavent, M. Application of in silico approaches for the generation of milk protein-derived bioactive peptides. J. Funct. Foods 2020, 64, 103636. [Google Scholar] [CrossRef]
- Patil, P.; Mandal, S.; Tomar, S.K.; Anand, S. Food protein-derived bioactive peptides in management of type 2 diabetes. Eur. J. Nutr. 2015, 54, 863–880. [Google Scholar] [CrossRef]
- Dowarah, J.; Singh, V.P. Anti-diabetic drugs recent approaches and advancements. Biorgan. Med. Chem. 2020, 28, 115263. [Google Scholar] [CrossRef] [PubMed]
- Brandt, S.J.; Götz, A.; Tschöp, M.H.; Müller, T.D. Gut hormone polyagonists for the treatment of type 2 diabetes. Peptides 2018, 100, 190–201. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Zhao, J.; Yang, R.; Zhao, W. Bioactive peptides with antidiabetic properties: A review. Int. J. Food Sci. Technol. 2019, 54, 1909–1919. [Google Scholar] [CrossRef] [Green Version]
- Cheung, B.M.; Li, C. Diabetes and hypertension: Is there a common metabolic pathway? Curr. Atheroscler. Rep. 2012, 14, 160–166. [Google Scholar] [CrossRef] [Green Version]
- Gourgari, E.; Wilhelm, E.E.; Hassanzadeh, H.; Aroda, V.R.; Shoulson, I. A comprehensive review of the FDA-approved labels of diabetes drugs: Indications, safety, and emerging cardiovascular safety data. J. Diabetes Complicat. 2017, 31, 1719–1727. [Google Scholar] [CrossRef]
- Hartmann-Boyce, J.; Morris, E.; Goyder, C.; Kinton, J.; Perring, J.; Nunan, D.; Mahtani, K.; Buse, J.B.; Del Prato, S.; Ji, L. Diabetes and COVID-19: Risks, management, and learnings from other national disasters. Diabetes Care 2020, 43, 1695–1703. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Gupta, R.; Ghosh, A.; Misra, A. Diabetes in COVID-19: Prevalence, pathophysiology, prognosis and practical considerations. Diabetes Metab. Syndr. 2020, 14, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Bloomgarden, Z.T. Diabetes and COVID-19. J. Diabetes 2020, 12, 347–348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Power, O.; Nongonierma, A.B.; Jakeman, P.; FitzGerald, R.J. Food protein hydrolysates as a source of dipeptidyl peptidase IV inhibitory peptides for the management of type 2 diabetes. Proc. Nutr. Soc. 2014, 73, 34–46. [Google Scholar] [CrossRef] [Green Version]
- Deacon, C.F. Peptide degradation and the role of DPP-4 inhibitors in the treatment of type 2 diabetes. Peptides 2018, 100, 150–157. [Google Scholar] [CrossRef] [PubMed]
- Caner, S.; Zhang, X.; Jiang, J.; Chen, H.M.; Nguyen, N.T.; Overkleeft, H.; Brayer, G.D.; Withers, S.G. Glucosyl epi-cyclophellitol allows mechanism-based inactivation and structural analysis of human pancreatic α-amylase. FEBS Lett. 2016, 590, 1143–1151. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Wu, T.; Fang, L.; Liu, C.; Liu, X.; Li, H.; Shi, J.; Li, M.; Min, W. Anti-diabetic effect by walnut (Juglans mandshurica Maxim.)-derived peptide LPLLR through inhibiting α-glucosidase and α-amylase, and alleviating insulin resistance of hepatic HepG2 cells. J. Funct. Foods 2020, 69, 103944. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, N.; Wang, W.; Wang, J.; Zhu, Z.; Li, X. Molecular mechanisms of novel peptides from silkworm pupae that inhibit α-glucosidase. Peptides 2016, 76, 45–50. [Google Scholar] [CrossRef]
- Ezzat, S.M.; Bishbishy, M.H.E.; Habtemariam, S.; Salehi, B.; Sharifi-Rad, M.; Martins, N.; Sharifi-Rad, J. Looking at marine-derived bioactive molecules as upcoming anti-diabetic agents: A special emphasis on PTP1B inhibitors. Molecules 2018, 23, 3334. [Google Scholar] [CrossRef] [Green Version]
- Zhao, W.; Zhang, D.; Yu, Z.; Ding, L.; Liu, J. Novel membrane peptidase inhibitory peptides with activity against angiotensin converting enzyme and dipeptidyl peptidase IV identified from hen eggs. J. Funct. Foods 2020, 64, 103649. [Google Scholar] [CrossRef]
- Zhao, B.; Su, K.; Mao, X.; Zhang, X. Separation and identification of enzyme inhibition peptides from dark tea protein. Bioorg. Chem. 2020, 99, 103772. [Google Scholar] [CrossRef] [PubMed]
- Sharkey, S.J.; Harnedy-Rothwell, P.A.; Allsopp, P.J.; Hollywood, L.E.; FitzGerald, R.J.; O’Harte, F.P. A Narrative Review of the Anti-Hyperglycemic and Satiating Effects of Fish Protein Hydrolysates and Their Bioactive Peptides. Mol. Nutr. Food Res. 2020, 64, 2000403. [Google Scholar] [CrossRef]
- Santos-Hernández, M.; Amigo, L.; Recio, I. Induction of CCK and GLP-1 release in enteroendocrine cells by egg white peptides generated during gastrointestinal digestion. Food Chem. 2020, 329, 127188. [Google Scholar] [CrossRef]
- Mora, L.; González-Rogel, D.; Heres, A.; Toldrá, F. Iberian dry-cured ham as a potential source of α-glucosidase-inhibitory peptides. J. Funct. Foods 2020, 67, 103840. [Google Scholar] [CrossRef]
- Jia, C.-l.; Hussain, N.; Obaroakpo, J.U.; Pang, X.-y.; Zhang, S.-w.; Lu, J.; Liu, L.; Lv, J.-p. Generation and characterization of dipeptidyl peptidase-IV inhibitory peptides from trypsin-hydrolyzed α-lactalbumin-rich whey proteins. Food Chem. 2020, 318, 126333. [Google Scholar] [CrossRef]
- Harnedy-Rothwell, P.A.; McLaughlin, C.M.; O’Keeffe, M.B.; Le Gouic, A.V.; Allsopp, P.J.; McSorley, E.M.; Sharkey, S.; Whooley, J.; McGovern, B.; O’Harte, F.P. Identification and characterisation of peptides from a boarfish (Capros aper) protein hydrolysate displaying in vitro dipeptidyl peptidase-IV (DPP-IV) inhibitory and insulinotropic activity. Food Res. Int. 2020, 131, 108989. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Gao, T.; Hou, Y.; Li, D.; Fu, L. Identification and characterization of two novel α-glucosidase inhibitory peptides from almond (Armeniaca sibirica) oil manufacture residue. LWT 2020, 134, 110215. [Google Scholar] [CrossRef]
- Gu, H.; Gao, J.; Shen, Q.; Gao, D.; Wang, Q.; Tangyu, M.; Mao, X. Dipeptidyl peptidase-IV inhibitory activity of millet protein peptides and the related mechanisms revealed by molecular docking. LWT 2021, 138, 110587. [Google Scholar]
- Omar, B.; Ahrén, B. Pleiotropic mechanisms for the glucose-lowering action of DPP-4 inhibitors. Diabetes 2014, 63, 2196–2202. [Google Scholar] [CrossRef] [Green Version]
- Zhong, J.; Gong, Q.; Goud, A.; Srinivasamaharaj, S.; Rajagopalan, S. Recent advances in dipeptidyl-peptidase-4 inhibition therapy: Lessons from the bench and clinical trials. J. Diabetes Res. 2015, 2015. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Ding, L.; Du, Z.; Yu, Z.; Liu, J. Hydrolysis and transport of egg white-derived peptides in Caco-2 cell monolayers and everted rat sacs. J. Agric. Food Chem. 2019, 67, 4839–4848. [Google Scholar] [CrossRef]
- Nongonierma, A.B.; Dellafiora, L.; Paolella, S.; Galaverna, G.; Cozzini, P.; FitzGerald, R.J. In silico approaches applied to the study of peptide analogs of Ile-Pro-Ile in relation to their dipeptidyl peptidase IV inhibitory properties. Front. Endocrinol. 2018, 9, 329. [Google Scholar] [CrossRef] [Green Version]
- Zhong, J.; Rao, X.; Rajagopalan, S. An emerging role of dipeptidyl peptidase 4 (DPP4) beyond glucose control: Potential implications in cardiovascular disease. Atherosclerosis 2013, 226, 305–314. [Google Scholar] [CrossRef]
- Kim, W.; Egan, J.M. The role of incretins in glucose homeostasis and diabetes treatment. Pharmacol. Rev. 2008, 60, 470–512. [Google Scholar] [CrossRef] [Green Version]
- Andersen, E.S.; Deacon, C.F.; Holst, J.J. Do we know the true mechanism of action of the DPP-4 inhibitors? Diabetes Obes. Metab. 2018, 20, 34–41. [Google Scholar] [CrossRef]
- Baggio, L.L.; Drucker, D.J. Biology of incretins: GLP-1 and GIP. Gastroenterology 2007, 132, 2131–2157. [Google Scholar] [CrossRef] [PubMed]
- Thoma, R.; Löffler, B.; Stihle, M.; Huber, W.; Ruf, A.; Hennig, M. Structural basis of proline-specific exopeptidase activity as observed in human dipeptidyl peptidase-IV. Structure 2003, 11, 947–959. [Google Scholar] [CrossRef] [Green Version]
- Nongonierma, A.B.; Paolella, S.; Mudgil, P.; Maqsood, S.; FitzGerald, R.J. Identification of novel dipeptidyl peptidase IV (DPP-IV) inhibitory peptides in camel milk protein hydrolysates. Food Chem. 2018, 244, 340–348. [Google Scholar] [CrossRef] [Green Version]
- Mudgil, P.; Kamal, H.; Yuen, G.C.; Maqsood, S. Characterization and identification of novel antidiabetic and anti-obesity peptides from camel milk protein hydrolysates. Food Chem. 2018, 259, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Ansari, P.; Azam, S.; Hannan, J.; Flatt, P.R.; Wahab, Y.H.A. Anti-hyperglycaemic activity of H. rosa-sinensis leaves is partly mediated by inhibition of carbohydrate digestion and absorption, and enhancement of insulin secretion. J. Ethnopharmacol. 2020, 253, 112647. [Google Scholar] [CrossRef] [PubMed]
- Mohanty, I.R.; Borde, M.; Maheshwari, U. Dipeptidyl peptidase IV Inhibitory activity of Terminalia arjuna attributes to its cardioprotective effects in experimental diabetes: In silico, in vitro and in vivo analyses. Phytomedicine 2019, 57, 158–165. [Google Scholar] [CrossRef] [PubMed]
- Cermeño, M.; Connolly, A.; O’Keeffe, M.B.; Flynn, C.; Alashi, A.M.; Aluko, R.E.; FitzGerald, R.J. Identification of bioactive peptides from brewers’ spent grain and contribution of Leu/Ile to bioactive potency. J. Funct. Foods 2019, 60, 103455. [Google Scholar] [CrossRef]
- Vilcacundo, R.; Martínez-Villaluenga, C.; Hernández-Ledesma, B. Release of dipeptidyl peptidase IV, α-amylase and α-glucosidase inhibitory peptides from quinoa (Chenopodium quinoa Willd.) during in vitro simulated gastrointestinal digestion. J. Funct. Foods 2017, 35, 531–539. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Chen, R.; Chen, X.; Zeng, Z.; Ma, H.; Chen, S. Dipeptidyl peptidase IV-inhibitory peptides derived from Silver Carp (Hypophthalmichthys molitrix Val.) proteins. J. Agric. Food Chem. 2016, 64, 831–839. [Google Scholar] [CrossRef]
- Wang, T.-Y.; Hsieh, C.-H.; Hung, C.-C.; Jao, C.-L.; Chen, M.-C.; Hsu, K.-C. Fish skin gelatin hydrolysates as dipeptidyl peptidase IV inhibitors and glucagon-like peptide-1 stimulators improve glycaemic control in diabetic rats: A comparison between warm-and cold-water fish. J. Funct. Foods 2015, 19, 330–340. [Google Scholar] [CrossRef]
- Pozharitskaya, O.N.; Shikov, A.N.; Laakso, I.; Seppänen-Laakso, T.; Makarenko, I.E.; Faustova, N.M.; Makarova, M.N.; Makarov, V.G. Bioactivity and chemical characterization of gonads of green sea urchin Strongylocentrotus droebachiensis from Barents Sea. J. Funct. Foods 2015, 17, 227–234. [Google Scholar] [CrossRef]
- Verma, M.; Gupta, S.J.; Chaudhary, A.; Garg, V.K. Protein tyrosine phosphatase 1B inhibitors as antidiabetic agents–A brief review. Bioorgan. Chem. 2017, 70, 267–283. [Google Scholar] [CrossRef]
- Abdjul, D.B.; Yamazaki, H.; Kanno, S.-I.; Wewengkang, D.S.; Rotinsulu, H.; Sumilat, D.A.; Ukai, K.; Kapojos, M.M.; Namikoshi, M. Furanoterpenes, new types of protein tyrosine phosphatase 1B inhibitors, from two Indonesian marine sponges, Ircinia and Spongia spp. Bioorgan. Med. Chem. Lett. 2017, 27, 1159–1161. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.-L.; Liao, X.-J.; Wang, Y.-H.; Si, X.; Shu, C.; Gong, E.-S.; Xie, X.; Ran, X.-L.; Li, B. Identification of Cyanidin-3-arabinoside Extracted from Blueberry as a Selective Protein Tyrosine Phosphatase 1B Inhibitor. J. Agric. Food Chem. 2019, 67, 13624–13634. [Google Scholar] [CrossRef]
- Rotinsulu, H.; Yamazaki, H.; Sugai, S.; Iwakura, N.; Wewengkang, D.S.; Sumilat, D.A.; Namikoshi, M. Cladosporamide A, a new protein tyrosine phosphatase 1B inhibitor, produced by an Indonesian marine sponge-derived Cladosporium sp. J. Nat. Med. 2018, 72, 779–783. [Google Scholar] [CrossRef]
- Seong, S.H.; Roy, A.; Jung, H.A.; Jung, H.J.; Choi, J.S. Protein tyrosine phosphatase 1B and α-glucosidase inhibitory activities of Pueraria lobata root and its constituents. J. Ethnopharmacol. 2016, 194, 706–716. [Google Scholar] [CrossRef]
- Zhang, X.-Y.; Wei, L.; Jian, W.; Ning, L.; Cheng, M.-S.; Koike, K. Protein tyrosine phosphatase 1B inhibitory activities of ursane-type triterpenes from Chinese raspberry, fruits of Rubus chingii. Chin. J. Nat. Med 2019, 17, 15–21. [Google Scholar] [CrossRef]
- Zhao, B.T.; Nguyen, D.H.; Le, D.D.; Choi, J.S.; Min, B.S.; Woo, M.H. Protein tyrosine phosphatase 1B inhibitors from natural sources. Arch. Pharmacal Res. 2018, 41, 130–161. [Google Scholar] [CrossRef]
- Chen, M.; Wang, K.; Zhang, Y.; Zhang, M.; Ma, Y.; Sun, H.; Jin, Z.; Zheng, H.; Jiang, H.; Yu, P. New insights into the biological activities of Chrysanthemum morifolium: Natural flavonoids alleviate diabetes by targeting α-glucosidase and the PTP-1B signaling pathway. Eur. J. Med. Chem. 2019, 178, 108–115. [Google Scholar] [CrossRef]
- Uddin, Z.; Song, Y.H.; Ullah, M.; Li, Z.; Kim, J.Y.; Park, K.H. Isolation and characterization of protein tyrosine phosphatase 1B (PTP1B) inhibitory polyphenolic compounds from Dodonaea viscosa and their kinetic analysis. Front. Chem. 2018, 6, 40. [Google Scholar] [CrossRef]
- Song, Y.H.; Uddin, Z.; Jin, Y.M.; Li, Z.; Curtis-Long, M.J.; Kim, K.D.; Cho, J.K.; Park, K.H. Inhibition of protein tyrosine phosphatase (PTP1B) and α-glucosidase by geranylated flavonoids from Paulownia tomentosa. J. Enzym. Inhib. Med. Chem. 2017, 32, 1195–1202. [Google Scholar] [CrossRef] [Green Version]
- Paudel, P.; Yu, T.; Seong, S.H.; Kuk, E.B.; Jung, H.A.; Choi, J.S. Protein tyrosine phosphatase 1B inhibition and glucose uptake potentials of mulberrofuran G, albanol B, and kuwanon G from root bark of Morus alba L. in insulin-resistant HepG2 cells: An in vitro and in silico study. Int. J. Mol. Sci. 2018, 19, 1542. [Google Scholar] [CrossRef] [Green Version]
- Ha, M.T.; Seong, S.H.; Nguyen, T.D.; Cho, W.-K.; Ah, K.J.; Ma, J.Y.; Woo, M.H.; Choi, J.S.; Min, B.S. Chalcone derivatives from the root bark of Morus alba L. act as inhibitors of PTP1B and α-glucosidase. Phytochemistry 2018, 155, 114–125. [Google Scholar] [CrossRef] [PubMed]
- Cerón-Romero, L.; Paoli, P.; Camici, G.; Flores-Morales, V.; Rios, M.Y.; Ramírez-Espinosa, J.J.; Hidalgo-Figueroa, S.; Navarrete-Vázquez, G.; Estrada-Soto, S. In vitro and in silico PTP-1B inhibition and in vivo antidiabetic activity of semisynthetic moronic acid derivatives. Bioorg. Med. Chem. Lett. 2016, 26, 2018–2022. [Google Scholar] [CrossRef] [PubMed]
- Valencia-Mejía, E.; Batista, K.A.; Fernández, J.J.A.; Fernandes, K.F. Antihyperglycemic and hypoglycemic activity of naturally occurring peptides and protein hydrolysates from easy-to-cook and hard-to-cook beans (Phaseolus vulgaris L.). Food Res. Int. 2019, 121, 238–246. [Google Scholar] [CrossRef] [PubMed]
- Karthiraj, T.; Babu, B.H.; Kumar, R.S. Task-specific deep eutectic solvent based extraction coupled cascade chromatography quantification of α-glucosidase inhibitory peptide from Ocimum tenuriflorum seeds. Microchem. J. 2020, 157, 104883. [Google Scholar] [CrossRef]
- González-Montoya, M.; Hernández-Ledesma, B.; Mora-Escobedo, R.; Martínez-Villaluenga, C. Bioactive peptides from germinated soybean with anti-diabetic potential by inhibition of dipeptidyl peptidase-IV, α-amylase, and α-glucosidase enzymes. Int. J. Mol. Sci. 2018, 19, 2883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uraipong, C.; Zhao, J. In vitro digestion of rice bran proteins produces peptides with potent inhibitory effects on α-glucosidase and angiotensin I converting enzyme. J. Sci. Food Agric. 2018, 98, 758–766. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Liang, K.; Jin, Y.; Zhang, M.; Chen, Y.; Wu, H.; Lai, F. Identification and characterization of two novel α-glucosidase inhibitory oligopeptides from hemp (Cannabis sativa L.) seed protein. J. Funct. Foods 2016, 26, 439–450. [Google Scholar] [CrossRef]
- Wang, R.; Zhao, H.; Pan, X.; Orfila, C.; Lu, W.; Ma, Y. Preparation of bioactive peptides with antidiabetic, antihypertensive, and antioxidant activities and identification of α-glucosidase inhibitory peptides from soy protein. Food Sci. Nutr. 2019, 7, 1848–1856. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, M.A.; Bester, M.J.; Neitz, A.W.; Gaspar, A.R. Rational in silico design of novel α-glucosidase inhibitory peptides and in vitro evaluation of promising candidates. Biomed. Pharmacother. 2018, 107, 234–242. [Google Scholar] [CrossRef] [Green Version]
- Obaroakpo, J.U.; Liu, L.; Zhang, S.; Lu, J.; Pang, X.; Lv, J. α-Glucosidase and ACE dual inhibitory protein hydrolysates and peptide fractions of sprouted quinoa yoghurt beverages inoculated with Lactobacillus casei. Food Chem. 2019, 299, 124985. [Google Scholar] [CrossRef]
- Ni, H.; Hayes, H.E.; Stead, D.; Raikos, V. Incorporating salal berry (Gaultheria shallon) and blackcurrant (Ribes nigrum) pomace in yogurt for the development of a beverage with antidiabetic properties. Heliyon 2018, 4, e00875. [Google Scholar] [CrossRef] [Green Version]
- Song, J.; Wang, Q.; Du, M.; Ji, X.; Mao, X. Identification of dipeptidyl peptidase-IV inhibitory peptides from mare whey protein hydrolysates. J. Dairy Sci. 2017, 100, 6885–6894. [Google Scholar] [CrossRef] [Green Version]
- Xu, F.; Yao, Y.; Xu, X.; Wang, M.; Pan, M.; Ji, S.; Wu, J.; Jiang, D.; Ju, X.; Wang, L. Identification and Quantification of DPP-IV-Inhibitory Peptides from Hydrolyzed-Rapeseed-Protein-Derived Napin with Analysis of the Interactions between Key Residues and Protein Domains. J. Agric. Food Chem. 2019, 67, 3679–3690. [Google Scholar] [CrossRef]
- Nongonierma, A.B.; Hennemann, M.; Paolella, S.; FitzGerald, R.J. Generation of wheat gluten hydrolysates with dipeptidyl peptidase IV (DPP-IV) inhibitory properties. Food Funct. 2017, 8, 2249–2257. [Google Scholar] [CrossRef] [Green Version]
- Raveschot, C.; Cudennec, B.; Coutte, F.; Flahaut, C.; Fremont, M.; Drider, D.; Dhulster, P. Production of bioactive peptides by Lactobacillus species: From gene to application. Front. Microbiol. 2018, 9, 2354. [Google Scholar] [CrossRef] [Green Version]
- Worsztynowicz, P.; Białas, W.; Grajek, W. Integrated approach for obtaining bioactive peptides from whey proteins hydrolysed using a new proteolytic lactic acid bacteria. Food Chem. 2020, 312, 126035. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Li, X.; Liu, H.; Li, Q.; Xiao, R.; Dudu, O.E.; Yang, L.; Ma, Y. The impact of multiple-species starters on the peptide profiles of yoghurts. Int. Dairy J. 2020, 106, 104684. [Google Scholar] [CrossRef]
- Mechmeche, M.; Kachouri, F.; Ksontini, H.; Hamdi, M. Production of bioactive peptides from tomato seed isolate by Lactobacillus plantarum fermentation and enhancement of antioxidant activity. Food Biotechnol. 2017, 31, 94–113. [Google Scholar] [CrossRef]
- Kęska, P.; Wójciak, K.M.; Stadnik, J. Bioactive peptides from beef products fermented by acid whey–in vitro and in silico study. Sci. Agric. 2019, 76, 311–320. [Google Scholar] [CrossRef]
- Li, C.; Kwok, L.-Y.; Mi, Z.; Bala, J.; Xue, J.; Yang, J.; Ma, Y.; Zhang, H.; Chen, Y. Characterization of the angiotensin-converting enzyme inhibitory activity of fermented milks produced with Lactobacillus casei. J. Dairy Sci. 2017, 100, 9495–9507. [Google Scholar] [CrossRef] [Green Version]
- Mudgil, P.; Baby, B.; Ngoh, Y.-Y.; Kamal, H.; Vijayan, R.; Gan, C.-Y.; Maqsood, S. Molecular binding mechanism and identification of novel anti-hypertensive and anti-inflammatory bioactive peptides from camel milk protein hydrolysates. LWT 2019, 112, 108193. [Google Scholar] [CrossRef]
- Liu, R.; Zhou, L.; Zhang, Y.; Sheng, N.-J.; Wang, Z.-K.; Wu, T.-Z.; Wang, X.-Z.; Wu, H. Rapid identification of dipeptidyl peptidase-IV (DPP-IV) inhibitory peptides from Ruditapes philippinarum hydrolysate. Molecules 2017, 22, 1714. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhang, Y.; Wang, Z.; Chen, S.; Luo, Y. Production and identification of antioxidant and angiotensin-converting enzyme inhibition and dipeptidyl peptidase IV inhibitory peptides from bighead carp (Hypophthalmichthys nobilis) muscle hydrolysate. J. Funct. Foods 2017, 35, 224–235. [Google Scholar] [CrossRef]
- Mune, M.A.M.; Minka, S.R.; Henle, T. Investigation on antioxidant, angiotensin converting enzyme and dipeptidyl peptidase IV inhibitory activity of Bambara bean protein hydrolysates. Food Chem. 2018, 250, 162–169. [Google Scholar] [CrossRef]
- Sila, A.; Alvarez, O.M.; Haddar, A.; Frikha, F.; Dhulster, P.; Nedjar-Arroume, N.; Bougatef, A. Purification, identification and structural modelling of DPP-IV inhibiting peptides from barbel protein hydrolysate. J. Chromatogr. B 2016, 1008, 260–269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rivero-Pino, F.; Espejo-Carpio, F.J.; Guadix, E.M. Production and identification of dipeptidyl peptidase IV (DPP-IV) inhibitory peptides from discarded Sardine pilchardus protein. Food Chem. 2020, 328, 127096. [Google Scholar] [CrossRef] [PubMed]
- Sato, K.; Miyasaka, S.; Tsuji, A.; Tachi, H. Isolation and characterization of peptides with dipeptidyl peptidase IV (DPPIV) inhibitory activity from natto using DPPIV from Aspergillus oryzae. Food Chem. 2018, 261, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Rutherfurd, S.M. Methodology for determining degree of hydrolysis of proteins in hydrolysates: A review. J. AOAC Int. 2010, 93, 1515–1522. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Liu, H.; Hong, H.; Luo, Y. Purification and identification of dipeptidyl peptidase IV and angiotensin-converting enzyme inhibitory peptides from silver carp (Hypophthalmichthys molitrix) muscle hydrolysate. Eur. Food Res. Technol. 2019, 245, 243–255. [Google Scholar] [CrossRef]
- Cermeño, M.; Stack, J.; Tobin, P.R.; O’Keeffe, M.B.; Harnedy, P.A.; Stengel, D.B.; FitzGerald, R.J. Peptide identification from a Porphyra dioica protein hydrolysate with antioxidant, angiotensin converting enzyme and dipeptidyl peptidase IV inhibitory activities. Food Funct. 2019, 10, 3421–3429. [Google Scholar] [CrossRef]
- Pujiastuti, D.Y.; Shih, Y.-H.; Chen, W.-L.; Hsu, J.-L. Screening of angiotensin-I converting enzyme inhibitory peptides derived from soft-shelled turtle yolk using two orthogonal bioassay-guided fractionations. J. Funct. Foods 2017, 28, 36–47. [Google Scholar] [CrossRef]
- Cottrell, J.S. Protein identification using MS/MS data. J. Proteomics 2011, 74, 1842–1851. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Hou, J.; Tanner, J.J.; Cheng, J. Bioinformatics methods for mass spectrometry-based proteomics data analysis. Int. J. Mol. Sci. 2020, 21, 2873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, A.L.-A.; Xiang, X.; Ong, P.S.; Mitchell, E.Q.Y.; Syn, N.; Wee, I.; Kumar, A.P.; Yong, W.P.; Sethi, G.; Goh, B.C. A review on liquid chromatography-tandem mass spectrometry methods for rapid quantification of oncology drugs. Pharmaceutics 2018, 10, 221. [Google Scholar] [CrossRef] [Green Version]
- Kapp, E.; Schütz, F. Overview of tandem mass spectrometry (MS/MS) database search algorithms. Curr. Protoc. Protein. Sci 2007, 49. [Google Scholar] [CrossRef]
- Allmer, J. Algorithms for the de novo sequencing of peptides from tandem mass spectra. Expert Rev. Proteom. 2011, 8, 645–657. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Xin, L.; Shan, B.; Chen, W.; Xie, M.; Yuen, D.; Zhang, W.; Zhang, Z.; Lajoie, G.A.; Ma, B. PEAKS DB: De novo sequencing assisted database search for sensitive and accurate peptide identification. Mol. Cell. Proteom. 2012, 11. [Google Scholar] [CrossRef] [Green Version]
- Medzihradszky, K.F.; Chalkley, R.J. Lessons in de novo peptide sequencing by tandem mass spectrometry. Mass Spectrom. Rev. 2015, 34, 43–63. [Google Scholar] [CrossRef] [Green Version]
- Seidler, J.; Zinn, N.; Boehm, M.E.; Lehmann, W.D. De novo sequencing of peptides by MS/MS. Proteomics 2010, 10, 634–649. [Google Scholar] [CrossRef] [PubMed]
- Vyatkina, K.; Wu, S.; Dekker, L.J.; VanDuijn, M.M.; Liu, X.; Tolic, N.; Dvorkin, M.; Alexandrova, S.; Luider, T.M.; Pasa-Tolic, L. De novo sequencing of peptides from top-down tandem mass spectra. J. Proteome Res. 2015, 14, 4450–4462. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Zhang, K.; Hendrie, C.; Liang, C.; Li, M.; Doherty-Kirby, A.; Lajoie, G. PEAKS: Powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2003, 17, 2337–2342. [Google Scholar] [CrossRef] [PubMed]
- Armirotti, A.; Millo, E.; Damonte, G. How to discriminate between leucine and isoleucine by low energy ESI-TRAP MS n. J. Am. Soc. Mass Spectrom. 2007, 18, 57–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bagal, D.; Kast, E.; Cao, P. Rapid distinction of leucine and isoleucine in monoclonal antibodies using nanoflow LCMSn. Anal. Chem. 2017, 89, 720–727. [Google Scholar] [CrossRef]
- Minkiewicz, P.; Iwaniak, A.; Darewicz, M. BIOPEP-UWM database of bioactive peptides: Current opportunities. Int. J. Mol. Sci. 2019, 20, 5978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwaniak, A.; Minkiewicz, P.; Darewicz, M.; Sieniawski, K.; Starowicz, P. BIOPEP database of sensory peptides and amino acids. Food Res. Int. 2016, 85, 155–161. [Google Scholar] [CrossRef]
- Uraipong, C.; Zhao, J. Rice bran protein hydrolysates exhibit strong in vitro α-amylase, β-glucosidase and ACE-inhibition activities. J. Sci. Food Agric. 2016, 96, 1101–1110. [Google Scholar] [CrossRef]
- Kumagai, Y.; Kitade, Y.; Kobayashi, M.; Watanabe, K.; Kurita, H.; Takeda, H.; Yasui, H.; Kishimura, H. Identification of ACE inhibitory peptides from red alga Mazzaella japonica. Eur. Food Res. Technol. 2020, 246, 2225–2231. [Google Scholar] [CrossRef]
- Parmar, H.; Hati, S.; Panchal, G.; Sakure, A.A. Purification and production of novel angiotensin I-converting enzyme (ACE) inhibitory bioactive peptides derived from fermented goat milk. Int. J. Pept. Res. Ther. 2020, 26, 997–1011. [Google Scholar] [CrossRef]
- Ji, D.; Udenigwe, C.C.; Agyei, D. Antioxidant peptides encrypted in flaxseed proteome: An in silico assessment. FSHW 2019, 8, 306–314. [Google Scholar] [CrossRef]
- e Silva, M.B.d.C.; da Cruz Souza, C.A.; Philadelpho, B.O.; da Cunha, M.M.N.; Batista, F.P.R.; da Silva, J.R.; Druzian, J.I.; Castilho, M.S.; Cilli, E.M.; Ferreira, E.S. In vitro and in silico studies of 3-hydroxy-3-methyl-glutaryl coenzyme A reductase inhibitory activity of the cowpea Gln-Asp-Phe peptide. Food Chem. 2018, 259, 270–277. [Google Scholar] [CrossRef]
- Gupta, S.; Kapoor, P.; Chaudhary, K.; Gautam, A.; Kumar, R.; Raghava, G.P.; Consortium, O.S.D.D. In silico approach for predicting toxicity of peptides and proteins. PLoS ONE 2013, 8, e73957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nongonierma, A.B.; FitzGerald, R.J. Learnings from quantitative structure–activity relationship (QSAR) studies with respect to food protein-derived bioactive peptides: A review. RCS Adv. 2016, 6, 75400–75413. [Google Scholar] [CrossRef]
- Nongonierma, A.B.; FitzGerald, R.J. Structure activity relationship modelling of milk protein-derived peptides with dipeptidyl peptidase IV (DPP-IV) inhibitory activity. Peptides 2016, 79, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nongonierma, A.B.; FitzGerald, R.J. Features of dipeptidyl peptidase IV (DPP-IV) inhibitory peptides from dietary proteins. J. Food Biochem. 2019, 43, e12451. [Google Scholar] [CrossRef] [Green Version]
- Liu, R.; Cheng, J.; Wu, H. Discovery of food-derived dipeptidyl peptidase IV inhibitory peptides: A review. Int. J. Mol. Sci. 2019, 20, 463. [Google Scholar] [CrossRef] [Green Version]
- Hu, S.; Fan, X.; Qi, P.; Zhang, X. Identification of anti-diabetes peptides from Spirulina platensis. J. Funct. Foods 2019, 56, 333–341. [Google Scholar] [CrossRef]
- Ibrahim, M.A.; Serem, J.C.; Bester, M.J.; Neitz, A.W.; Gaspar, A.R. Multiple antidiabetic effects of three α-glucosidase inhibitory peptides, PFP, YPL and YPG: Dipeptidyl peptidase–IV inhibition, suppression of lipid accumulation in differentiated 3T3-L1 adipocytes and scavenging activity on methylglyoxal. Int. J. Biol. Macromol. 2019, 122, 104–114. [Google Scholar] [CrossRef] [Green Version]
- Nongonierma, A.B.; FitzGerald, R.J. Inhibition of dipeptidyl peptidase IV (DPP-IV) by proline containing casein-derived peptides. J. Funct. Foods 2013, 5, 1909–1917. [Google Scholar] [CrossRef] [Green Version]
- Umezawa, H.; Aoyagi, T.; Ogawa, K.; Naganawa, H.; Hamada, M.; Takeuchi, T. Diprotins A and B, inhibitors of dipeptidyl aminopeptidase IV, produced by bacteria. J. Antibiot. 1984, 37, 422–425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nongonierma, A.B.; FitzGerald, R.J. An in silico model to predict the potential of dietary proteins as sources of dipeptidyl peptidase IV (DPP-IV) inhibitory peptides. Food Chem. 2014, 165, 489–498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lan, V.T.T.; Ito, K.; Ohno, M.; Motoyama, T.; Ito, S.; Kawarasaki, Y. Analyzing a dipeptide library to identify human dipeptidyl peptidase IV inhibitor. Food Chem. 2015, 175, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Ouertani, A.; Neifar, M.; Ouertani, R. Effectiveness of enzyme inhibitors in biomedicine and pharmacotherapy. Adv. Tissue Eng. Regen. Med. Open Access 2019, 5, 85–90. [Google Scholar]
- Rahfeld, J.; Schierborn, M.; Hartrodt, B.; Neubert, K.; Heins, J. Are diprotin A (Ile-Pro-Ile) and diprotin B (Val-Pro-Leu) inhibitors or substrates of dipeptidyl peptidase IV? Biochim. Biophys. Acta. Protein Struct. Mol. Enzymol. 1991, 1076, 314–316. [Google Scholar] [CrossRef]
- Nongonierma, A.B.; FitzGerald, R.J. Inhibition of dipeptidyl peptidase IV (DPP-IV) by tryptophan containing dipeptides. Food Funct. 2013, 4, 1843–1849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lan, V.T.T.; Ito, K.; Ito, S.; Kawarasaki, Y. Trp-Arg-Xaa tripeptides act as uncompetitive-type inhibitors of human dipeptidyl peptidase IV. Peptides 2014, 54, 166–170. [Google Scholar] [CrossRef] [PubMed]
- Nongonierma, A.B.; FitzGerald, R.J. Susceptibility of milk protein-derived peptides to dipeptidyl peptidase IV (DPP-IV) hydrolysis. Food Chem. 2014, 145, 845–852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nongonierma, A.B.; FitzGerald, R.J. Prospects for the management of type 2 diabetes using food protein-derived peptides with dipeptidyl peptidase IV (DPP-IV) inhibitory activity. Curr. Opin. Food. Sci. 2016, 8, 19–24. [Google Scholar] [CrossRef] [Green Version]
- Liebler, D.C.; Zimmerman, L.J. Targeted quantitation of proteins by mass spectrometry. Biochemistry 2013, 52, 3797–3806. [Google Scholar] [CrossRef]
- Priyanto, A.D.; Doerksen, R.J.; Chang, C.-I.; Sung, W.-C.; Widjanarko, S.B.; Kusnadi, J.; Lin, Y.-C.; Wang, T.-C.; Hsu, J.-L. Screening, discovery, and characterization of angiotensin-I converting enzyme inhibitory peptides derived from proteolytic hydrolysate of bitter melon seed proteins. J. Proteom. 2015, 128, 424–435. [Google Scholar] [CrossRef]
- Shih, Y.-H.; Chen, F.-A.; Wang, L.-F.; Hsu, J.-L. Discovery and study of novel antihypertensive peptides derived from Cassia obtusifolia seeds. J. Agric. Food Chem. 2019, 67, 7810–7820. [Google Scholar] [CrossRef]
- Hatanaka, T.; Inoue, Y.; Arima, J.; Kumagai, Y.; Usuki, H.; Kawakami, K.; Kimura, M.; Mukaihara, T. Production of dipeptidyl peptidase IV inhibitory peptides from defatted rice bran. Food Chem. 2012, 134, 797–802. [Google Scholar] [CrossRef]
- Rudolph, S.; Lunow, D.; Kaiser, S.; Henle, T. Identification and quantification of ACE-inhibiting peptides in enzymatic hydrolysates of plant proteins. Food Chem. 2017, 224, 19–25. [Google Scholar] [CrossRef]
- Salmaso, V.; Moro, S. Bridging molecular docking to molecular dynamics in exploring ligand-protein recognition process: An overview. Front. Pharmacol. 2018, 9, 923. [Google Scholar] [CrossRef] [Green Version]
- Meduru, H.; Wang, Y.-T.; Tsai, J.J.; Chen, Y.-C. Finding a potential dipeptidyl peptidase-4 (DPP-4) inhibitor for type-2 diabetes treatment based on molecular docking, pharmacophore generation, and molecular dynamics simulation. Int. J. Mol. Sci. 2016, 17, 920. [Google Scholar] [CrossRef] [Green Version]
- Meng, X.-Y.; Zhang, H.-X.; Mezei, M.; Cui, M. Molecular docking: A powerful approach for structure-based drug discovery. Curr. Comput. Aided Drug Des. 2011, 7, 146–157. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Wang, L.-J.; Jiang, B.; Li, X.-q.; Guo, C.-l.; Guo, S.-j.; Shi, D.-Y. Recent progress of the development of dipeptidyl peptidase-4 inhibitors for the treatment of type 2 diabetes mellitus. Eur. J. Med. Chem. 2018, 151, 145–157. [Google Scholar] [CrossRef] [Green Version]
- Hiramatsu, H.; Yamamoto, A.; Kyono, K.; Higashiyama, Y.; Fukushima, C.; Shima, H.; Sugiyama, S.; Inaka, K.; Shimizu, R. The crystal structure of human dipeptidyl peptidase IV (DPPIV) complex with diprotin A. Biol. Chem. 2004, 385, 561–564. [Google Scholar] [CrossRef] [PubMed]
- Nongonierma, A.B.; Mooney, C.; Shields, D.C.; FitzGerald, R.J. In silico approaches to predict the potential of milk protein-derived peptides as dipeptidyl peptidase IV (DPP-IV) inhibitors. Peptides 2014, 57, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Chen, X.; Wu, J.; Zhou, Y.; Qian, Y.; Fang, M.; Xie, J.; Wei, D. Dipeptidyl peptidase IV inhibitory peptides from Chlorella vulgaris: In silico gastrointestinal hydrolysis and molecular mechanism. Eur. Food Res. Technol. 2017, 243, 1739–1748. [Google Scholar] [CrossRef]
- He, J.; Chu, Y. Small-molecule GLP-1 secretagogs: Challenges and recent advances. Drug Discov. Today 2020, 25, 2023–2029. [Google Scholar] [CrossRef] [PubMed]
- Kuhre, R.E.; Albrechtsen, N.J.W.; Deacon, C.F.; Balk-Møller, E.; Rehfeld, J.F.; Reimann, F.; Gribble, F.M.; Holst, J.J. Peptide production and secretion in GLUTag, NCI-H716 and STC-1 cells: A comparison to native L-cells. J. Mol. Endocrinol. 2016, 56, 201. [Google Scholar] [CrossRef] [Green Version]
- Kamakura, R.; Raza, G.S.; Prasannan, A.; Walkowiak, J.; Herzig, K.-H. Dipeptidyl peptidase 4 and GLP-1 interplay in STC-1 and GLUTag cell lines. Peptides 2020, 134, 170419. [Google Scholar] [CrossRef]
- Diakogiannaki, E.; Gribble, F.M.; Reimann, F. Nutrient detection by incretin hormone secreting cells. Physiol. Behav. 2012, 106, 387–393. [Google Scholar] [CrossRef] [Green Version]
- Verhoeckx, K.; Cotter, P.; López-Expósito, I.; Kleiveland, C.; Lea, T.; Mackie, A.; Requena, T.; Swiatecka, D.; Wichers, H. The Impact of Food Bioactives on Health: In Vitro and Ex Vivo Models; Springer Nature: London, UK, 2015. [Google Scholar]
- Brubaker, P.; Schloos, J.; Drucker, D. Regulation of glucagon-like peptide-1 synthesis and secretion in the GLUTag enteroendocrine cell line. Endocrinology 1998, 139, 4108–4114. [Google Scholar] [CrossRef]
- Komatsu, Y.; Wada, Y.; Izumi, H.; Shimizu, T.; Takeda, Y.; Hira, T.; Hara, H. Casein materials show different digestion patterns using an in vitro gastrointestinal model and different release of glucagon-like peptide-1 by enteroendocrine GLUTag cells. Food Chem. 2019, 277, 423–431. [Google Scholar] [CrossRef]
- Geraedts, M.C.; Troost, F.J.; Fischer, M.A.; Edens, L.; Saris, W.H. Direct induction of CCK and GLP-1 release from murine endocrine cells by intact dietary proteins. Mol. Nutr. Food Res. 2011, 55, 476–484. [Google Scholar] [CrossRef] [PubMed]
- Gillespie, A.L.; Green, B.D. The bioactive effects of casein proteins on enteroendocrine cell health, proliferation and incretin hormone secretion. Food Chem. 2016, 211, 148–159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Halloran, F.; Bruen, C.; McGrath, B.; Schellekens, H.; Murray, B.; Cryan, J.F.; Kelly, A.L.; McSweeney, P.L.; Giblin, L. A casein hydrolysate increases GLP-1 secretion and reduces food intake. Food Chem. 2018, 252, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Lacroix, I.M.; Li-Chan, E.C. Inhibition of dipeptidyl peptidase (DPP)-IV and α-glucosidase activities by pepsin-treated whey proteins. J. Agric. Food Chem. 2013, 61, 7500–7506. [Google Scholar] [CrossRef] [PubMed]
- Nongonierma, A.B.; FitzGerald, R.J. Dipeptidyl peptidase IV inhibitory and antioxidative properties of milk protein-derived dipeptides and hydrolysates. Peptides 2013, 39, 157–163. [Google Scholar] [CrossRef] [Green Version]
- Gillespie, A.L.; Calderwood, D.; Hobson, L.; Green, B.D. Whey proteins have beneficial effects on intestinal enteroendocrine cells stimulating cell growth and increasing the production and secretion of incretin hormones. Food Chem. 2015, 189, 120–128. [Google Scholar] [CrossRef]
- Power-Grant, O.; Bruen, C.; Brennan, L.; Giblin, L.; Jakeman, P.; FitzGerald, R. In vitro bioactive properties of intact and enzymatically hydrolysed whey protein: Targeting the enteroinsular axis. Food Funct. 2015, 6, 972–980. [Google Scholar] [CrossRef]
- Sánchez-Moya, T.; Planes-Muñoz, D.; Frontela-Saseta, C.; Ros-Berruezo, G.; López-Nicolás, R. Milk whey from different animal species stimulates the in vitro release of CCK and GLP-1 through a whole simulated intestinal digestion. Food Funct. 2020, 11, 7208–7216. [Google Scholar] [CrossRef]
- Tomaro-Duchesneau, C.; LeValley, S.L.; Röth, D.; Sun, L.; Horrigan, F.T.; Kalkum, M.; Hyser, J.M.; Britton, R.A. Discovery of a bacterial peptide as a modulator of GLP-1 and metabolic disease. Sci. Rep. 2020, 10, 4922. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le Nevé, B.; Daniel, H. Selected tetrapeptides lead to a GLP-1 release from the human enteroendocrine cell line NCI-H716. Regul. Pept. 2011, 167, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Caron, J.; Cudennec, B.; Domenger, D.; Belguesmia, Y.; Flahaut, C.; Kouach, M.; Lesage, J.; Goossens, J.-F.; Dhulster, P.; Ravallec, R. Simulated GI digestion of dietary protein: Release of new bioactive peptides involved in gut hormone secretion. Food Res. Int. 2016, 89, 382–390. [Google Scholar] [CrossRef]
- Domenger, D.; Caron, J.; Belguesmia, Y.; Lesage, J.; Dhulster, P.; Ravallec, R.; Cudennec, B. Bioactivities of hemorphins released from bovine haemoglobin gastrointestinal digestion: Dual effects on intestinal hormones and DPP-IV regulations. J. Funct. Foods 2017, 36, 9–17. [Google Scholar] [CrossRef]
- Ojo, O.; Conlon, J.; Flatt, P.; Abdel-Wahab, Y. Frog skin peptides (tigerinin-1R, magainin-AM1,-AM2, CPF-AM1, and PGla-AM1) stimulate secretion of glucagon-like peptide 1 (GLP-1) by GLUTag cells. Biochem. Biophys. Res. Commun. 2013, 431, 14–18. [Google Scholar] [CrossRef] [PubMed]
- McLaughlin, C.M.; Lampis, S.; Mechkarska, M.; Coquet, L.; Jouenne, T.; King, J.D.; Mangoni, M.L.; Lukic, M.L.; Scorciapino, M.A.; Conlon, J.M. Purification, conformational analysis, and properties of a family of tigerinin peptides from skin secretions of the crowned bullfrog Hoplobatrachus occipitalis. J. Nat. Prod. 2016, 79, 2350–2356. [Google Scholar] [CrossRef] [PubMed]
- Owolabi, B.O.; Ojo, O.O.; Srinivasan, D.K.; Conlon, J.M.; Flatt, P.R.; Abdel-Wahab, Y.H. In vitro and in vivo insulinotropic properties of the multifunctional frog skin peptide hymenochirin-1B: A structure–activity study. Amino Acids 2016, 48, 535–547. [Google Scholar] [CrossRef]
- Diakogiannaki, E.; Pais, R.; Tolhurst, G.; Parker, H.E.; Horscroft, J.; Rauscher, B.; Zietek, T.; Daniel, H.; Gribble, F.M.; Reimann, F. Oligopeptides stimulate glucagon-like peptide-1 secretion in mice through proton-coupled uptake and the calcium-sensing receptor. Diabetologia 2013, 56, 2688–2696. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.; Reimer, R.A. Dairy protein and leucine alter GLP-1 release and mRNA of genes involved in intestinal lipid metabolism in vitro. Nutrition 2009, 25, 340–349. [Google Scholar] [CrossRef] [Green Version]
- Pais, R.; Gribble, F.M.; Reimann, F. Signalling pathways involved in the detection of peptones by murine small intestinal enteroendocrine L-cells. Peptides 2016, 77, 9–15. [Google Scholar] [CrossRef] [Green Version]
- Reimann, F.; Williams, L.; da Silva Xavier, G.; Rutter, G.; Gribble, F. Glutamine potently stimulates glucagon-like peptide-1 secretion from GLUTag cells. Diabetologia 2004, 47, 1592–1601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Srinivasan, K.; Viswanad, B.; Asrat, L.; Kaul, C.; Ramarao, P. Combination of high-fat diet-fed and low-dose streptozotocin-treated rat: A model for type 2 diabetes and pharmacological screening. Pharmacol. Res. 2005, 52, 313–320. [Google Scholar] [CrossRef] [PubMed]
- Skovsø, S. Modeling type 2 diabetes in rats using high fat diet and streptozotocin. J. Diabetes Investig. 2014, 5, 349–358. [Google Scholar] [CrossRef]
- Goyal, S.N.; Reddy, N.M.; Patil, K.R.; Nakhate, K.T.; Ojha, S.; Patil, C.R.; Agrawal, Y.O. Challenges and issues with streptozotocin-induced diabetes—A clinically relevant animal model to understand the diabetes pathogenesis and evaluate therapeutics. Chem.-Biol. Interact. 2016, 244, 49–63. [Google Scholar] [CrossRef]
- Mahmood, R.; Kayani, W.K.; Ahmed, T.; Malik, F.; Hussain, S.; Ashfaq, M.; Ali, H.; Rubnawaz, S.; Green, B.D.; Calderwood, D. Assessment of antidiabetic potential and phytochemical profiling of Rhazya stricta root extracts. BMC Complement. Altern. Med. 2020, 20, 293. [Google Scholar] [CrossRef]
- Borde, M.; Mohanty, I.; Maheshwari, U.; Suman, R.; Deshmukh, Y. Natural dipeptidyl peptidase-4 inhibitor Terminalia arjuna mitigates myocardial infarction co-existing with diabetes in experimental rats. J. Diabetes Metab. Disord. Control 2018, 5, 48–56. [Google Scholar]
- Srivastava, S.; Shree, P.; Tripathi, Y.B. Active phytochemicals of Pueraria tuberosa for DPP-IV inhibition: In silico and experimental approach. J. Diabetes Metab. Disord. 2017, 16, 46. [Google Scholar] [CrossRef] [Green Version]
- Velarde-Salcedo, A.J.; Regalado-Rentería, E.; Velarde-Salcedo, R.; Juárez-Flores, B.I.; Barrera-Pacheco, A.; De Mejía, E.G.; De La Rosa, A.P.B. Consumption of amaranth induces the accumulation of the antioxidant protein paraoxonase/arylesterase 1 and modulates dipeptidyl peptidase iv activity in plasma of streptozotocin-induced hyperglycemic rats. Lifestyle Genom. 2017, 10, 181–193. [Google Scholar] [CrossRef]
- González-Abuín, N.; Martínez-Micaelo, N.; Blay, M.; Pujadas, G.; Garcia-Vallvé, S.; Pinent, M.; Ardévol, A. Grape seed-derived procyanidins decrease dipeptidyl-peptidase 4 activity and expression. J. Agric. Food Chem. 2012, 60, 9055–9061. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.-L.; Hung, C.-C.; Jao, C.-L.; Tung, Y.-S.; Hsu, K.-C. Porcine skin gelatin hydrolysate as a dipeptidyl peptidase IV inhibitor improves glycemic control in streptozotocin-induced diabetic rats. J. Funct. Foods 2014, 11, 235–242. [Google Scholar] [CrossRef]
- Hsieh, C.; Wang, T.; Hung, C.; Chen, M.; Hsu, K. Improvement of glycemic control in streptozotocin-induced diabetic rats by Atlantic salmon skin gelatin hydrolysate as the dipeptidyl-peptidase IV inhibitor. Food Funct. 2015, 6, 1887–1892. [Google Scholar] [CrossRef]
- Ishikawa, Y.; Hira, T.; Inoue, D.; Harada, Y.; Hashimoto, H.; Fujii, M.; Kadowaki, M.; Hara, H. Rice protein hydrolysates stimulate GLP-1 secretion, reduce GLP-1 degradation, and lower the glycemic response in rats. Food Funct. 2015, 6, 2525–2534. [Google Scholar] [CrossRef]
- Mochida, T.; Hira, T.; Hara, H. The corn protein, zein hydrolysate, administered into the ileum attenuates hyperglycemia via its dual action on glucagon-like peptide-1 secretion and dipeptidyl peptidase-IV activity in rats. Endocrinology 2010, 151, 3095–3104. [Google Scholar] [CrossRef] [Green Version]
- Hsieh, C.-H.; Wang, T.-Y.; Hung, C.-C.; Jao, C.-L.; Hsieh, Y.-L.; Wu, S.-X.; Hsu, K.-C. In silico, in vitro and in vivo analyses of dipeptidyl peptidase IV inhibitory activity and the antidiabetic effect of sodium caseinate hydrolysate. Food Funct. 2016, 7, 1122–1128. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, N.; Kolibabka, M.; Busch, S.; Bugert, P.; Kaiser, U.; Lin, J.; Fleming, T.; Morcos, M.; Klein, T.; Schlotterer, A. The DPP4 inhibitor linagliptin protects from experimental diabetic retinopathy. PLoS ONE 2016, 11, e0167853. [Google Scholar] [CrossRef]
- Uenishi, H.; Kabuki, T.; Seto, Y.; Serizawa, A.; Nakajima, H. Isolation and identification of casein-derived dipeptidyl-peptidase 4 (DPP-4)-inhibitory peptide LPQNIPPL from gouda-type cheese and its effect on plasma glucose in rats. Int. Dairy J. 2012, 22, 24–30. [Google Scholar] [CrossRef]
- Lu, Y.; Lu, P.; Wang, Y.; Fang, X.; Wu, J.; Wang, X. A Novel Dipeptidyl Peptidase IV Inhibitory Tea Peptide Improves Pancreatic β-Cell Function and Reduces α-Cell Proliferation in Streptozotocin-Induced Diabetic Mice. Int. J. Mol. Sci. 2019, 20, 322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sartorius, T.; Weidner, A.; Dharsono, T.; Boulier, A.; Wilhelm, M.; Schön, C. Postprandial Effects of a Proprietary Milk Protein Hydrolysate Containing Bioactive Peptides in Prediabetic Subjects. Nutrients 2019, 11, 1700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amigo-Benavent, M.; Power-Grant, O.; FitzGerald, R.J.; Jakeman, P. The insulinotropic and incretin response to feeding a milk based protein matrix in healthy young women. J. Funct. Foods 2020, 72, 104056. [Google Scholar] [CrossRef]
- Hovland, I.H.; Leikanger, I.S.; Stokkeland, O.; Waage, K.H.; Mjøs, S.A.; Brokstad, K.A.; McCann, A.; Ueland, P.M.; Slizyte, R.; Carvajal, A. Effects of low doses of fish and milk proteins on glucose regulation and markers of insulin sensitivity in overweight adults: A randomised, double blind study. Eur. J. Nutr. 2019, 59, 1013–1029. [Google Scholar] [CrossRef]
- Drummond, E.; Flynn, S.; Whelan, H.; Nongonierma, A.B.; Holton, T.r.s.A.; Robinson, A.; Egan, T.; Cagney, G.; Shields, D.C.; Gibney, E.R. Casein hydrolysate with glycemic control properties: Evidence from cells, animal models, and humans. J. Agric. Food Chem. 2018, 66, 4352–4363. [Google Scholar] [CrossRef] [PubMed]
- Devasia, S.; Kumar, S.; Stephena, P.; Inoue, N.; Sugihara, F.; Suzuki, K. Double blind, randomized clinical study to evaluate efficacy of collagen peptide as add on nutritional supplement in Type 2 diabetes. J. Clin. Nutr. Food Sci. 2018, 1, 6–11. [Google Scholar]
- Saleh, L.; Schrier, N.L.; Bruins, M.J.; Steegers, E.A.; van den Meiracker, A.H.; Visser, W. Effect of oral protein hydrolysate on glucose control in patients with gestational diabetes. Clin. Nutr. 2018, 37, 878–883. [Google Scholar] [CrossRef]
- Gallego, M.; Mora, L.; Hayes, M.; Reig, M.; Toldrá, F. Peptides with potential cardioprotective effects derived from dry-cured ham byproducts. J. Agric. Food Chem. 2019, 67, 1115–1126. [Google Scholar] [CrossRef] [PubMed]
- Valencia, I.; Peiró, C.; Lorenzo, Ó.; Sánchez-Ferrer, C.F.; Eckel, J.; Romacho, T. DPP4 and ACE2 in diabetes and COVID-19: Therapeutic targets for cardiovascular complications? Front. Pharmacol. 2020, 11, 1161. [Google Scholar] [CrossRef] [PubMed]

| Precursor Protein | Sequence | IC50 | Mode of Inhibition | Peptide Hydrolyzed by DPP-IV | Docking | Cell Model/In Vivo | Reference |
|---|---|---|---|---|---|---|---|
| Boarfish protein hydrolysate | IPVDM | 21.72 µM | nd | nd | nd | BRIN-BD11 cells Caco-2 cell | [36] |
| APIT | 34.73 µM | nd | nd | nd | |||
| VPTP | 38.93 µM | nd | nd | nd | |||
| GPIN | 48.96 µM | nd | nd | nd | |||
| LPVYD | 51.36 µM | nd | nd | nd | |||
| LPVDM | 53.50 µM | nd | nd | nd | |||
| APLER | 63.67 µM | nd | nd | nd | |||
| IPGA | 66.37 µM | nd | nd | nd | |||
| GPSL | 68.13 µM | nd | nd | nd | |||
| GPSI | 73.15 µM | nd | nd | nd | |||
| APVP | 79.10 µM | nd | nd | nd | |||
| VPDPR | 90.37 µM | nd | nd | nd | |||
| APLT | 91.10 µM | nd | nd | nd | |||
| APLT | 115.27 µM | nd | nd | nd | |||
| MPAVP | 116.27 µM | nd | nd | nd | |||
| GPGI | 116.37 µM | nd | nd | nd | |||
| GPLN | 126.51 µM | nd | nd | nd | |||
| PAVP | 131.90 µM | nd | nd | nd | |||
| GPGL | 154.12 µM | nd | nd | nd | |||
| LPGA | 164.37 µM | nd | nd | nd | |||
| AALP | 164.52 µM | nd | nd | nd | |||
| TPTV | 251.58 µM | nd | nd | nd | |||
| AAIP | 261.58 µM | nd | nd | nd | |||
| YPL(pS)L | 261.46 µM | nd | nd | nd | |||
| TPGI | 282.16 µM | nd | nd | nd | |||
| TPGL | 297.41 µM | nd | nd | nd | |||
| YPII(pS) | 300.67 µM | nd | nd | nd | |||
| YPIL(pS) | 302.47 µM | nd | nd | nd | |||
| ISAP | 393.88 µM | nd | nd | nd | |||
| YPL(pT)V | 511.47 µM | nd | nd | nd | |||
| YPLV(pT) | 508.37 µM | nd | nd | nd | |||
| α-lactalbumin-rich whey protein | LDQWLCEKL | 131 μM | Non-competitive | nd | nd | nd | [35] |
| EQLTKCEVFR | 883 μM | nd | nd | nd | nd | ||
| KILDKVGINYWLAHK | 930 μM | nd | nd | nd | nd | ||
| ILDKVGINYWLAHK | 456 μM | nd | nd | nd | nd | ||
| VGINYWLAHK | 765 μM | Mixed-type | nd | nd | nd | ||
| Atlantic salmon (Salmo salar) skin | YYGYTGAFR | 91.24 µM (0.1 mg/mL) | Mixed-type | nd | Yes | nd | [3] |
| LDKVFR | 231.95 µM (0.18 mg/mL) | Competitive | nd | Yes | nd | ||
| VLATSGPG | 1.73 mM (1.21 mg/mL) | Non-competitive | nd | Yes | nd | ||
| Porphyra dioica protein | YLVA | 439.5 μM | nd | nd | nd | nd | [97] |
| Soft-shelled turtle yolk hydrolysate | VPGLAL | 289.2 μM | Competitive | Yes | nd | nd | [2] |
| LPSW | 269.7 μM | Competitive | Yes | nd | nd | ||
| LPLF | 463.6 μM | Competitive | Yes | nd | nd | ||
| WLQL | 432.5 μM | Uncompetitive | No | nd | nd | ||
| Egg white ovalbumin | CF | 2.99 mM | nd | nd | nd | nd | [1] |
| KM | 2.22 mM | nd | nd | nd | nd | ||
| ELPF | 9.92 mM | nd | nd | nd | nd | ||
| AM | 2.79 mM | nd | nd | nd | nd | ||
| ADHPF | 1.66 mM | nd | nd | nd | nd | ||
| LPR | 1.43 mM | nd | nd | nd | nd | ||
| PR | 4.11 mM | nd | nd | nd | nd | ||
| FR | 2.47 mM | nd | nd | nd | nd | ||
| PRM | 2.50 mM | nd | nd | nd | nd | ||
| GR | 2.83 mM | nd | nd | nd | nd | ||
| Hen egg proteins | ADF | 16.83 mM | nd | nd | Yes | nd | [30] |
| MIR | 4.86 mM | nd | nd | Yes | nd | ||
| FGR | 46.22 mM | nd | nd | Yes | nd | ||
| CDR | 24.49 mM | nd | nd | nd | nd | ||
| Dark tea protein | MSLYPR | 1.76 mM (1.35 mg/mL) | nd | nd | Yes | nd | [31] |
| QGQELLPSDFK | 3.08 mM (3.89 mg/mL) | nd | nd | Yes | nd | ||
| Brewers’ spent grain | APLP | 122.45 µM | nd | nd | nd | nd | [52] |
| IPIPQ | 75.70 µM | nd | nd | nd | nd | ||
| IPLQP | 105.45 µM | nd | nd | nd | nd | ||
| IPVP | 38.67 µM | nd | nd | nd | nd | ||
| IPY | 52.15 µM | nd | nd | nd | nd | ||
| LAVP | 98.76 µM | nd | nd | nd | nd | ||
| LPIA | 45.07 µM | nd | nd | nd | nd | ||
| LPVP | 105.25 µM | nd | nd | nd | nd | ||
| LPY | 87.15 µM | nd | nd | nd | nd | ||
| PAIP | 111.18 µM | nd | nd | nd | nd | ||
| S. platensis protein | LRSELAAWSR | 140.94 µM (167.3 µg/mL) | nd | nd | Yes | HepG2 cell | [123] |
| Camel whey protein | VPV | 6.6 µM | Competitive | nd | nd | nd | [8] |
| YPI | 35.0 µM | Competitive | nd | nd | nd | ||
| VPF | 55.1 µM | Competitive | nd | nd | nd | ||
| EPVK | 330.1 µM | Competitive | nd | nd | nd | ||
| LAHKPL | 239.7 µM | Competitive | nd | nd | nd | ||
| YPLR | 360.1 µM | Competitive | nd | nd | nd | ||
| Camel milk protein | ILDKEGIDY | 347.8 µM | Mixed-type | No | nd | nd | [48] |
| ILDKVGINY | 321.5 µM | Mixed-type | No | nd | nd | ||
| ILELA | 721.1 µM | Competitive | No | nd | nd | ||
| LLQLEAIR | 177.8 µM | Competitive | No | nd | nd | ||
| LPVP | 87.0 µM | Competitive | No | nd | nd | ||
| MPVQA | 93.3 µM | Competitive | Yes | nd | nd | ||
| SPVVPF | 214.1 µM | Mixed-type | Yes | nd | nd | ||
| YPVEPF | 138.0 µM | Mixed-type | Yes | nd | nd | ||
| Silver carp (Hypophthalmichthys molitrix) muscle hydrolysate | LLDLGVP | 2665.46 µM | nd | nd | nd | nd | [96] |
| AALEQTER | 647.02 µM | nd | nd | nd | nd | ||
| ILYGDFK | 2614.14 µM | nd | nd | nd | nd | ||
| KAVGEPPLF | 1317.39 µM | nd | nd | nd | nd | ||
| GPAGPQGPR | 842.10 µM | nd | nd | nd | nd | ||
| Synthetic | PFP | 1389.14 µM | Uncompetitive | nd | Yes | 3T3-L1 | [124] |
| YPL | 364.62 µM | Uncompetitive | nd | Yes | |||
| YPG | 173.96 µM | Uncompetitive | nd | Yes | |||
| Egg white | IRDLLER | 186.23 μM | nd | nd | Yes | Caco-2 cell/Wistar male rats | [41] |
| YAEERYP | 340.62 μM | nd | nd | Yes | |||
| IRNVLQPS | 598.28 μM | nd | nd | Yes | |||
| Napin of rapeseed (Brassica napus) | PAGPF | 135.70 μM | Mixed-type | nd | Yes | nd | [80] |
| IPQVS | 52.16 μM | Competitive | nd | Yes | nd | ||
| ELHQEEPL | 78.46 μM | Mixed-type | nd | Yes | nd | ||
| KTMPGP | 162.73 μM | Uncompetitive | nd | Yes | nd | ||
| Analogs of ile-Pro-ile | APA | 43.3 μM | Competitive | No | Yes | nd | [42] |
| APF | 65.8 μM | Competitive | No | Yes | nd | ||
| APR | 119.7 μM | Competitive | No | Yes | nd | ||
| IPA | 28.3 μM | Competitive | No | Yes | nd | ||
| KPA | 74.5 μM | Competitive | No | Yes | nd | ||
| FPF | 247 μM | Competitive | Yes | Yes | nd | ||
| FPI | 45.2 μM | Competitive | No | Yes | nd | ||
| FPW | 54.9 μM | Competitive | Yes | Yes | nd | ||
| IPF | 47.3 μM | Competitive | Yes | Yes | nd | ||
| IPW | 175.3 μM | Competitive | Yes | Yes | nd | ||
| WPF | 159.8 μM | Mixed-type | Yes | Yes | nd | ||
| WPT | 133 μM | Mixed-type | No | Yes | nd | ||
| WPW | 120.1 μM | Mixed-type | Yes | Yes | nd | ||
| Water-soluble extracts from natto | KL | 159.84 μM (41.40 µg/mL) | nd | nd | nd | nd | [94] |
| LR | 2.08 mM (598.02 µg/mL) | nd | nd | nd | nd |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nong, N.T.P.; Hsu, J.-L. Characteristics of Food Protein-Derived Antidiabetic Bioactive Peptides: A Literature Update. Int. J. Mol. Sci. 2021, 22, 9508. https://doi.org/10.3390/ijms22179508
Nong NTP, Hsu J-L. Characteristics of Food Protein-Derived Antidiabetic Bioactive Peptides: A Literature Update. International Journal of Molecular Sciences. 2021; 22(17):9508. https://doi.org/10.3390/ijms22179508
Chicago/Turabian StyleNong, Nhung Thi Phuong, and Jue-Liang Hsu. 2021. "Characteristics of Food Protein-Derived Antidiabetic Bioactive Peptides: A Literature Update" International Journal of Molecular Sciences 22, no. 17: 9508. https://doi.org/10.3390/ijms22179508
APA StyleNong, N. T. P., & Hsu, J.-L. (2021). Characteristics of Food Protein-Derived Antidiabetic Bioactive Peptides: A Literature Update. International Journal of Molecular Sciences, 22(17), 9508. https://doi.org/10.3390/ijms22179508







