Protein Carbonylation: Emerging Roles in Plant Redox Biology and Future Prospects
Abstract
1. Introduction
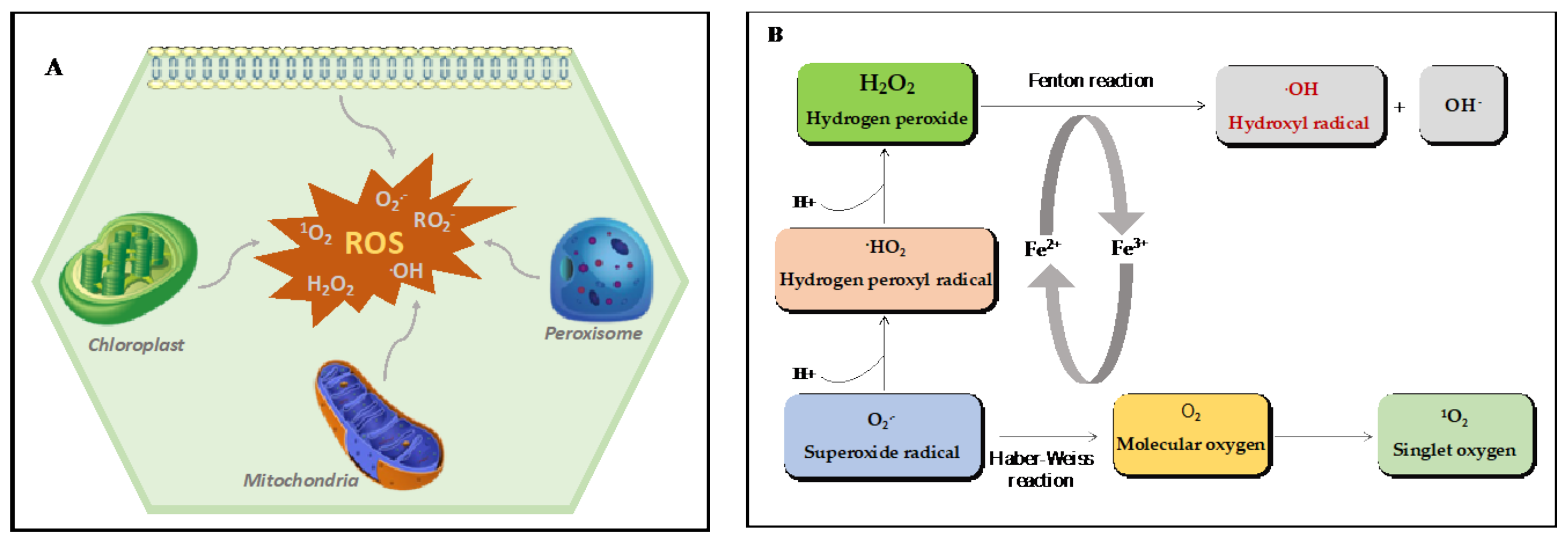
2. ROS: Diversity, Reactivity, and Sites of Production in Plants
2.1. ROS Diversity and Reactivity
2.1.1. Singlet oxygen (1O2)
2.1.2. Superoxide Anion (O2•−)
2.1.3. Hydrogen Peroxide (H2O2)
2.1.4. Hydroxyl Radical (HO•)
2.2. ROS Production Sites in Plants
2.2.1. Chloroplasts
2.2.2. Mitochondria
2.2.3. Peroxisomes
2.2.4. Apoplasts (Plasma Membrane and the Cell Wall)
3. Common ROS-Mediated Post-Translational Modification (PTMs)
3.1. Methionine Oxidation
3.2. Cysteinylation (Cysteine Oxidation) and Glutathionylation
3.3. Nitrosylation
3.4. Persulfidation
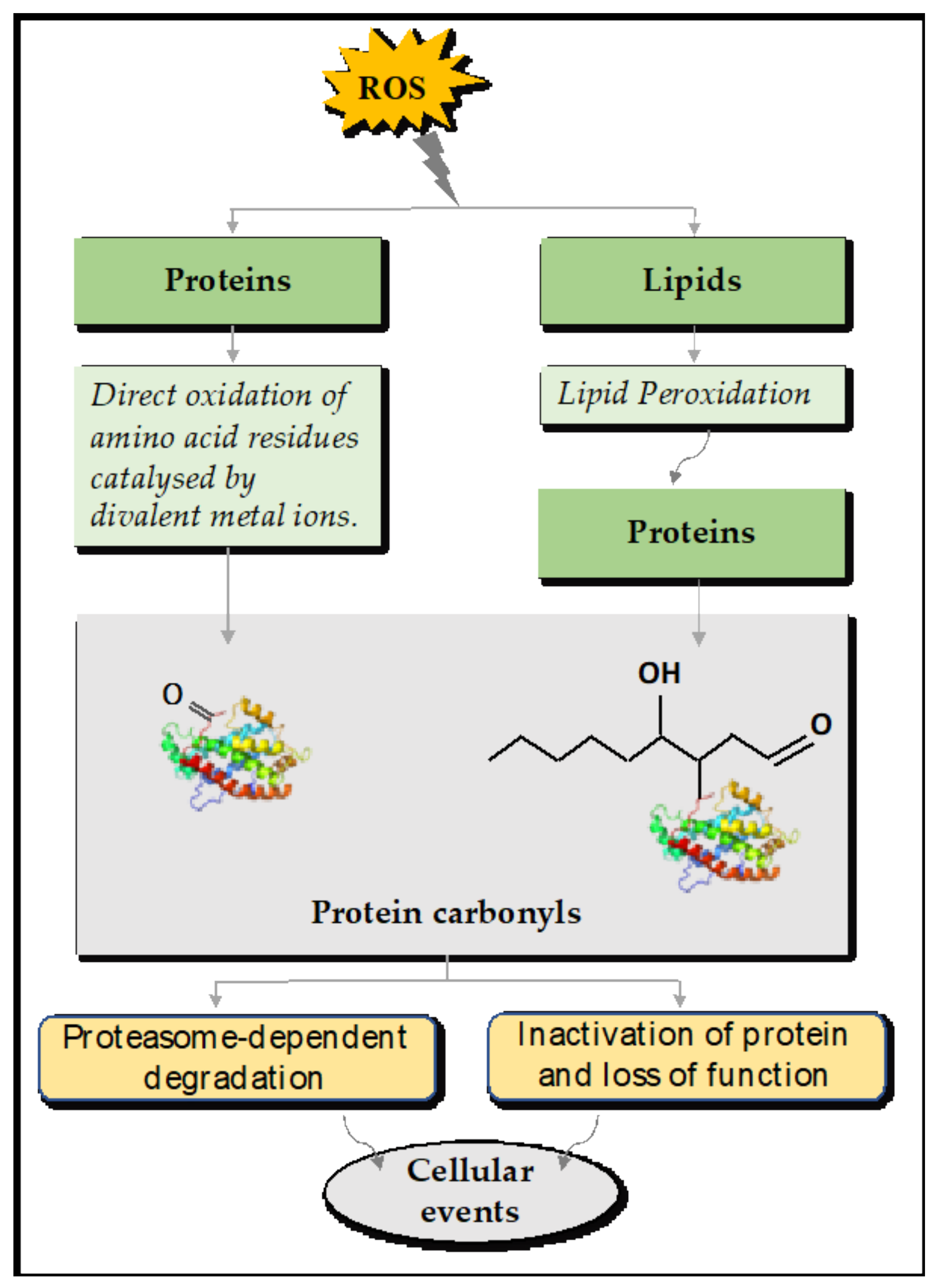
4. Details of Protein Carbonylation
4.1. Direct and Indirect Reactions of Protein Carbonylation
4.2. The Fates of Carbonylated Proteins
5. Importance of Protein Carbonylation in Seed After-Ripening and Germination
6. Importance of Protein Carbonylation in Proteome Remodeling under Nutrient Starvation and Stress Conditions
7. Protein Carbonylation Serves as a Signal Transduction Mechanism in Bacteria and Mammalian Cells
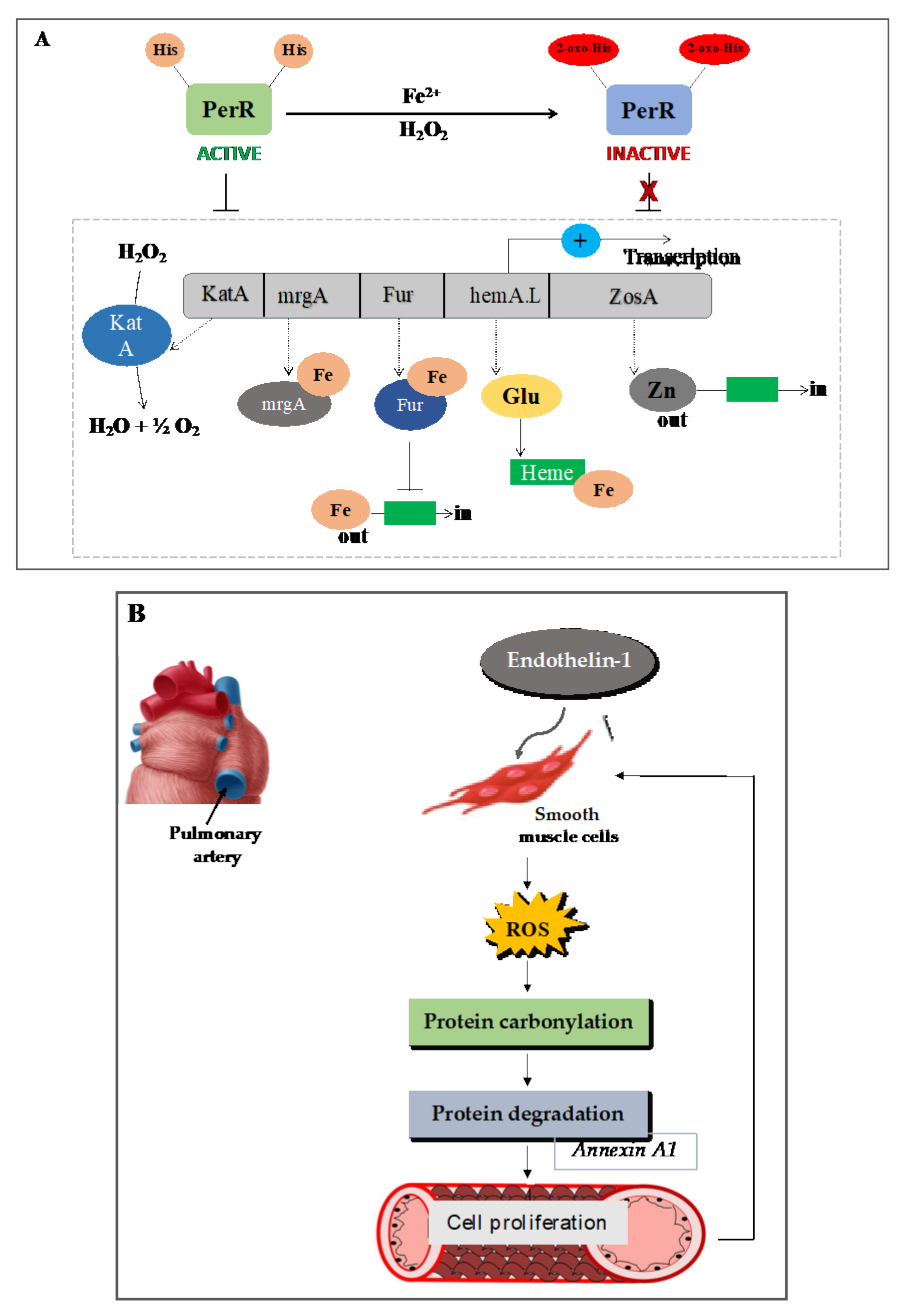
7.1. Carbonylation of the Transcription Repressor PerR Facilitates H2O2 Sensing and the Expression of Oxidative Response Genes in Prokaryotes
7.2. In Animals: Mammalian Cell
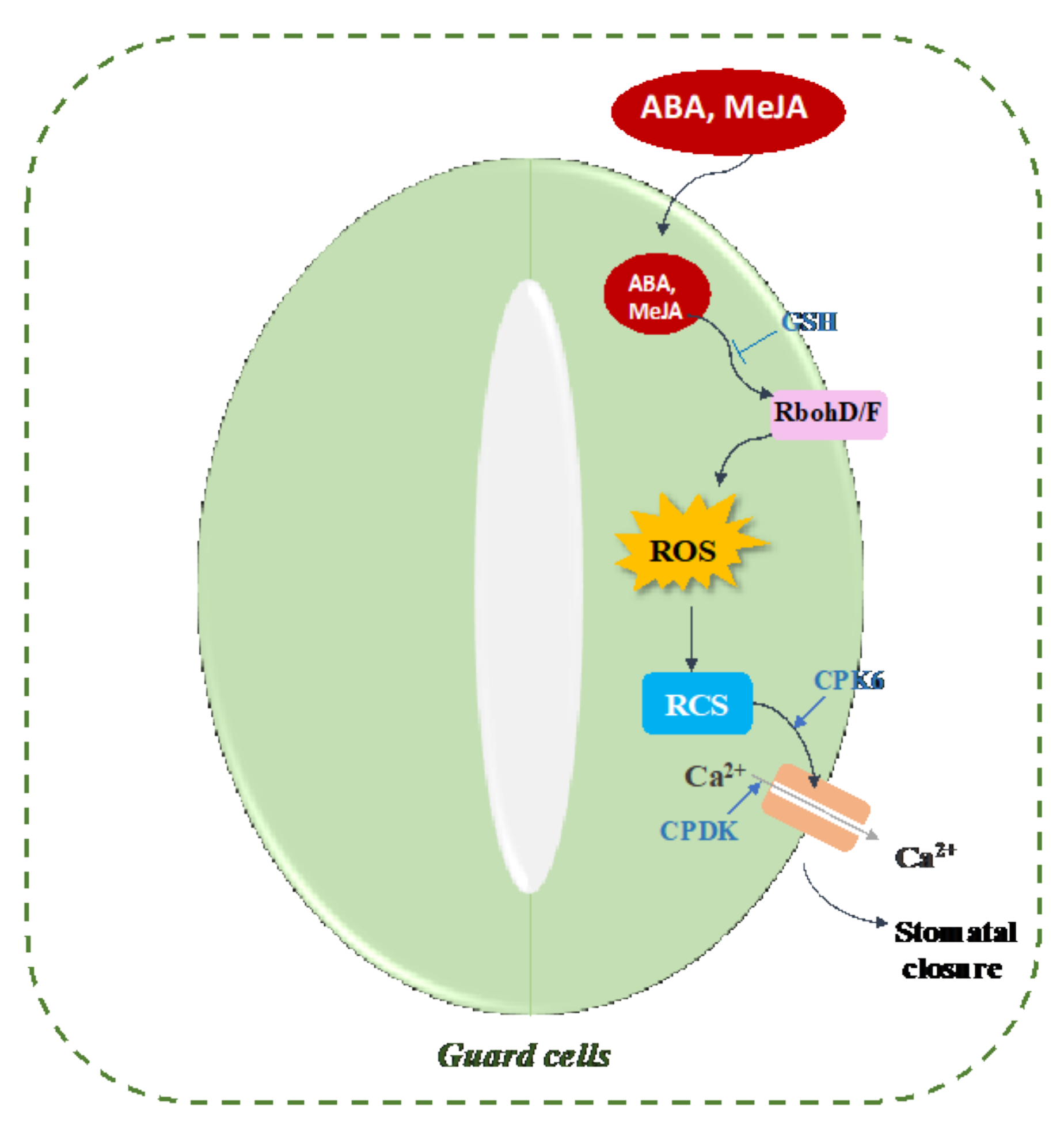
8. The Implication of Protein Carbonylation in Phytohormone Signaling Pathways
9. Crosstalk Between Carbonylation and Other PTMs
10. Target Specificity in Protein Carbonylation
11. Challenges and Approaches for Studying the Roles of Protein Carbonylation in Plants: Lessons from Studies in Mammalians
12. Conclusion and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mittler, R. ROS Are Good. Trends Plant Sci. 2017, 22, 11–19. [Google Scholar] [CrossRef]
- Friso, G.; Van Wijk, K.J. Posttranslational protein modifications in plant metabolism. Plant Physiol. 2015, 169, 1469–1487. [Google Scholar] [CrossRef]
- Waszczak, C.; Akter, S.; Eeckhout, D.; Persiau, G.; Wahni, K.; Bodra, N.; Van Molle, I.; De Smet, B.; Vertommen, D.; Gevaert, K.; et al. Sulfenome mining in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2014, 111, 11545–11550. [Google Scholar] [CrossRef] [PubMed]
- Waszczak, C.; Carmody, M.; Kangasjärvi, J. Reactive Oxygen Species in Plant Signaling. Annu. Rev. Plant Biol. 2018, 69, 209–236. [Google Scholar] [CrossRef]
- Johansson, E.; Olsson, O.; Nyström, T. Progression and specificity of protein oxidation in the life cycle of Arabidopsis thaliana. J. Biol. Chem. 2004, 279, 22204–22208. [Google Scholar] [CrossRef] [PubMed]
- Lounifi, I.; Arc, E.; Molassiotis, A.; Job, D.; Rajjou, L.; Tanou, G. Interplay between protein carbonylation and nitrosylation in plants. Proteomics 2013, 13, 568–578. [Google Scholar] [CrossRef]
- Ayala, A.; Muñoz, M.F.; Argüelles, S. Lipid peroxidation: Production, metabolism, and signaling mechanisms of malondialdehyde and 4-hydroxy-2-nonenal. Oxid. Med. Cell. Longev. 2014, 2014, 360438. [Google Scholar] [CrossRef]
- Scott, W.; Sattler, E.; Mè Ne-Saffrané, L.; Farmer, E.E.; Krischke, M.; Mueller, M.J.; Dellapenna, D. Nonenzymatic Lipid Peroxidation Reprograms Gene Expression and Activates Defense Markers in Arabidopsis Tocopherol-Deficient Mutants. Plant Cell 2006, 18, 3706–3720. [Google Scholar] [CrossRef]
- Weber, H.; Chételat, A.; Reymond, P.; Farmer, E.E. Selective and powerful stress gene expression in Arabidopsis in response to malondialdehyde. Plant J. 2004, 37, 877–888. [Google Scholar] [CrossRef]
- Dueckershoff, K.; Mueller, S.; Mueller, M.J.; Reinders, J. Impact of cyclopentenone-oxylipins on the proteome of Arabidopsis thaliana. BBA Proteins Proteom. 2008, 1784, 1975–1985. [Google Scholar] [CrossRef]
- Job, C.; Rajjou, L.; Lovigny, Y.; Belghazi, M.; Job, D. Patterns of Protein Oxidation in Arabidopsis Seeds and during Germination. Plant Physiol. 2005, 138, 790–802. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.-L.; Yu, H.-D.; Guan, Y.; Li, J.-K.; Guo, F.-Q. Carbonylation and loss-of-function analyses of SBPase reveal its metabolic interface role in oxidative stress, carbon assimilation, and multiple aspects of growth and development in Arabidopsis. Mol. Plant 2012, 5, 1082–1099. [Google Scholar] [CrossRef]
- Nystrom, T. Role of oxidative carbonylation in protein quality control and senescence. EMBO J 2005, 24, 1311–1317. [Google Scholar] [CrossRef] [PubMed]
- Mène-Saffrané, L.; Davoine, C.; Stolz, S.; Majcherczyk, P.; Farmer, E.E. Genetic removal of tri-unsaturated fatty acids suppresses developmental and molecular phenotypes of an Arabidopsis tocopherol-deficient mutant: Whole-body mapping of malondialdehyde pools in a complex eukaryote. J. Biol. Chem. 2007, 282, 35749–35756. [Google Scholar] [CrossRef] [PubMed]
- Levine, R.L. Carbonyl modified proteins in cellular regulation, aging, and disease. Free Radic. Biol. Med. 2002, 32, 790–796. [Google Scholar] [CrossRef]
- Biswas, M.S.; Fukaki, H.; Mori, I.C.; Nakahara, K.; Mano, J. Reactive oxygen species and reactive carbonyl species constitute a feed-forward loop in auxin signaling for lateral root formation. Plant J. 2019, 100, 536–548. [Google Scholar] [CrossRef]
- Biswas, S.; Mano, J. Lipid peroxide-derived short-chain carbonyls mediate hydrogen peroxide-induced and salt-induced programmed cell death in plants. Plant Physiol. 2015, 168, 885–898. [Google Scholar] [CrossRef]
- Islam, M.M.; Ye, W.; Matsushima, D.; Munemasa, S.; Okuma, E.; Nakamura, Y.; Biswas, M.S.; Mano, J.; Murata, Y. Reactive carbonyl species mediate ABA signaling in guard cells. Plant Cell Physiol. 2016, 57, 2552–2563. [Google Scholar] [CrossRef]
- Islam, M.M.; Ye, W.; Akter, F.; Rhaman, M.S.; Matsushima, D.; Munemasa, S.; Okuma, E.; Nakamura, Y.; Biswas, M.S.; Mano, J.; et al. Reactive Carbonyl Species Mediate Methyl Jasmonate-Induced Stomatal Closure. Plant Cell Physiol. 2020, 61, 1788–1797. [Google Scholar] [CrossRef]
- Halliwell, B.; Gutteridge, J.M. Free Radicals in Biology and Medicine; Oxford University Press: Oxford, UK, 1990; Volume 58. [Google Scholar]
- Foyer, C.H.; Noctor, G. Redox sensing and signalling associated with reactive oxygen in chloroplasts, peroxisomes and mitochondria. Physiol. Plant. 2003, 119, 355–364. [Google Scholar] [CrossRef]
- Halliwell, B. Reactive Species and Antioxidants. Redox Biology Is a Fundamental Theme of Aerobic Life. Plant Physiol. 2006, 141, 312–322. [Google Scholar] [CrossRef]
- König, J.; Jung, T.; Grune, T. Protein Carbonylation in Aging and Senescence. Protein Carbonylation Princ. Anal. Biol. Implic. 2017, 272–290. [Google Scholar] [CrossRef]
- Mignolet-Spruyt, L.; Xu, E.; Idänheimo, N.; Hoeberichts, F.A.; Mühlenbock, P.; Brosché, M.; Van Breusegem, F.; Kangasjärvi, J. Spreading the news: Subcellular and organellar reactive oxygen species production and signalling. J. Exp. Bot. 2016, 67, 3831–3844. [Google Scholar] [CrossRef]
- Vaahtera, L.; Brosché, M.; Wrzaczek, M.; Kangasjärvi, J. Specificity in ROS signaling and transcript signatures. Antioxid. Redox Signal. 2014, 21, 1422–1441. [Google Scholar] [CrossRef] [PubMed]
- Davies, M.J. Reactive species formed on proteins exposed to singlet oxygen. Photochem. Photobiol. Sci. 2004, 3, 17–25. [Google Scholar] [CrossRef]
- Triantaphylidès, C.; Krischke, M.; Hoeberichts, F.A.; Ksas, B.; Gresser, G.; Havaux, M.; Van Breusegem, F.; Mueller, M.J. Singlet oxygen is the major reactive oxygen species involved in photooxidative damage to plants. Plant Physiol. 2008, 148, 960–968. [Google Scholar] [CrossRef] [PubMed]
- Fischer, B.B.; Hideg, É.; Krieger-Liszkay, A. Production, detection, and signaling of singlet oxygen in photosynthetic organisms. Antioxid. Redox Signal. 2013, 18, 2145–2162. [Google Scholar] [CrossRef] [PubMed]
- Ruban, A.V.; Johnson, M.P.; Duffy, C.D.P. The photoprotective molecular switch in the photosystem II antenna. Biochim. Biophys. Acta Bioenerg. 2012, 1817, 167–181. [Google Scholar] [CrossRef]
- Farmer, E.E.; Mueller, M.J. ROS-Mediated Lipid Peroxidation and RES-Activated Signaling. Annu. Rev. Plant Biol. 2013, 64, 429–450. [Google Scholar] [CrossRef] [PubMed]
- Havaux, M. Carotenoid oxidation products as stress signals in plants. Plant J. 2014, 79, 597–606. [Google Scholar] [CrossRef] [PubMed]
- Fridovich, I. Superoxide anion radical (O2−·), superoxide dismutases, and related matters. J. Biol. Chem. 1997, 272, 18515–18517. [Google Scholar] [CrossRef] [PubMed]
- Egorov, S.Y.; Kamalov, V.F.; Koroteev, N.I.; Krasnovsky, A.A.; Toleutaev, B.N.; Zinukov, S.V. Rise and decay kinetics of photosensitized singlet oxygen luminescence in water. Measurements with nanosecond time-correlated single photon counting technique. Chem. Phys. Lett. 1989, 163, 421–424. [Google Scholar] [CrossRef]
- Skovsen, E.; Snyder, J.W.; Lambert, J.D.C.; Ogilby, P.R. Lifetime and diffusion of singlet oxygen in a cell. J. Phys. Chem. B 2005, 109, 8570–8573. [Google Scholar] [CrossRef] [PubMed]
- Henzler, T.; Steudle, E. Transport and metabolic degradation of hydrogen peroxide in Chara corallina: Model calculations and measurements with the pressure probe suggest transport of H2O2 across water channels. J. Exp. Bot. 2000, 51, 2053–2066. [Google Scholar] [CrossRef]
- Mittler, R.; Berkowitz, G. Hydrogen peroxide, a messenger with too many roles? Redox Rep. 2001, 6, 69–72. [Google Scholar] [CrossRef] [PubMed]
- Haber, F.; Weiss, J.; Seph, J.O.; Eiss, W. The Catalytic Decom position o f Hydrogen Peroxide by Iron Salts. Proc. R. Soc. Lond. Ser. A Math. Phys. Sci. 1934, 147, 332–351. [Google Scholar]
- Kehrer, J.P. The Haber-Weiss reaction and mechanisms of toxicity. Toxicology 2000, 149, 43–50. [Google Scholar] [CrossRef]
- Halliwell, B.; Gutteridge, J.M.C.; Aruoma, O.I. The deoxyribose method: A simple “test-tube” assay for determination of rate constants for reactions of hydroxyl radicals. Anal. Biochem. 1987, 165, 215–219. [Google Scholar] [CrossRef]
- Betteridge, D.J. What is oxidative stress? Metabolism 2000, 49, 3–8. [Google Scholar] [CrossRef]
- Carter, R. Oxygen: The Molecule that made the World. JRSM 2003, 96, 46–47. [Google Scholar] [CrossRef][Green Version]
- Das, K.; Roychoudhury, A. Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants. Front. Environ. Sci. 2014, 2, 1–13. [Google Scholar] [CrossRef]
- Harrison, P.M.; Bauminger, E.R.; Hechel, D.; Hodson, N.W.; Nowik, I.; Treffry, A.; Yewdall, S.J. Mechanism of Fe(II) oxidation and core formation in ferritin. In Progress in Iron Research; Hershko, C., Konijn, A.M., Aisen, P., Eds.; Advances in Experimental Medicine and Biology; Springer: Boston, MA, USA, 1994; Volume 356, pp. 1–12. [Google Scholar] [CrossRef]
- Sweetlove, L.J.; Heazlewood, J.L.; Herald, V.; Holtzapffel, R.; Day, D.A.; Leaver, C.J.; Millar, A.H. The impact of oxidative stress on Arabidopsis mitochondria. Plant J. 2002, 32, 891–904. [Google Scholar] [CrossRef]
- Asada, K. Production and Scavenging of Reactive Oxygen Species in Chloroplasts and Their Functions. Plant Physiol. 2006, 141, 391–396. [Google Scholar] [CrossRef]
- Corpas, F.J.; Barroso, J.B.; Palma, J.M.; Rodriguez-Ruiz, M. Plant peroxisomes: A nitro-oxidative cocktail. Redox Biol. 2017, 11, 535–542. [Google Scholar] [CrossRef]
- Daudi, A.; Cheng, Z.; O’brien, J.A.; Mammarella, N.; Khan, S.; Ausubel, F.M.; Paul Bolwell, G. The Apoplastic Oxidative Burst Peroxidase in Arabidopsis Is a Major Component of Pattern-Triggered Immunity. Plant Cell 2012, 24, 275–287. [Google Scholar] [CrossRef]
- Qi, J.; Wang, J.; Gong, Z.; Zhou, J.M. Apoplastic ROS signaling in plant immunity. Curr. Opin. Plant Biol. 2017, 38, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Asada, K.; Kiso, K.; Yoshikawa, K. Univalent reduction of molecular oxygen by spinach chloroplasts on illumination. J. Biol. Chem. 1974, 249, 2175–2181. [Google Scholar] [CrossRef]
- Telfer, A.; Bishop, S.M.; Phillips, D.; Barber, J. Isolated Photosynthetic Reaction Center of Photosystem II as a Sensitizer for the Formation of Singlet Oxygen. J. Biol. Chem. 1994, 269, 13244–13253. [Google Scholar] [CrossRef]
- Triantaphylidès, C.; Havaux, M. Singlet oxygen in plants: Production, detoxification and signaling. Trends Plant Sci. 2009, 14, 219–228. [Google Scholar] [CrossRef]
- Pospíšil, P. Production of reactive oxygen species by photosystem II as a response to light and temperature stress. Front. Plant Sci. 2016, 7, 1950. [Google Scholar] [CrossRef] [PubMed]
- Mehler, A.H. Studies on reactions of illuminated chloroplasts. II. Stimulation and inhibition of the reaction with molecular oxygen. Arch. Biochem. Biophys. 1951, 34, 339–351. [Google Scholar] [CrossRef]
- Asada, K.; Allen, J.; Foyer, C.H.; Matthijs, H.C.P. The water-water cycle as alternative photon and electron sinks. Philos. Trans. R. Soc. B Biol. Sci. 2000, 355, 1419–1431. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.C.C.; Šlesak, I.; Jordá, L.; Sotnikov, A.; Melzer, M.; Miszalski, Z.; Mullineaux, P.M.; Parker, J.E.; Karpińska, B.; Karpiñski, S. Arabidopsis chloroplastic glutathione peroxidases play a role in cross talk between photooxidative stress and immune responses. Plant Physiol. 2009, 150, 670–683. [Google Scholar] [CrossRef] [PubMed]
- Møller, I.M. Plant mitochondria and oxidative stress: Electron transport, NADPH turnover, and metabolism of reactive oxygen species. Annu. Rev. Plant Biol. 2001, 52, 561–591. [Google Scholar] [CrossRef] [PubMed]
- Cvetkovska, M.; Alber, N.A.; Vanlerberghe, G.C. The signaling role of a mitochondrial superoxide burst during stress. Plant Signal. Behav. 2013, 8, e22749. [Google Scholar] [CrossRef] [PubMed]
- Popov, V.N.; Simonian, R.A.; Skulachev, V.P.; Starkov, A.A. Inhibition of the alternative oxidase stimulates H2O2 production in plant mitochondria. FEBS Lett. 1997, 415, 87–90. [Google Scholar] [CrossRef]
- Petrov, V.; Hille, J.; Mueller-Roeber, B.; Gechev, T.S. ROS-mediated abiotic stress-induced programmed cell death in plants. Front. Plant Sci. 2015, 6, 69. [Google Scholar] [CrossRef]
- Del Rio, L.A.; Fernandez, V.M.; Ruperez, F.L.; Sandalio, L.M.; Palma, J.M. NADH Induces the Generation of Superoxide Radicals in Leaf Peroxisomes’. Plant Physiol. 1989, 89, 728–731. [Google Scholar] [CrossRef]
- Foyer, C.H.; Bloom, A.J.; Queval, G.; Noctor, G. Photorespiratory metabolism: Genes, mutants, energetics, and redox signaling. Annu. Rev. Plant Biol. 2009, 60, 455–484. [Google Scholar] [CrossRef]
- del Río, L.A.; Palma, J.M.; Sandalio, L.M.; Corpas, F.J.; Pastori, G.M.; Bueno, P. Peroxisomes as a source of superoxide and hydrogen peroxide in stressed plants Superoxide dismutases (SODs) and superoxide radicals in peroxisomes Metabolism of activated oxygen in leaf peroxisomes from stressed plants. Biochem. Soc. Trans. 1996, 24, 434–438. [Google Scholar]
- Xu, H.; Zhang, J.; Zeng, J.; Jiang, L.; Liu, E.; Peng, C.; He, Z.; Peng, X. Inducible antisense suppression of glycolate oxidase reveals its strong regulation over photosynthesis in rice. J. Exp. Bot. 2009, 60, 1799–1809. [Google Scholar] [CrossRef] [PubMed]
- del Río, L.A.; Corpas, F.J.; Sandalio, L.M.; Palma, J.M.; Gómez, M.; Barroso, J.B. Reactive oxygen species, antioxidant systems and nitric oxide in peroxisomes. J. Exp. Bot. 2002, 53, 1255–1272. [Google Scholar] [CrossRef] [PubMed]
- Marino, D.; Dunand, C.; Puppo, A.; Pauly, N. A burst of plant NADPH oxidases. Trends Plant Sci. 2012, 17, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Karpinska, B.; Karlsson, M.; Schinkel, H.; Streller, S.; Süss, K.-H.; Melzer, M.; Wingsle, G. A Novel Superoxide Dismutase with a High Isoelectric Point in Higher Plants. Expression, Regulation, and Protein Localization. Plant Physiol. 2001, 126, 1668–1677. [Google Scholar] [CrossRef] [PubMed]
- Camejo, D.; Guzm, A.; Moreno, A. Reactive oxygen species, essential molecules, during plantepathogen interactions. Plant Physiol. Biochem. 2016, 103, 10–23. [Google Scholar] [CrossRef]
- Berlett, B.S.; Stadtman, E.R. Protein Oxidation in Aging, Disease, and Oxidative Stress. J. Biol. Chem. 1997, 272, 20313–20316. [Google Scholar] [CrossRef] [PubMed]
- Dean, R.T.; Fu, S.; Stocker, R.; Davies, M.J. Biochemistry and pathology of radical-mediated protein oxidation. Biochem. J. 1997, 324, 1–18. [Google Scholar] [CrossRef]
- Rhoads, D.M.; Umbach, A.L.; Subbaiah, C.C.; Siedow, J.N. Mitochondrial Reactive Oxygen Species. Contribution to Oxidative Stress and Interorganellar Signaling. Plant Physiol. 2006, 141, 357–366. [Google Scholar] [CrossRef] [PubMed]
- Davies, M.J. The oxidative environment and protein damage. Biochim. Biophys. Acta Proteins Proteom. 2005, 1703, 93–109. [Google Scholar] [CrossRef]
- Ghezzi, P.; Bonetto, V. Redox proteomics: Identification of oxidatively modified proteins. Proteomics 2003, 3, 1145–1153. [Google Scholar] [CrossRef]
- Gustavsson, N.; Kokke, B.P.; Härndahl, U.; Silow, M.; Bechtold, U.; Poghosyan, Z.; Murphy, D.; Boelens, W.C.; Sundby, C. A peptide methionine sulfoxide reductase highly expressed in photosynthetic tissue in Arabidopsis thaliana can protect the chaperone-like activity of a chloroplast-localized small heat shock protein. Plant J. 2002, 29, 545–553. [Google Scholar] [CrossRef] [PubMed]
- Levine, R.L.; Mosoni, L.; Berlett, B.S.; Stadtman, E.R. Methionine residues as endogenous antioxidants in proteins. Proc. Natl. Acad. Sci. USA 1996, 93, 15036–15040. [Google Scholar] [CrossRef] [PubMed]
- Smirnoff, N.; Cumbes, Q.J. Hydroxyl radical scavenging activity of compatible solutes. Phytochemistry 1989, 28, 1057–1060. [Google Scholar] [CrossRef]
- Møller, I.M.; Jensen, P.E.; Hansson, A. Oxidative modifications to cellular components in plants. Annu. Rev. Plant Biol. 2007, 58, 459–481. [Google Scholar] [CrossRef] [PubMed]
- Buchanan-Wollaston, V.; Page, T.; Harrison, E.; Breeze, E.; Pyung, O.L.; Hong, G.N.; Lin, J.F.; Wu, S.H.; Swidzinski, J.; Ishizaki, K.; et al. Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J. 2005, 42, 567–585. [Google Scholar] [CrossRef]
- Rey, P.; Bécuwe, N.; Barrault, M.B.; Rumeau, D.; Havaux, M.; Biteau, B.; Toledano, M.B. The Arabidopsis thaliana sulfiredoxin is a plastidic cysteine-sulfinic acid reductase involved in the photooxidative stress response. Plant J. 2007, 49, 505–514. [Google Scholar] [CrossRef]
- Zaffagnini, M.; Bedhomme, M.; Groni, H.; Marchand, C.H.; Puppo, C.; Gontero, B.; Cassier-Chauvat, C.; Decottignies, P.; Lemaire, S.D. Glutathionylation in the Photosynthetic Model Organism Chlamydomonas reinhardtii: A Proteomic Survey. Mol. Cell. Proteom. 2012, 11, M111.014142. [Google Scholar] [CrossRef]
- Yu, M.; Lamattina, L.; Spoel, S.H.; Loake, G.J. Nitric oxide function in plant biology: A redox cue in deconvolution. New Phytol. 2014, 202, 1142–1156. [Google Scholar] [CrossRef]
- Navrot, N.; Finnie, C.; Svensson, B. Plant redox proteomics. J. Proteom. 2011, 74, 1450–1462. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Loake, G.J.; Chu, C.; Levine, A. Cross-talk of nitric oxide and reactive oxygen species in plant programed cell death. Front. Plant Sci. 2013, 4, 314. [Google Scholar] [CrossRef] [PubMed]
- Trapet, P.; Kulik, A.; Lamotte, O.; Jeandroz, S.; Bourque, S.; Nicolas-Francès, V.; Rosnoblet, C.; Besson-Bard, A.; Wendehenne, D. NO signaling in plant immunity: A tale of messengers. Phytochemistry 2015, 112, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Costa, N.J.; Dahm, C.C.; Hurrell, F.; Taylor, E.R.; Murphy, M.P. Interactions of Mitochondrial Thiols with Nitric Oxide. Antioxid. Redox Signal. 2003, 5, 291–305. [Google Scholar] [CrossRef]
- Dahm, C.C.; Moore, K.; Murphy, M.P. Persistent S-nitrosation of complex I and other mitochondrial membrane proteins by S-nitrosothiols but not nitric oxide or peroxynitrite: Implications for the interaction of nitric oxide with mitochondria. J. Biol. Chem. 2006, 281, 10056–10065. [Google Scholar] [CrossRef]
- Cuevasanta, E.; Lange, M.; Bonanata, J.; Laura Coitiñ, E.; Ferrer-Sueta, G.; Filipovic, M.R.; Alvarez, B. Reaction of Hydrogen Sulfide with Disulfide and Sulfenic Acid to Form the Strongly Nucleophilic Persulfide. J. Biol. Chem. 2015, 290, 26866–26880. [Google Scholar] [CrossRef]
- Aroca, A.; Schneider, M.; Scheibe, R.; Gotor, C.; Romero, L.C. Hydrogen Sulfide Regulates the Cytosolic/Nuclear Partitioning of Glyceraldehyde-3-Phosphate Dehydrogenase by Enhancing its Nuclear Localization. Plant Cell Physiol. 2017, 58, 983–992. [Google Scholar] [CrossRef]
- Aroca, A.; Gotor, C.; Romero, L.C. Hydrogen Sulfide Signaling in Plants: Emerging Roles of Protein Persulfidation. Front. Plant Sci. 2018, 9, 1369. [Google Scholar] [CrossRef]
- Shen, J.; Zhang, J.; Zhou, M.; Zhou, H.; Cui, B.; Gotor, C.; Romero, L.C.; Fu, L.; Yang, J.; Foyer, C.H.; et al. Persulfidation-based modification of cysteine desulfhydrase and the NADPH oxidase RBOHD controls guard cell abscisic acid signaling. Plant Cell 2020, 32, 1000–1017. [Google Scholar] [CrossRef] [PubMed]
- Stadtman, E.R. Metal ion-catalyzed oxidation of proteins: Biochemical mechanism and biological consequences. Free Radic. Biol. Med. 1990, 9, 315–325. [Google Scholar] [CrossRef]
- Colombo, G.; Clerici, M.; Garavaglia, M.E.; Giustarini, D.; Rossi, R.; Milzani, A.; Dalle-Donne, I. A step-by-step protocol for assaying protein carbonylation in biological samples. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2016, 1019, 178–190. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.; Suzuki, Y. Protein Carbonylation Signaling in Pulmonary Hypertension. Am. Thorac. Soc. Int. Conf. 2009, A2482. [Google Scholar] [CrossRef]
- Fedorova, M. Diversity of Protein Carbonylation Pathways. Protein Carbonylation 2017, 48–82. [Google Scholar] [CrossRef]
- Biswas, M.S.; Terada, R.; Mano, J. Inactivation of carbonyl-detoxifying enzymes by H2O2 is a trigger to increase carbonyl load for initiating programmed cell death in plants. Antioxidants 2020, 9, 141. [Google Scholar] [CrossRef]
- Requena, J.R.; Levine, R.L.; Stadtman, E.R. Recent advances in the analysis of oxidized proteins Review Article. Amino Acids 2003, 25, 221–226. [Google Scholar] [CrossRef] [PubMed]
- Jung, T.; Höhn, A.; Grune, T. The proteasome and the degradation of oxidized proteins: Part II—Protein oxidation and proteasomal degradation. Redox Biol. 2014, 2, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Maisonneuve, E.; Ducret, A.; Khoueiry, P.; Lignon, S.; Longhi, S.; Talla, E.; Dukan, S. Rules governing selective protein carbonylation. PLoS ONE 2009, 4, e7269. [Google Scholar] [CrossRef]
- Møller, I.M.; Rogowska-Wrzesinska, A.; Rao, R.S.P. Protein carbonylation and metal-catalyzed protein oxidation in a cellular perspective. J. Proteom. 2011, 74, 2228–2242. [Google Scholar] [CrossRef]
- Valentine, J.S.; Gralla, E.B. Introduction: Reactive Oxygen Species Special Feature. Proc. Natl. Acad. Sci. USA 2008, 105, 8178. [Google Scholar] [CrossRef]
- Requena, J.R.; Groth, D.; Legname, G.; Stadtman, E.R.; Prusiner, S.B.; Levine, R.L. Copper-catalyzed oxidation of the recombinant SHa(29-231) prion protein. Proc. Natl. Acad. Sci. USA 2001, 98, 7170–7175. [Google Scholar] [CrossRef]
- Aldini, G.; Dalle-Donne, I.; Facino, R.M.; Milzani, A.; Carini, M. Intervention strategies to inhibit protein carbonylation by lipoxidation-derived reactive carbonyls. Med. Res. Rev. 2007, 27, 817–868. [Google Scholar] [CrossRef] [PubMed]
- Perluigi, M.; Coccia, R.; Allan Butterfield, D. 4-hydroxy-2-nonenal, a reactive product of lipid peroxidation, and neurodegenerative diseases: A toxic combination illuminated by redox proteomics studies. Antioxidants Redox Signal. 2012, 17, 1590–1609. [Google Scholar] [CrossRef]
- West, J.D.; Marnett, L.J. Endogenous reactive intermediates as modulators of cell signaling and cell death. Chem. Res. Toxicol. 2006, 19, 173–194. [Google Scholar] [CrossRef]
- Mano, J. Reactive carbonyl species: Their production from lipid peroxides, action in environmental stress, and the detoxification mechanism. Plant Physiol. Biochem. 2012, 59, 90–97. [Google Scholar] [CrossRef] [PubMed]
- Milic, I.; Kipping, M.; Hoffmann, R.; Fedorova, M. Separation and characterization of oxidized isomeric lipid-peptide adducts by ion mobility mass spectrometry. J. Mass Spectrom. 2015, 50, 1386–1392. [Google Scholar] [CrossRef] [PubMed]
- Rogowska-Wrzesinska, A.; Wojdyla, K.; Nedić, O.; Baron, C.P.; Griffiths, H.R. Analysis of protein carbonylation—Pitfalls and promise in commonly used methods. Free Radic. Res. 2014, 48, 1145–1162. [Google Scholar] [CrossRef]
- Mano, J.; Tokushige, K.; Mizoguchi, H.; Fujii, H.; Khorobrykh, S. Accumulation of lipid peroxide-derived, toxic α, β-unsaturated aldehydes (E)-2-pentenal, acrolein and (E)-2-hexenal in leaves under photoinhibitory illumination. Plant Biotechnol. 2010, 27, 193–197. [Google Scholar] [CrossRef]
- Kristensen, B.K.; Askerlund, P.; Bykova, N.V.; Egsgaard, H.; Møller, I.M. Identification of oxidised proteins in the matrix of rice leaf mitochondria by immunoprecipitation and two-dimensional liquid chromatography-tandem mass spectrometry. Phytochemistry 2004, 65, 1839–1851. [Google Scholar] [CrossRef]
- Aldini, G.; Carini, M.; Yeum, K.-J.; Vistoli, G. Novel molecular approaches for improving enzymatic and nonenzymatic detoxification of 4-hydroxynonenal: Toward the discovery of a novel class of bioactive compounds. Free Radic. Biol. Med. 2014, 69, 145–156. [Google Scholar] [CrossRef]
- Chen, Z.H.; Niki, E. 4-Hydroxynonenal (4-HNE) has been widely accepted as an inducer of oxidative stress. Is this the whole truth about it or can 4-HNE also exert protective effects? IUBMB Life 2006, 58, 372–373. [Google Scholar] [CrossRef] [PubMed]
- Guéraud, F.; Atalay, M.; Bresgen, N.; Cipak, A.; Eckl, P.M.; Huc, L.; Jouanin, I.; Siems, W.; Uchida, K. Chemistry and biochemistry of lipid peroxidation products. Free Radic. Res. 2010, 44, 1098–1124. [Google Scholar] [CrossRef]
- Roede, J.R.; Jones, D.P. Reactive Species and Mitochondrial Dysfunction: Mechanistic Significance of 4-Hydroxynonenal. Environ. Mol. Mutagen. 2010, 51, 380. [Google Scholar] [CrossRef]
- Zhao, Y.; Miriyala, S.; Miao, L.; Mitov, M.; Schnell, D.; Dhar, S.; Cai, J.; Klein, J.; Sultana, R.; Butterfield, D.; et al. Redox proteomic identification of HNE-bound mitochondrial proteins in cardiac tissues reveals a systemic effect on energy metabolism after doxorubicin treatment. Free. Radic. Biol. Med. 2014, 72, 55–65. [Google Scholar] [CrossRef]
- Traverso, N.; Menini, S.; Maineri, E.P.; Patriarca, S.; Odetti, P.; Cottalasso, D.; Marinari, U.M.; Pronzato, M.A. Malondialdehyde, a Lipoperoxidation-Derived Damage To Proteins. America 2004, 59, 890–895. [Google Scholar] [CrossRef]
- Pizzimenti, S.; Ciamporcero, E.; Daga, M.; Pettazzoni, P.; Arcaro, A.; Cetrangolo, G.; Minelli, R.; Dianzani, C.; Lepore, A.; Gentile, F.; et al. Interaction of aldehydes derived from lipid peroxidation and membrane proteins. Front. Physiol. 2013, 4, 242. [Google Scholar] [CrossRef]
- Levine, R.L.; Oliver, C.N.; Fulks, R.M.; Stadtman, E.R. Turnover of bacterial glutamine synthetase: Oxidative inactivation precedes proteolysis. Proc. Natl. Acad. Sci. USA 1981, 78, 2120–2124. [Google Scholar] [CrossRef] [PubMed]
- Grune, T.; Catalgol, B.; Licht, A.; Ermak, G.; Pickering, A.M.; Ngo, J.K.; Davies, K.J.A. HSP70 mediates dissociation and reassociation of the 26S proteasome during adaptation to oxidative stress. Free Radic. Biol. Med. 2011, 51, 1355–1364. [Google Scholar] [CrossRef]
- Gili, B.-N.; Sharon, M. Regulating the 20S Proteasome Ubiquitin-Independent Degradation Pathway. Biomolecules 2014, 4, 862–884. [Google Scholar] [CrossRef]
- Yu, F.; Xie, Q. Non-26S Proteasome Endomembrane Trafficking Pathways in ABA Signaling. Trends Plant Sci. 2017, 22, 976–985. [Google Scholar] [CrossRef]
- Signorelli, S.; Tarkowski, Ł.P.; Van Den Ende, W.; Bassham, D.C. Linking Autophagy to Abiotic and Biotic Stress Responses. Trends Plant Sci. 2019, 24, 413–430. [Google Scholar] [CrossRef]
- Avin-Wittenberg, T.; Bajdzienko, K.; Wittenberg, G.; Alseekh, S.; Tohge, T.; Bock, R.; Giavalisco, P.; Fernie, A.R. Global Analysis of the Role of Autophagy in Cellular Metabolism and Energy Homeostasis in Arabidopsis Seedlings under Carbon Starvation. Plant Cell 2015, 27, 306–322. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Contento, A.L.; Bassham, D.C. Disruption of Autophagy Results in Constitutive Oxidative Stress in Arabidopsis. Autophagy 2007, 3, 257–258. [Google Scholar] [CrossRef] [PubMed]
- Rachakonda, G.; Xiong, Y.; Sekhar, K.R.; Stamer, S.L.; Liebler, D.C.; Freeman, M.L. Covalent Modification at Cys151 Dissociates the Electrophile Sensor Keap1 from the Ubiquitin Ligase CUL3. Chem. Res. Toxicol. 2008, 21, 705–710. [Google Scholar] [CrossRef]
- Farré, J.-C.; Subramani, S. Mechanistic insights into selective autophagy pathways: Lessons from yeast. Nat. Rev. Mol. Cell Biol. 2016, 17, 537–552. [Google Scholar] [CrossRef] [PubMed]
- Svenning, S.; Lamark, T.; Krause, K.; Johansen, T. Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy 2011, 7, 993–1010. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Wang, J.; Cheng, Y.; Chi, Y.-J.; Fan, B. NBR1-Mediated Selective Autophagy Targets Insoluble Ubiquitinated Protein Aggregates in Plant Stress Responses. PLoS Genet. 2013, 9, 1003196. [Google Scholar] [CrossRef] [PubMed]
- Carrera, E.; Holman, T.; Medhurst, A.; Dietrich, D.; Footitt, S.; Theodoulou, F.L.; Holdsworth, M.J. Seed after-ripening is a discrete developmental pathway associated with specific gene networks in Arabidopsis. Plant J. 2008, 53, 214–224. [Google Scholar] [CrossRef] [PubMed]
- Holdsworth, M.J.; Bentsink, L.; Soppe, W.J.J. Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol. 2008, 179, 33–54. [Google Scholar] [CrossRef]
- Oracz, K.; Bouteau, H.E.M.; Farrant, J.M.; Cooper, K.; Belghazi, M.; Job, C.; Job, D.; Corbineau, F.; Bailly, C. ROS production and protein oxidation as a novel mechanism for seed dormancy alleviation. Plant J. 2007, 50, 452–465. [Google Scholar] [CrossRef] [PubMed]
- Leymarie, J.; Vitkauskaité, G.; Hoang, H.H.; Gendreau, E.; Chazoule, V.; Meimoun, P.; Corbineau, F.; El-Maarouf-Bouteau, H.; Bailly, C. Role of Reactive Oxygen Species in the Regulation of Arabidopsis Seed Dormancy. Plant Cell Physiol. 2012, 53, 96–106. [Google Scholar] [CrossRef]
- Müller, K.; Carstens, A.C.; Linkies, A.; Torres, M.A.; Leubner-Metzger, G. The NADPH-oxidase AtrbohB plays a role in Arabidopsis seed after-ripening. New Phytol. 2009, 184, 885–897. [Google Scholar] [CrossRef]
- Tesnier, K.; Strookman-Donkers, H.M.; Pijlen, J.G.V.; Geest, A.V.D.; Bino, R.; Groot, S. A controlled deterioration test for Arabidopsis thaliana reveals genetic variation in seed quality. Seed Sci. Technol. 2002, 30, 149–166. [Google Scholar]
- Skirycz, A.; De Bodt, S.; Obata, T.; De Clercq, I.; Claeys, H.; De Rycke, R.; Andriankaja, M.; Van Aken, O.; Van Breusegem, F.; Fernie, A.R.; et al. Developmental Stage Specificity and the Role of Mitochondrial Metabolism in the Response of Arabidopsis Leaves to Prolonged Mild Osmotic Stress. Plant Physiol. 2010, 152, 226–244. [Google Scholar] [CrossRef] [PubMed]
- Seki, M.; Umezawa, T.; Urano, K.; Shinozaki, K. Regulatory metabolic networks in drought stress responses. Curr. Opin. Plant Biol. 2007, 10, 296–302. [Google Scholar] [CrossRef]
- Contento, A.L.; Kim, S.J.; Bassham, D.C. Transcriptome profiling of the response of arabidopsis suspension culture cells to Suc starvation. Plant Physiol. 2004, 135, 2330–2347. [Google Scholar] [CrossRef]
- Buchanan, B.B.; Balmer, Y. Redox regulation: A broadening horizon. Annu. Rev. Plant Biol. 2005, 56, 187–220. [Google Scholar] [CrossRef]
- Ishizaki, K.; Larson, T.R.; Schauer, N.; Fernie, A.R.; Graham, I.A.; Leaver, C.J. The critical role of Arabidopsis electron-transfer flavoprotein:ubiquinone oxidoreductase during dark-induced starvation. Plant Cell 2005, 17, 2587–2600. [Google Scholar] [CrossRef]
- Brouquisse, R.; Gaudillère, J.P.; Raymond, P. Induction of a carbon-starvation-related proteolysis in whole maize plants submitted to light/dark cycles and to extended darkness. Plant Physiol. 1998, 117, 1281–1291. [Google Scholar] [CrossRef] [PubMed]
- Desnues, B.; Cuny, C.; Grégori, G.; Dukan, S.; Aguilaniu, H.; Nyström, T. Differential oxidative damage and expression of stress defence regulons in culturable and non-culturable Escherichia coli cells. EMBO Rep. 2003, 4, 400–404. [Google Scholar] [CrossRef]
- Noda, Y.; Berlett, B.S.; Stadtman, E.R.; Aponte, A.; Morgan, M.; Shen, R.F. Identification of enzymes and regulatory proteins in Escherichia coli that are oxidized under nitrogen, carbon, or phosphate starvation. Proc. Natl. Acad. Sci. USA 2007, 104, 18456–18460. [Google Scholar] [CrossRef]
- Fredriksson, Å.; Ballesteros, M.; Dukan, S.; Nyström, T. Defense against Protein Carbonylation by DnaK/DnaJ and Proteases of the Heat Shock Regulon. J. Bacteriol. 2005, 187, 4207–4213. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, K.; Chio, T.I.; Gu, H.; Banerjee, A.; Sorrentino, A.M.; Sackett, D.L.; Bane, S.L. Benzocoumarin Hydrazine: A Large Stokes Shift Fluorogenic Sensor for Detecting Carbonyls in Isolated Biomolecules and in Live Cells. ACS Sens. 2017, 2, 128–134. [Google Scholar] [CrossRef]
- Basset, G.; Raymond, P.; Malek, L.; Brouquisse, R. Changes in the expression and the enzymic properties of the 20S proteasome in sugar-starved maize roots. Evidence for an in vivo oxidation of the proteasome. Plant Physiol. 2002, 128, 1149–1162. [Google Scholar] [CrossRef]
- Kumsta, C.; Thamsen, M.; Jakob, U. Effects of Oxidative Stress on Behavior, Physiology, and the Redox Thiol Proteome of Caenorhabditis elegans. Antioxid. Redox Signal. 2011, 14, 1023–1037. [Google Scholar] [CrossRef]
- Winter, J.; Linke, K.; Jatzek, A.; Jakob, U. Severe oxidative stress causes inactivation of DnaK and activation of the redox-regulated chaperone Hsp33. Mol. Cell 2005, 17, 381–392. [Google Scholar] [CrossRef]
- Huang, Q.; Wang, H.; Perry, S.W.; Figueiredo-Pereira, M.E. Negative Regulation of 26S Proteasome Stability via Calpain-mediated Cleavage of Rpn10 Subunit upon Mitochondrial Dysfunction in Neurons *. J. Biol. Chem. 2013, 288, 12161–12174. [Google Scholar] [CrossRef] [PubMed]
- Shringarpure, R.; Grune, T.; Mehlhase, J.; Davies, K.J.A. Ubiquitin conjugation is not required for the degradation of oxidized proteins by proteasome. J. Biol. Chem. 2003, 278, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Reichmann, D.; Voth, W.; Jakob, U. Maintaining a Healthy Proteome during Oxidative Stress. Mol. Cell 2018, 69, 203–213. [Google Scholar] [CrossRef] [PubMed]
- Imai, J.; Maruya, M.; Yashiroda, H.; Yahara, I.; Tanaka, K. The molecular chaperone Hsp90 plays a role in the assembly and maintenance of the 26S proteasome. EMBO J. 2003, 22, 3557–3567. [Google Scholar] [CrossRef]
- Cabiscol, E.; Tamarit, J.; Ros, J. Protein carbonylation: Proteomics, specificity and relevance to aging. Mass Spectrom. Rev. 2014, 33, 21–48. [Google Scholar] [CrossRef] [PubMed]
- Deshmukh, F.K.; Yaffe, D.; Olshina, M.A.; Gili, B.-N.; Sharon, M. The Contribution of the 20S Proteasome to Proteostasis. Biomolecules 2019, 9, 190. [Google Scholar] [CrossRef] [PubMed]
- Kurepa, J.; Toh-E, A.; Smalle, J.A. 26S proteasome regulatory particle mutants have increased oxidative stress tolerance. Plant J. 2008, 53, 102–114. [Google Scholar] [CrossRef] [PubMed]
- Kurepa, J.; Wang, S.; Li, Y.; Smalle, J. Proteasome regulation, plant growth and stress tolerance. Plant Signal. Behav. 2009, 4, 924–927. [Google Scholar] [CrossRef]
- Mongkolsuk, S.; Helmann, J.D. Regulation of inducible peroxide stress responses. Mol. Microbiol. 2002, 45, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Storz, G.; Imlay, J.A. Oxidative stress. Curr. Opin. Microbiol. 1999, 2, 188–194. [Google Scholar] [CrossRef]
- Carmel-Harel, O.; Storz, G. Roles of the Glutathione- and Thioredoxin-Dependent Reduction Systems in the Escherichia Coli and Saccharomyces Cerevisiae Responses to Oxidative Stress. Annu. Rev. Microbiol. 2000, 54, 439–461. [Google Scholar] [CrossRef]
- Zheng, M.; Storz, G. Redox sensing by prokaryotic transcription factors. Biochem. Pharmacol. 2000, 59, 1–6. [Google Scholar] [CrossRef]
- Helmann, J.D.; Wu, M.F.W.; Gaballa, A.; Kobel, P.A.; Morshedi, M.M.; Fawcett, P.; Paddon, C. The global transcriptional response of Bacillus subtilis to peroxide stress is coordinated by three transcription factors. J. Bacteriol. 2003, 185, 243–253. [Google Scholar] [CrossRef] [PubMed]
- Bsat, N.; Herbig, A.; Casillas-Martinez, L.; Setlow, P.; Helmann, J.D. Bacillus subtilis contains multiple Fur homologues: Identification of the iron uptake (Fur) and peroxide regulon (PerR) repressors. Mol. Microbiol. 1998, 29, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Keramati, L.; Helmann, J.D. Coordinate regulation of Bacillus subtilis peroxide stress genes by hydrogen peroxide and metal ions. Proc. Natl. Acad. Sci. USA 1995, 92, 8190–8194. [Google Scholar] [CrossRef]
- Kiley, P.J.; Storz, G. Exploiting Thiol Modifications. PLoS Biol. 2004, 2, e400. [Google Scholar] [CrossRef]
- Herbig, A.F.; Helmann, J.D. Roles of metal ions and hydrogen peroxide in modulating the interaction of the Bacillus subtilis PerR peroxide regulon repressor with operator DNA. Mol. Microbiol. 2001, 41, 849–859. [Google Scholar] [CrossRef]
- Ortiz de Orué Lucana, D.; Wedderhoff, I.; Groves, M.R. ROS-Mediated Signalling in Bacteria: Zinc-Containing Cys-X-X-Cys Redox Centres and Iron-Based Oxidative Stress. J. Signal Transduct. 2012, 2012, 1–9. [Google Scholar] [CrossRef]
- Lee, J.W.; Helmann, J.D. The PerR transcription factor senses H2O2 by metal-catalysed histidine oxidation. Nature 2006, 440, 363–367. [Google Scholar] [CrossRef] [PubMed]
- Curtis, J.M.; Hahn, W.S.; Long, E.K.; Burrill, J.S.; Arriaga, E.A.; Bernlohr, D.A. Protein carbonylation and metabolic control systems. Trends Endocrinol. Metab. 2012, 23, 399–406. [Google Scholar] [CrossRef]
- Hassoun, P.M.; Thappa, V.; Landman, M.J.; Fanburg, B.L. Endothelin 1: Mitogenic Activity on Pulmonary Artery Smooth Muscle Cells and Release from Hypoxic Endothelial Cells. Proc. Soc. Exp. Biol. Med. 1992, 199, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Janakidevi, K.; Fisher, M.A.; Del Vecchio, P.J.; Tiruppathi, C.; Figge, J.; Malik, A.B. Endothelin-1 stimulates DNA synthesis and proliferation of pulmonary artery smooth muscle cells. Am. J. Physiol. Cell Physiol. 1992, 263, C1295–C1301. [Google Scholar] [CrossRef]
- Wong, C.M.; Marcocci, L.; Liu, L.; Suzuki, Y.J. Cell signaling by protein carbonylation and decarbonylation. Antioxidants Redox Signal. 2010, 12, 393–404. [Google Scholar] [CrossRef]
- Wedgwood, S.; Black, S.M. Role of Reactive Oxygen Species in Vascular Remodeling Associated with Pulmonary Hypertension. Antioxidants Redox Signal. 2003, 5, 759–769. [Google Scholar] [CrossRef]
- Wedgwood, S.; Dettman, R.W.; Black, S.M. ET-1 stimulates pulmonary arterial smooth muscle cell proliferation via induction of reactive oxygen species. Am. J. Physiol. Lung Cell. Mol. Physiol. 2001, 281, L1058–L1067. [Google Scholar] [CrossRef]
- Davie, N.; Haleen, S.J.; Upton, P.D.; Polak, J.M.; Yacoub, M.H.; Morrell, N.W.; Wharton, J. ETA and ETB receptors modulate the proliferation of human pulmonary artery smooth muscle cells. Am. J. Respir. Crit. Care Med. 2002, 165, 398–405. [Google Scholar] [CrossRef]
- Miyauchi, T.; Yorikane, R.; Sakai, S.; Sakurai, T.; Okada, M.; Nishikibe, M.; Yano, M.; Yamaguchi, I.; Sugishita, Y.; Goto, K. Contribution of endogenous endothelin-1 to the progression of cardiopulmonary alterations in rats with monocrotaline-induced pulmonary hypertension. Circ. Res. 1993, 73, 887–897. [Google Scholar] [CrossRef] [PubMed]
- Zamora, M.R.; Stelzner, T.J.; Webb, S.; Panos, R.J.; Ruff, L.J.; Dempsey, E.C. Overexpression of endothelin-1 and enhanced growth of pulmonary artery smooth muscle cells from fawn-hooded rats. Am. J. Physiol. Lung Cell. Mol. Physiol. 1996, 270, L101–L109. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.M.; Cheema, A.K.; Zhang, L.; Suzuki, Y.J. Protein carbonylation as a novel mechanism in redox signaling. Circ. Res. 2008, 102, 310–318. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Jia, Z.; Zhang, L.; Yamamoto, M.; Misra, H.P.; Trush, M.A.; Li, Y. Antioxidants and Phase 2 Enzymes in Macrophages: Regulation by Nrf2 Signaling and Protection Against Oxidative and Electrophilic Stress. Exp. Biol. Med. 2008, 233, 463–474. [Google Scholar] [CrossRef]
- McMahon, M.; Itoh, K.; Yamamoto, M.; Hayes, J.D. Keap1-dependent proteasomal degradation of transcription factor Nrf2 contributes to the negative regulation of antioxidant response element-driven gene expression. J. Biol. Chem. 2003, 278, 21592–21600. [Google Scholar] [CrossRef]
- Fang, J.; Holmgren, A. Inhibition of thioredoxin and thioredoxin reductase by 4-hydroxy-2-nonenal in vitro and in vivo. J. Am. Chem. Soc. 2006, 128, 1879–1885. [Google Scholar] [CrossRef]
- Sharma, R.; Sharma, A.; Dwivedi, S.; Zimniak, P.; Awasthi, S.; Awasthi, Y.C. 4-Hydroxynonenal self-limits Fas-mediated DISC-independent apoptosis by promoting export of Daxx from the nucleus to the cytosol and its binding to Fas. Biochemistry 2008, 47, 143–156. [Google Scholar] [CrossRef]
- Fujino, G.; Noguchi, T.; Matsuzawa, A.; Yamauchi, S.; Saitoh, M.; Takeda, K.; Ichijo, H. Thioredoxin and TRAF Family Proteins Regulate Reactive Oxygen Species-Dependent Activation of ASK1 through Reciprocal Modulation of the N-Terminal Homophilic Interaction of ASK1. Mol. Cell. Biol. 2007, 27, 8152–8163. [Google Scholar] [CrossRef]
- Bai, X.; Yang, L.; Tian, M.; Chen, J.; Shi, J. Nitric Oxide Enhances Desiccation Tolerance of Recalcitrant Antiaris toxicaria Seeds via Protein S-Nitrosylation and Carbonylation. PLoS ONE 2011, 6, 20714. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.M.; Ye, W.; Matsushima, D.; Rhaman, M.S.; Munemasa, S.; Okuma, E.; Nakamura, Y.; Biswas, M.S.; Mano, J.; Murata, Y. Reactive carbonyl species function as signal mediators downstream of H2O2 production and regulate [Ca2+]cyt elevation in ABA signal pathway in Arabidopsis guard cells. Plant Cell Physiol. 2019, 60, 1146–1159. [Google Scholar] [CrossRef]
- Pel, Z.M.; Murata, Y.; Benning, G.; Thomine, S.; Klüsener, B.; Allen, G.J.; Grill, E.; Schroeder, J.I. Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature 2000, 406, 731–734. [Google Scholar] [CrossRef]
- Kwak, J.M.; Mori, I.C.; Pei, Z.M.; Leonhard, N.; Angel Torres, M.; Dangl, J.L.; Bloom, R.E.; Bodde, S.; Jones, J.D.G.; Schroeder, J.I. NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in arabidopsis. EMBO J. 2003, 22, 2623–2633. [Google Scholar] [CrossRef]
- Suhita, D.; Raghavendra, A.S.; Kwak, J.M.; Vavasseur, A. Cytoplasmic alkalization precedes reactive oxygen species production during methyl jasmonate- and abscisic acid-induced stomatal closure. Plant Physiol. 2004, 134, 1536–1545. [Google Scholar] [CrossRef]
- Munemasa, S.; Oda, K.; Watanabe-Sugimoto, M.; Nakamura, Y.; Shimoishi, Y.; Murata, Y. The coronatine-insensitive 1 mutation reveals the hormonal signaling interaction between abscisic acid and methyl jasmonate in Arabidopsis guard cells. Specific impairment of ion channel activation and second messenger production. Plant Physiol. 2007, 143, 1398–1407. [Google Scholar] [CrossRef]
- Islam, M.M.; Hossain, M.A.; Jannat, R.; Munemasa, S.; Nakamura, Y.; Mori, I.C.; Murata, Y. Cytosolic alkalization and cytosolic calcium oscillation in Arabidopsis guard cells Response to ABA and MeJA. Plant Cell Physiol. 2010, 51, 1721–1730. [Google Scholar] [CrossRef]
- Akter, N.; Sobahan, M.A.; Uraji, M.; Ye, W.; Hossain, M.A.; Mori, I.C.; Nakamura, Y.; Murata, Y. Effects of depletion of glutathione on abscisic acidand methyl jasmonate-induced stomatal closure in arabidopsis thaliana. Biosci. Biotechnol. Biochem. 2012, 76, 2032–2037. [Google Scholar] [CrossRef]
- Akter, N.; Okuma, E.; Sobahan, M.A.; Uraji, M.; Munemasa, S.; Nakamura, Y.; Mori, I.C.; Murata, Y. Negative Regulation of Methyl Jasmonate-Induced Stomatal Closure by Glutathione in Arabidopsis. J. Plant Growth Regul. 2013, 32, 208–215. [Google Scholar] [CrossRef]
- McAinsh, M.R.; Brownlee, C.; Hetherington, A.M. Abscisic acid-induced elevation of guard cell cytosolic Ca2+ precedes stomatal closure. Nature 1990, 343, 186–188. [Google Scholar] [CrossRef]
- McAinsh, M.R.; Brownlee, C.; Hetherington, A.M. Visualizing Changes in Cytosolic-Free Ca2+ during the Response of Stomatal Guard Cells to Abscisic Acid. Plant Cell 1992, 4, 1113–1122. [Google Scholar] [CrossRef]
- Staxén, I.; Pical, C.; Montgomery, L.T.; Gray, J.E.; Hetherington, A.M.; Mcainsh, M.R. Abscisic acid induces oscillations in guard-cell cytosolic free calcium that involve phosphoinositide-specific phospholipase C. Proc. Natl. Acad. Sci. USA 1999, 96, 1779–1784. [Google Scholar] [CrossRef]
- Murata, Y.; Pei, Z.-M.; Mori, I.C.; Schroeder, J. Abscisic Acid Activation of Plasma Membrane Ca2+ Channels in Guard Cells Requires Cytosolic NAD(P)H and Is Differentially Disrupted Upstream and Downstream of Reactive Oxygen Species Production in abi1-1 and abi2-1 Protein Phosphatase 2C Mutants. Plant Cell 2001, 13, 2513–2523. [Google Scholar] [CrossRef]
- Munemasa, S.; Muroyama, D.; Nagahashi, H.; Nakamura, Y.; Mori, I.C.; Murata, Y. Regulation of reactive oxygen species-mediated abscisic acid signaling in guard cells and drought tolerance by glutathione. Front. Plant Sci. 2013, 4, 472. [Google Scholar] [CrossRef]
- Rodríguez, A.A.; Grunberg, K.A.; Taleisnik, E.L. Reactive oxygen species in the elongation zone of maize leaves are necessary for leaf extension. Plant Physiol. 2002, 129, 1627–1632. [Google Scholar] [CrossRef] [PubMed]
- Ros Barceló, A. Xylem parenchyma cells deliver the H2O2 necessary for lignification in differentiating xylem vessels. Planta 2005, 220, 747–756. [Google Scholar] [CrossRef]
- Li, S.; Xue, L.; Xu, S.; Feng, H.; An, L. Hydrogen peroxide involvement in formation and development of adventitious roots in cucumber. Plant Growth Regul. 2007, 52, 173–180. [Google Scholar] [CrossRef]
- Foreman, J.; Demidchik, V.; Bothwell, J.H.F.; Mylona, P.; Miedema, H.; Angel Torres, M.; Linstead, P.; Costa, S.; Brownlee, C.; Jones, J.D.G.; et al. Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 2003, 422, 442–446. [Google Scholar] [CrossRef]
- Su, G.-X.; Zhang, W.-H.; Liu, Y.-L. Involvement of Hydrogen Peroxide Generated by Polyamine Oxidative Degradation in the Development of Lateral Roots in Soybean. J. Integr. Plant Biol. 2006, 48, 426–432. [Google Scholar] [CrossRef]
- Chen, Y.H.; Chao, Y.Y.; Hsu, Y.Y.; Kao, C.H. Heme oxygenase is involved in H2O2-induced lateral root formation in apocynin-treated rice. Plant Cell Rep. 2013, 32, 219–226. [Google Scholar] [CrossRef]
- Montillet, J.-L.; Leonhardt, N.; Mondy, S.; Tranchimand, S.; Rumeau, D.; Boudsocq, M.; Garcia, A.V.; Douki, T.; Bigeard, J.; Laurière, C.; et al. An Abscisic Acid-Independent Oxylipin Pathway Controls Stomatal Closure and Immune Defense in Arabidopsis. PLoS Biol. 2013, 11, e1001513. [Google Scholar] [CrossRef]
- Li, N.; Sun, L.; Zhang, L.; Song, Y.; Hu, P.; Li, C.; Hao, F.S. AtrbohD and AtrbohF negatively regulate lateral root development by changing the localized accumulation of superoxide in primary roots of Arabidopsis. Planta 2015, 241, 591–602. [Google Scholar] [CrossRef]
- Orman-Ligeza, B.; Parizot, B.; de Rycke, R.; Fernandez, A.; Himschoot, E.; van Breusegem, F.; Bennett, M.J.; Périlleux, C.; Beeckman, T.; Draye, X. RBOH-mediated ROS production facilitates lateral root emergence in Arabidopsis. Development 2016, 143, 3328–3339. [Google Scholar] [CrossRef]
- Kim, M.; McLoughlin, F.; Basha, E.; Vierling, E. Assessing Plant Tolerance to Acute Heat Stress. Bio-Protocol 2017, 7, 1–12. [Google Scholar] [CrossRef]
- Ma, F.; Wang, L.; Li, J.; Samma, M.K.; Xie, Y.; Wang, R.; Wang, J.; Zhang, J.; Shen, W. Interaction between HY1 and H2O2 in auxin-induced lateral root formation in Arabidopsis. Plant Mol. Biol. 2014, 85, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, T.; Suzuki, T.; Kobayashi, A.; Wakabayashi, J.; Maher, J.; Motohashi, H.; Yamamoto, M. Physiological Significance of Reactive Cysteine Residues of Keap1 in Determining Nrf2 Activity. Mol. Cell. Biol. 2008, 28, 2758–2770. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Liu, H.; Iles, K.E.; Liu, R.M.; Postlethwait, E.M.; Laperche, Y.; Forman, H.J. 4-Hydroxynonenal induces rat γ-glutamyl transpeptidase through mitogen-activated protein kinase-mediated electrophile response element/nuclear factor erythroid 2-related factor 2 signaling. Am. J. Respir. Cell Mol. Biol. 2006, 34, 174–181. [Google Scholar] [CrossRef]
- Rudolph, T.K.; Freeman, B.A. Transduction of Redox Signaling by Electrophile-Protein Reactions. Sci. Signal. 2009, 2, re7. [Google Scholar] [CrossRef]
- Tanou, G.; Job, C.; Rajjou, L.; Arc, E.; Belghazi, M.; Diamantidis, G.; Molassiotis, A.; Job, D. Proteomics reveals the overlapping roles of hydrogen peroxide and nitric oxide in the acclimation of citrus plants to salinity. Plant J. 2009, 60, 795–804. [Google Scholar] [CrossRef]
- Tanou, G.; Filippou, P.; Belghazi, M.; Job, D.; Diamantidis, G.; Fotopoulos, V.; Molassiotis, A. Oxidative and nitrosative-based signaling and associated post-translational modifications orchestrate the acclimation of citrus plants to salinity stress. Plant J. 2012, 72, 585–599. [Google Scholar] [CrossRef]
- Kakizawa, S.; Shibazaki, M.; Mori, N. Protein oxidation inhibits NO-mediated signaling pathway for synaptic plasticity. Neurobiol. Aging 2012, 33, 535–545. [Google Scholar] [CrossRef] [PubMed]
- Krasuska, U.; Ciacka, K.; Ebska, K.D.; Bogatek, R.; Gniazdowska, A. Dormancy alleviation by NO or HCN leading to decline of protein carbonylation levels in apple (Malus domestica Borkh.) embryos. J. Plant Physiol. 2014, 171, 1132–1141. [Google Scholar] [CrossRef]
- Galligan, J.J.; Rose, K.L.; Beavers, W.N.; Hill, S.; Tallman, K.A.; Tansey, W.P.; Marnett, L.J. Stable Histone Adduction by 4-Oxo-2-nonenal: A Potential Link between Oxidative Stress and Epigenetics. J. Am. Chem. Soc. 2014, 136, 11864–11866. [Google Scholar] [CrossRef]
- Chavez, J.D.; Wu, J.; Bisson, W.; Maier, C.S. Site-specific proteomic analysis of lipoxidation adducts in cardiac mitochondria reveals chemical diversity of 2-alkenal adduction. J. Proteom. 2011, 74, 2417–2429. [Google Scholar] [CrossRef] [PubMed]
- Lv, H.; Han, J.; Liu, J.; Zheng, J.; Liu, R. CarSPred: A Computational Tool for Predicting Carbonylation Sites of Human Proteins. PLoS ONE 2014, 9, 111478. [Google Scholar] [CrossRef] [PubMed]
- Weng, S.L.; Huang, K.Y.; Kaunang, F.J.; Huang, C.H.; Kao, H.J.; Chang, T.H.; Wang, H.Y.; Lu, J.J.; Lee, T.Y. Investigation and identification of protein carbonylation sites based on position-specific amino acid composition and physicochemical features. BMC Bioinform. 2017, 18, 66. [Google Scholar] [CrossRef] [PubMed]
- Prasad Rao, R.S.; Zhang, N.; Xu, D.; MaxMoøller, I. CarbonylDB: A curated data-resource of protein carbonylation sites. Bioinformatics 2018, 34, 2518–2520. [Google Scholar] [CrossRef]
- Shyama Prasad Rao, R.; Møller, I.M. Pattern of occurrence and occupancy of carbonylation sites in proteins. Proteomics 2011, 11, 4166–4173. [Google Scholar] [CrossRef]
- Yang, J.; Tallman, K.A.; Porter, N.A.; Liebler, D.C. Quantitative chemoproteomics for site-specific analysis of protein alkylation by 4-hydroxy-2-nonenal in cells. Anal. Chem. 2015, 87, 2535–2541. [Google Scholar] [CrossRef] [PubMed]
- Levine, R.L.; Williams, J.A.; Stadtman, E.P.; Shacter, E. Carbonyl assays for determination of oxidatively modified proteins. Methods Enzymol. 1994, 233, 346–357. [Google Scholar] [CrossRef] [PubMed]
- Reznick, A.Z.; Packer, L. Oxidative damage to proteins: Spectrophotometric method for carbonyl assay. Methods Enzymol. 1994, 233, 357–363. [Google Scholar] [CrossRef]
- Chavez, J.; Wu, J.; Han, B.; Chung, W.G.; Maier, C.S. New role for an old probe: Affinity labeling of oxylipid protein conjugates by N′-aminooxymethylcarbonylhydrazino D-biotin. Anal. Chem. 2006, 78, 6847–6854. [Google Scholar] [CrossRef]
- Madian, A.G.; Regnier, F.E. Proteomic Identification of Carbonylated Proteins and Their Oxidation Sites. J. Proteome Res. 2010, 9, 3766–3780. [Google Scholar] [CrossRef]
- Carini, M.; Aldini, G.; Facino, R.M. Mass spectrometry for detection of 4-hydroxy-trans-2-nonenal (HNE) adducts with peptides and proteins. Mass Spectrom. Rev. 2004, 23, 281–305. [Google Scholar] [CrossRef]
- Bollineni, R.C.; Hoffmann, R.; Fedorova, M. Identification of protein carbonylation sites by two-dimensional liquid chromatography in combination with MALDI- and ESI-MS. J. Proteom. 2011, 74, 2338–2350. [Google Scholar] [CrossRef]
- Tzeng, S.C.; Maier, C.S. Label-free proteomics assisted by affinity enrichment for elucidating the chemical reactivity of the liver mitochondrial proteome toward adduction by the lipid electrophile 4-hydroxy-2-nonenal (HNE). Front. Chem. 2016, 4, 2. [Google Scholar] [CrossRef] [PubMed]
- Matamoros, M.A.; Kim, A.; Peñuelas, M.; Ihling, C.; Griesser, E.; Hoffmann, R.; Fedorova, M.; Frolov, A.; Becana, M. Protein carbonylation and glycation in legume nodules. Plant Physiol. 2018, 177, 1510–1528. [Google Scholar] [CrossRef] [PubMed]
- Yin, G.; Xin, X.; Fu, S.; An, M.; Wu, S.; Chen, X.; Zhang, J.; He, J.; Whelan, J.; Lu, X. Proteomic and carbonylation profile analysis at the critical node of seed ageing in Oryza sativa. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Arnett, D.; Quillin, A.; Geldenhuys, W.J.; Menze, M.A.; Konkle, M. 4-Hydroxynonenal and 4-Oxononenal Differentially Bind to the Redox Sensor MitoNEET. Chem. Res. Toxicol. 2019, 32, 977–981. [Google Scholar] [CrossRef]
- Han, B.; Hare, M.; Wickramasekara, S.; Fang, Y.; Maier, C.S. A comparative ‘bottom up’ proteomics strategy for the site-specific identification and quantification of protein modifications by electrophilic lipids. J. Proteom. 2012, 75, 5724–5733. [Google Scholar] [CrossRef]
- Codreanu, S.G.; Ullery, J.C.; Zhu, J.; Tallman, K.A.; Beavers, W.N.; Porter, N.A.; Marnett, L.J.; Zhang, B.; Liebler, D.C. Alkylation Damage by Lipid Electrophiles Targets Functional Protein Systems. Mol. Cell. Proteom. 2014, 13, 849–859. [Google Scholar] [CrossRef]
- Weerapana, E.; Wang, C.; Simon, G.M.; Richter, F.; Khare, S.; Dillon, M.B.D.; Bachovchin, D.A.; Mowen, K.; Baker, D.; Cravatt, B.F. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature 2010, 468, 790–795. [Google Scholar] [CrossRef]
- Wang, C.; Weerapana, E.; Blewett, M.M.; Cravatt, B.F. A chemoproteomic platform to quantitatively map targets of lipid-derived electrophiles. Nat. Methods 2014, 11, 79–85. [Google Scholar] [CrossRef]
- Matthews, M.L.; He, L.; Horning, B.D.; Olson, E.J.; Correia, B.E.; Yates, J.R.; Dawson, P.E.; Cravatt, B.F. Chemoproteomic profiling and discovery of protein electrophiles in human cells. Nat. Chem. 2017, 9, 234–243. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, Y.; Lan, T.; Qin, W.; Zhu, Y.; Qin, K.; Gao, J.; Wang, H.; Hou, X.; Chen, N.; et al. Quantitative Profiling of Protein Carbonylations in Ferroptosis by an Aniline-Derived Probe. J. Am. Chem. Soc. 2018, 140, 4712–4720. [Google Scholar] [CrossRef]
- Parvez, S.; Long, M.J.C.; Lin, H.Y.; Zhao, Y.; Haegele, J.A.; Pham, V.N.; Lee, D.K.; Aye, Y. T-REX on-demand redox targeting in live cells. Nat. Protoc. 2016, 11, 2328–2356. [Google Scholar] [CrossRef]
- Schmidt, R.; Schippers, J.H.M. ROS-mediated redox signaling during cell differentiation in plants. Biochim. Biophys. Acta Gen. Subj. 2015, 1850, 1497–1508. [Google Scholar] [CrossRef]
- Wrzaczek, M.; Brosché, M.; Kangasjä Rvi, J. ROS signaling loops-production, perception, regulation. Curr. Opin. Plant Biol. 2013, 16, 575–582. [Google Scholar] [CrossRef] [PubMed]
- Smirnoff, N.; Arnaud, D. Hydrogen peroxide metabolism and functions in plants. New Phytol. 2019, 221, 1197–1214. [Google Scholar] [CrossRef] [PubMed]
- Mueller, S.; Hilbert, B.; Dueckershoff, K.; Roitsch, T.; Krischke, M.; Mueller, M.J.; Berger, S. General detoxification and stress responses are mediated by oxidized lipids through TGA transcription factors in arabidopsis. Plant Cell 2008, 20, 768–785. [Google Scholar] [CrossRef] [PubMed]
- Mueller, M.J.; Berger, S. Reactive electrophilic oxylipins: Pattern recognition and signalling. Phytochemistry 2009, 70, 1511–1521. [Google Scholar] [CrossRef] [PubMed]
- Ichi Mano, J.; Biswas, M.S.; Sugimoto, K. Reactive Carbonyl Species: A Missing Link in ROS Signaling. Plants 2019, 8, 391. [Google Scholar] [CrossRef]
- Vollenweider, S.; Weber, H.; Stolz, S.; Chételat, A.; Farmer, E.E. Fatty acid ketodienes and fatty acid ketotrienes: Michael addition acceptors that accumulate in wounded and diseased Arabidopsis leaves. Plant J. 2000, 24, 467–476. [Google Scholar] [CrossRef]
- Améras, E.; Stolz, S.; Vollenweider, S.; Reymond, P.; Mène-Saffrané, L.; Farmer, E.E. Reactive electrophile species activate defense gene expression in Arabidopsis. Plant J. 2003, 34, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Winger, A.M.; Taylor, N.L.; Heazlewood, J.L.; Day, D.A.; Millar, A.H. The Cytotoxic Lipid Peroxidation Product 4-Hydroxy-2-nonenal Covalently Modifies a Selective Range of Proteins Linked to Respiratory Function in Plant Mitochondria. J. Biol. Chem. 2007, 282, 37436–37447. [Google Scholar] [CrossRef] [PubMed]
- Poganik, J.R.; Long, M.J.C.; Aye, Y. Getting the Message? Native Reactive Electrophiles Pass Two Out of Three Thresholds to be Bona Fide Signaling Mediators. BioEssays 2018, 40, 1700240. [Google Scholar] [CrossRef] [PubMed]

| Lipid Peroxide-Derived Reactive Carbonyl Species | Predominant PUFAs | Preference of Amino Acids for Modification | Type of Reaction with Amino Acids | References |
|---|---|---|---|---|
| 4-Hydroxy-(E)-2-nonenal (4-HNE) | Linoleic acid (LA: 18:2ω-6) Arachidonic acid (AA: 20:4, ω-6) | Cys >> His > Lys | Michael addition | [108,109,110,111,112,113] |
| Malondialdehyde (MDA) | Arachidonic acid (AA: 20:4, ω-6) | Lys >> His > Arg | Michael addition | [7,113,114] |
| Acrolein | Linoleic acid (LA: 18:2ω-6) | Cys >> His > Lys | Michael addition or Schiff-base formation | [108,115] |
| 4-Oxo-nonenal (4-ONE) | Linoleic acid (LA: 18:2ω-6) Arachidonic acid (AA: 20:4, ω-6) | Lys >> Cys > His > Arg | Schiff-base formation | [110,113] |
| Hormonal Signaling | Physiological Processes | Forms of RCS Involved | References |
|---|---|---|---|
| Auxin signaling | Lateral root formation | HNE, acrolein, crotonaldehyde, butyraldehyde | [16] |
| ABA signaling | Stomatal closure | HNE, MDA | [17,18,183] |
| MeJA signaling | Stomatal closure | HNE, MDA | [19] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tola, A.J.; Jaballi, A.; Missihoun, T.D. Protein Carbonylation: Emerging Roles in Plant Redox Biology and Future Prospects. Plants 2021, 10, 1451. https://doi.org/10.3390/plants10071451
Tola AJ, Jaballi A, Missihoun TD. Protein Carbonylation: Emerging Roles in Plant Redox Biology and Future Prospects. Plants. 2021; 10(7):1451. https://doi.org/10.3390/plants10071451
Chicago/Turabian StyleTola, Adesola J., Amal Jaballi, and Tagnon D. Missihoun. 2021. "Protein Carbonylation: Emerging Roles in Plant Redox Biology and Future Prospects" Plants 10, no. 7: 1451. https://doi.org/10.3390/plants10071451
APA StyleTola, A. J., Jaballi, A., & Missihoun, T. D. (2021). Protein Carbonylation: Emerging Roles in Plant Redox Biology and Future Prospects. Plants, 10(7), 1451. https://doi.org/10.3390/plants10071451







