Global Transcriptional Analyses of the Wnt-Induced Development of Neural Stem Cells from Human Pluripotent Stem Cells
Abstract
:1. Introduction
2. Results
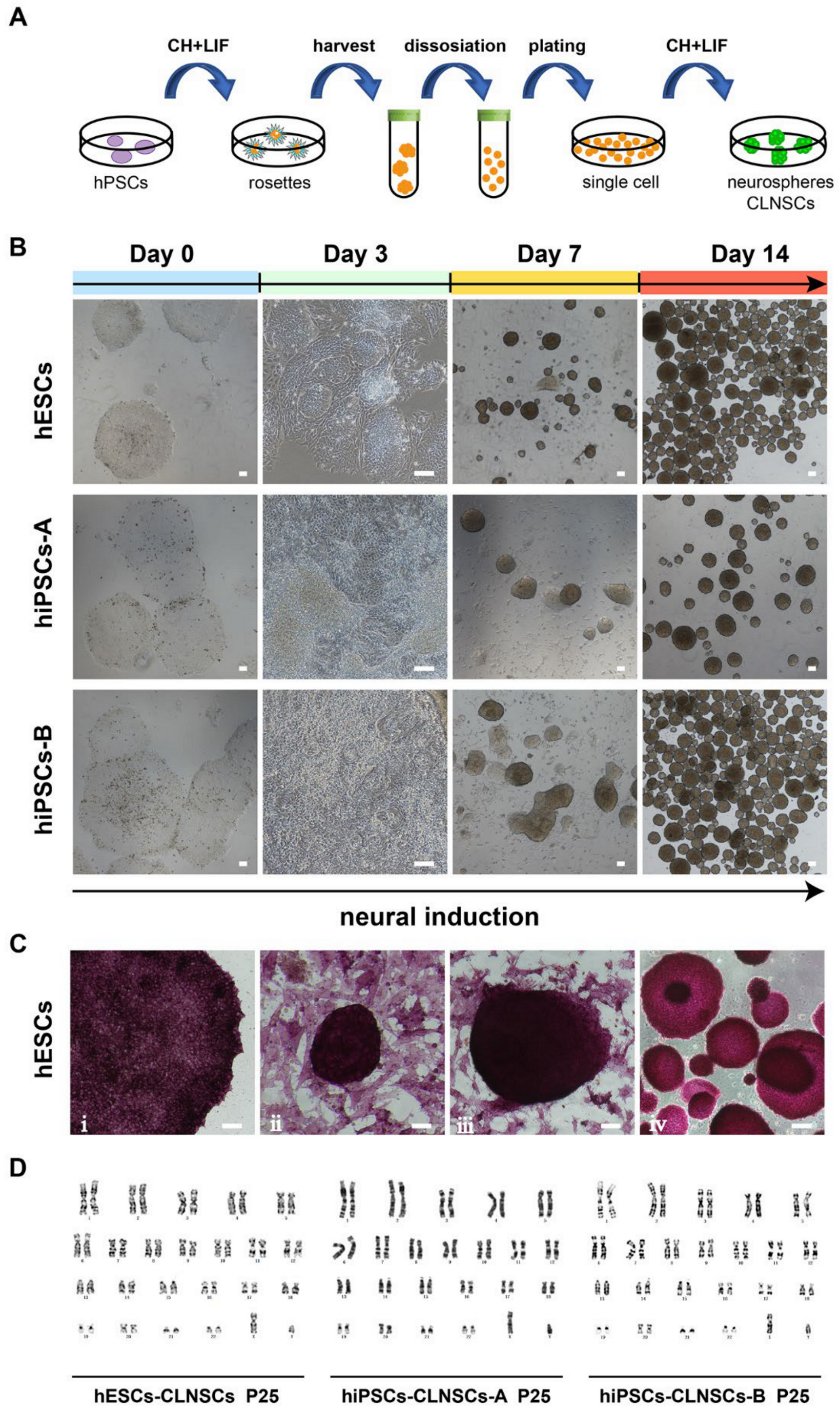
2.1. Combination of CHIR-99021 and LIF Can Generate NSCs from hPSCs
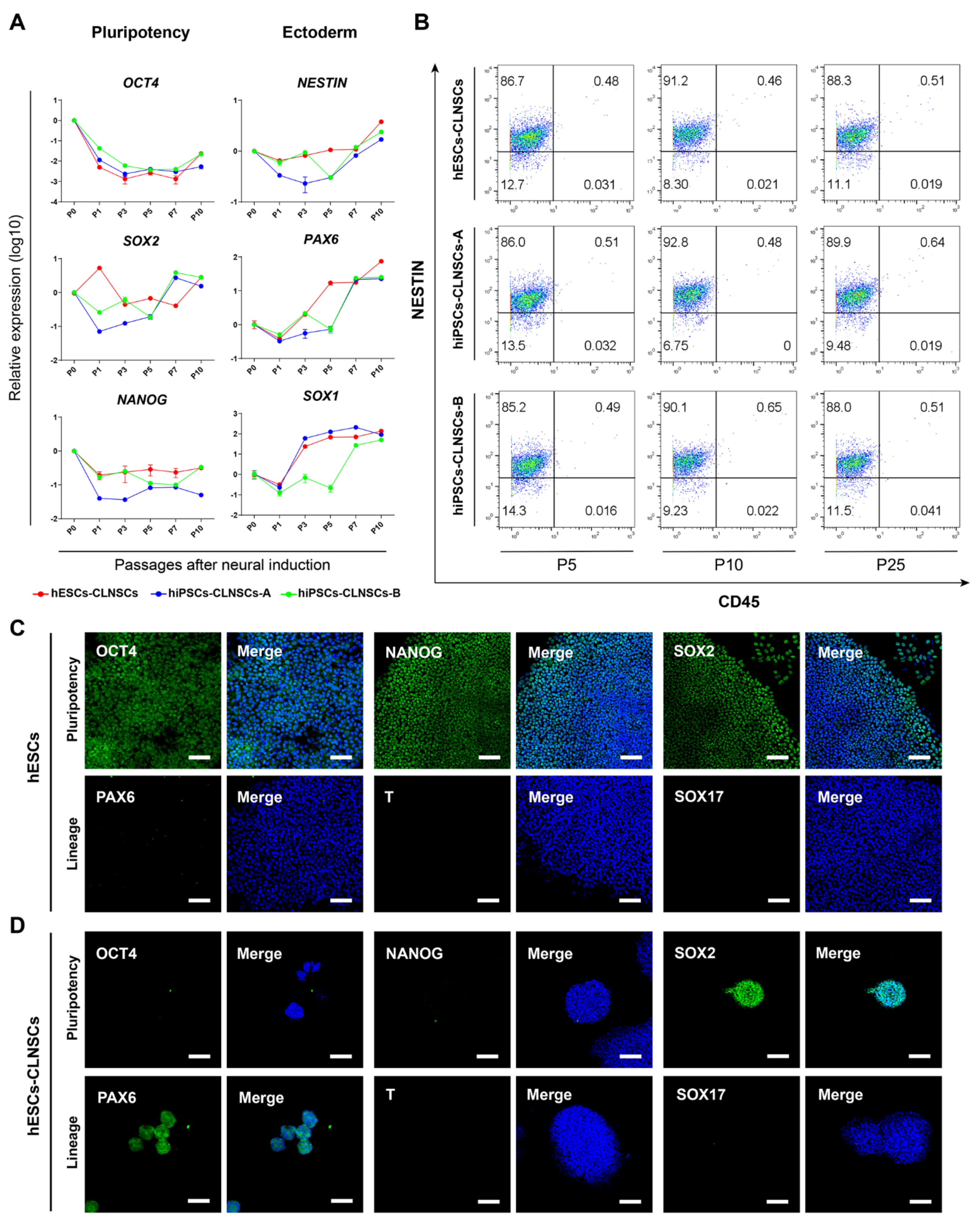
2.2. Characteristic of NSCs Derived from hPSCs
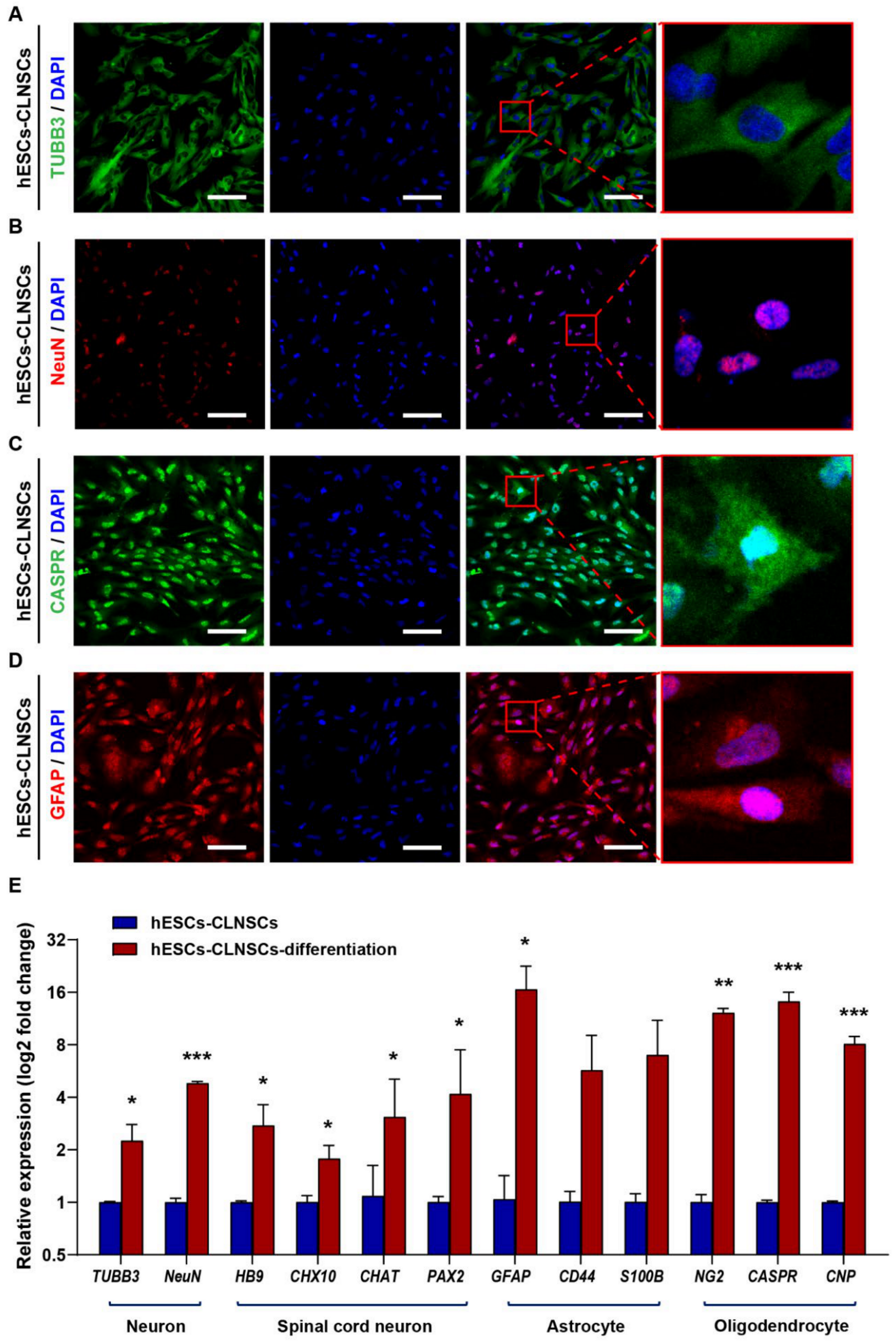
2.3. CLNSCs Display Multipotency during Differentiation
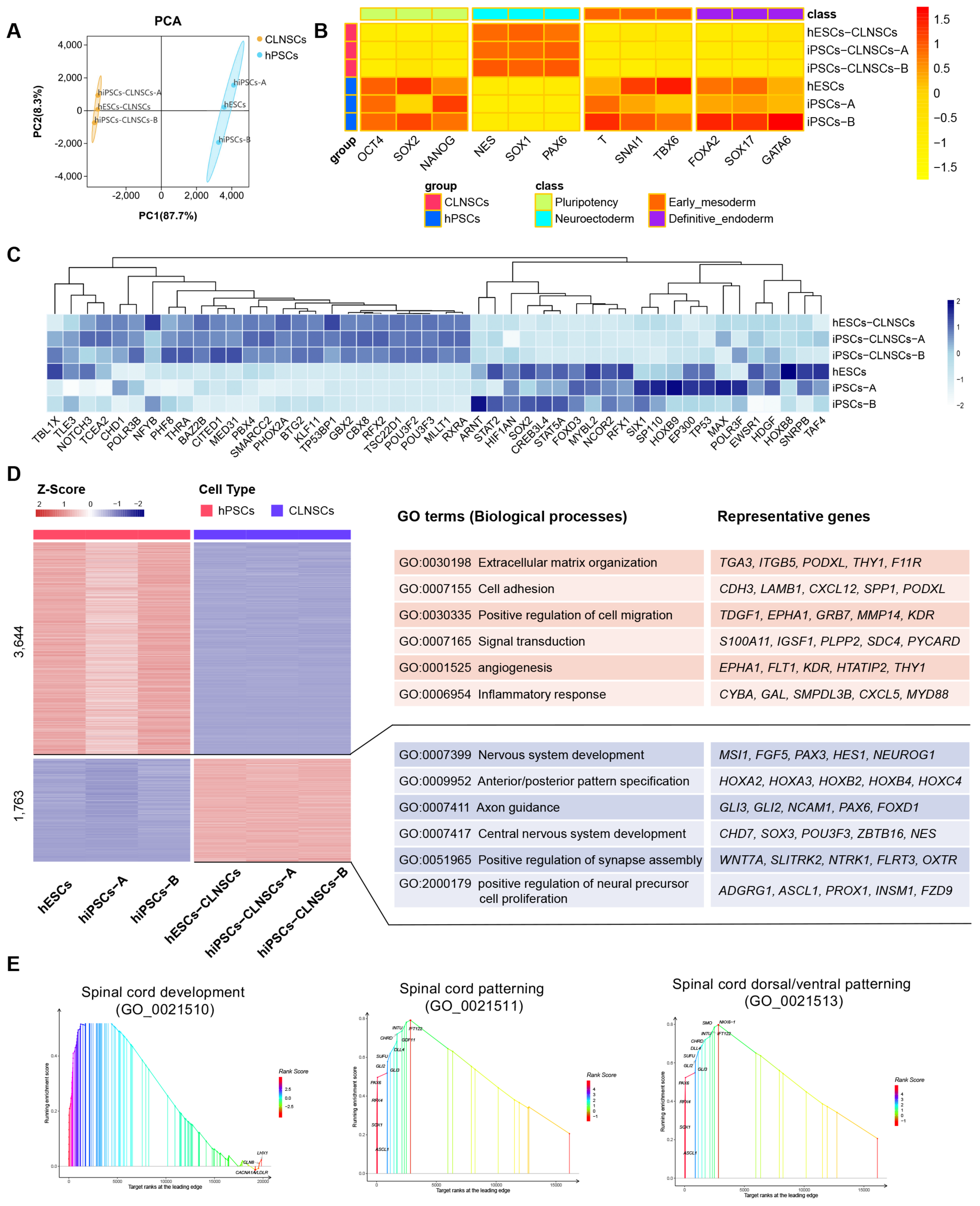
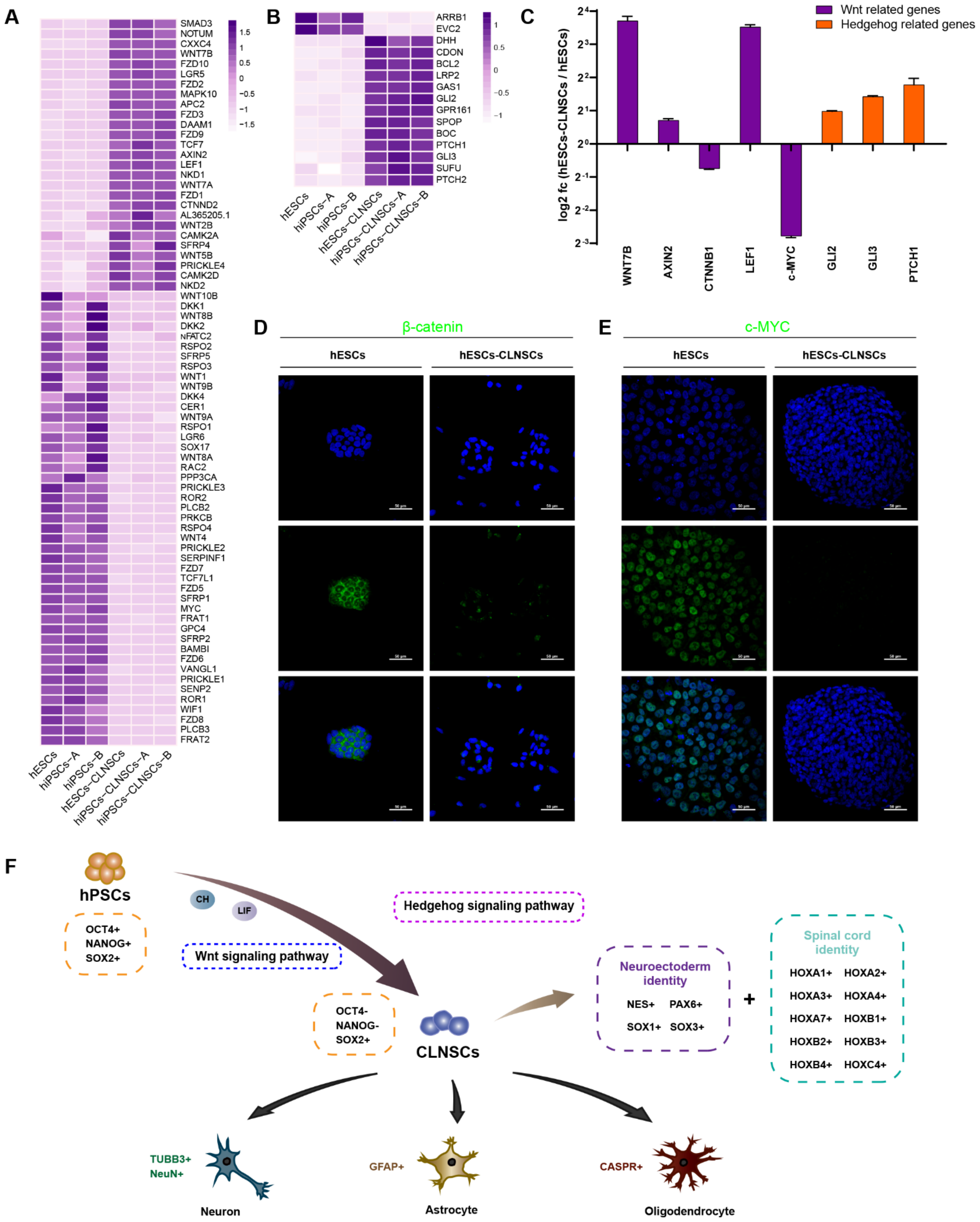
2.4. Global Transcriptional Features of CLNSCs
2.5. Spinal Cord Region-Specific Gene Expression Pattern of CLNSCs
2.6. Gene Network and Core Pathways in Self-Renewing CLNSCs
3. Discussion
4. Materials and Methods
4.1. Human Pluripotent Stem Cells Culture
4.2. Neural Induction and Neural Stem Cells Maintenance
4.3. Differentiation of Neural Stem Cells
4.4. Alkaline Phosphatase Staining
4.5. Karyotype
4.6. Immunocytochemistry
4.7. Real-Time PCR
4.8. Flow Cytometry
4.9. RNA-seq and Analysis
4.10. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Muir, K.W.; Bulters, D.; Willmot, M.; Sprigg, N.; Dixit, A.; Ward, N.; Tyrrell, P.; Majid, A.; Dunn, L.; Bath, P.; et al. Intracerebral implantation of human neural stem cells and motor recovery after stroke: Multicentre prospective single-arm study (PISCES-2). J. Neurol. Neurosurg. Psychiatry 2020, 91, 396–401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalladka, D.; Sinden, J.; McLean, J.; Moreton, F.C.; Huang, X.; Muir, K.W. Increased deep grey matter functional connectivity of poststroke hNSC implanted ipsilesional putamen. J. Neurol. Neurosurg. Psychiatry 2019, 90, 959–960. [Google Scholar] [CrossRef] [PubMed]
- Mazzini, L.; Gelati, M.; Profico, D.C.; Sorarù, G.; Ferrari, D.; Copetti, M.; Muzi, G.; Ricciolini, C.; Carletti, S.; Giorgi, C.; et al. Results from Phase I Clinical Trial with Intraspinal Injection of Neural Stem Cells in Amyotrophic Lateral Sclerosis: A Long-Term Outcome. Stem Cells Transl. Med. 2019, 8, 887–897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levi, A.; Anderson, K.; Okonkwo, D.; Park, P.; Bryce, T.N.; Kurpad, S.N.; Aarabi, B.; Hsieh, J.; Gant, K. Clinical Outcomes from a Multi-Center Study of Human Neural Stem Cell Transplantation in Chronic Cervical Spinal Cord Injury. J. Neurotrauma 2019, 36, 891–902. [Google Scholar] [CrossRef] [PubMed]
- Curtis, E.; Martin, J.; Gabel, B.; Sidhu, N.; Rzesiewicz, T.K.; Mandeville, R.; Van Gorp, S.; Leerink, M.; Tadokoro, T.; Marsala, S.; et al. A First-in-Human, Phase I Study of Neural Stem Cell Transplantation for Chronic Spinal Cord Injury. Cell Stem Cell 2018, 22, 941–950.e6. [Google Scholar] [CrossRef] [Green Version]
- Portnow, J.; Synold, T.; Badie, B.; Tirughana, R.; Lacey, S.F.; D’Apuzzo, M.; Metz, M.Z.; Najbauer, J.; Bedell, V.; Vo, T.; et al. Neural Stem Cell–Based Anticancer Gene Therapy: A First-in-Human Study in Recurrent High-Grade Glioma Patients. Clin. Cancer Res. 2017, 23, 2951–2960. [Google Scholar] [CrossRef] [Green Version]
- Bohaciakova, D.; Hruska-Plochan, M.; Tsunemoto, R.; Gifford, W.D.; Driscoll, S.P.; Glenn, T.D.; Wu, S.; Marsala, S.; Navarro, M.; Tadokoro, T.; et al. A scalable solution for isolating human multipotent clinical-grade neural stem cells from ES precursors. Stem Cell Res. Ther. 2019, 10, 83. [Google Scholar] [CrossRef]
- Lu, B.; Morgans, C.W.; Girman, S.; Luo, J.; Zhao, J.; Du, H.; Lim, S.; Ding, S.; Svendsen, C.; Zhang, K.; et al. Neural Stem Cells Derived by Small Molecules Preserve Vision. Transl. Vis. Sci. Technol. 2013, 2, 1. [Google Scholar] [CrossRef] [Green Version]
- Han, X.; Yu, L.; Ren, J.; Wang, M.; Liu, Z.; Hu, X.; Hu, D.; Chen, Y.; Chen, L.; Zhang, Y.; et al. Efficient and Fast Differentiation of Human Neural Stem Cells from Human Embryonic Stem Cells for Cell Therapy. Stem Cells Int. 2017, 2017, 9405204. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, Y.; Okada, Y.; Itakura, G.; Iwai, H.; Nishimura, S.; Yasuda, A.; Nori, S.; Hikishima, K.; Konomi, T.; Fujiyoshi, K.; et al. Pre-evaluated safe human iPSC-derived neural stem cells promote functional recovery after spinal cord injury in common marmoset without tumorigenicity. PLoS ONE 2012, 7, e52787. [Google Scholar] [CrossRef] [Green Version]
- Fujimoto, Y.; Abematsu, M.; Falk, A.; Tsujimura, K.; Sanosaka, T.; Juliandi, B.; Semi, K.; Namihira, M.; Komiya, S.; Smith, A.; et al. Treatment of a Mouse Model of Spinal Cord Injury by Transplantation of Human Induced Pluripotent Stem Cell-Derived Long-Term Self-Renewing Neuroepithelial-Like Stem Cells. Stem Cells 2012, 30, 1163–1173. [Google Scholar] [CrossRef]
- Salewski, R.P.; Mitchell, R.A.; Li, L.; Shen, C.; Milekovskaia, M.; Nagy, A.; Fehlings, M.G. Transplantation of Induced Pluripotent Stem Cell-Derived Neural Stem Cells Mediate Functional Recovery Following Thoracic Spinal Cord Injury Through Remyelination of Axons. Stem Cells Transl. Med. 2015, 4, 743–754. [Google Scholar] [CrossRef]
- Kojima, K.; Miyoshi, H.; Nagoshi, N.; Kohyama, J.; Itakura, G.; Kawabata, S.; Ozaki, M.; Iida, T.; Sugai, K.; Ito, S.; et al. Selective Ablation of Tumorigenic Cells Following Human Induced Pluripotent Stem Cell-Derived Neural Stem/Progenitor Cell Transplantation in Spinal Cord Injury. Stem Cells Transl. Med. 2018, 8, 260–270. [Google Scholar] [CrossRef] [Green Version]
- Tanimoto, Y.; Yamasaki, T.; Nagoshi, N.; Nishiyama, Y.; Nori, S.; Nishimura, S.; Iida, T.; Ozaki, M.; Tsuji, O.; Ji, B.; et al. In vivo monitoring of remnant undifferentiated neural cells following human induced pluripotent stem cell-derived neural stem/progenitor cells transplantation. Stem Cells Transl. Med. 2020, 9, 465–477. [Google Scholar] [CrossRef] [Green Version]
- Sheng, C.; Zheng, Q.; Wu, J.; Xu, Z.; Wang, L.; Li, W.; Zhang, H.; Zhao, X.-Y.; Liu, L.; Wang, Z.; et al. Direct reprogramming of Sertoli cells into multipotent neural stem cells by defined factors. Cell Res. 2011, 22, 208–218. [Google Scholar] [CrossRef] [Green Version]
- Ring, K.L.; Tong, L.M.; Balestra, M.E.; Javier, R.; Andrews-Zwilling, Y.; Li, G.; Walker, D.; Zhang, W.R.; Kreitzer, A.C.; Huang, Y. Direct Reprogramming of Mouse and Human Fibroblasts into Multipotent Neural Stem Cells with a Single Factor. Cell Stem Cell 2012, 11, 100–109. [Google Scholar] [CrossRef] [Green Version]
- Liao, W.; Huang, N.; Yu, J.; Jares, A.; Yang, J.; Zieve, G.; Avila, C.; Jiang, X.; Zhang, X.-B.; Ma, Y. Direct Conversion of Cord Blood CD34+ Cells Into Neural Stem Cells by OCT4. Stem Cells Transl. Med. 2015, 4, 755–763. [Google Scholar] [CrossRef]
- Kim, J.-J.; Shin, J.-H.; Yu, K.-R.; Lee, B.-C.; Kang, I.; Lee, J.Y.; Kim, D.-H.; Seo, Y.; Kim, H.-S.; Choi, S.W.; et al. Direct Conversion of Human Umbilical Cord Blood into Induced Neural Stem Cells with SOX2 and HMGA2. Int. J. Stem Cells 2017, 10, 227–234. [Google Scholar] [CrossRef] [Green Version]
- Flax, J.D.; Aurora, S.; Yang, C.; Simonin, C.; Wills, A.M.; Billinghurst, L.L.; Jendoubi, M.; Sidman, R.L.; Wolfe, J.H.; Kim, S.U.; et al. Engraftable human neural stem cells respond to developmental cues, replace neurons, and express foreign genes. Nat. Biotechnol. 1998, 16, 1033–1039. [Google Scholar] [CrossRef]
- Reynolds, B.; Weiss, S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science 1992, 255, 1707–1710. [Google Scholar] [CrossRef] [Green Version]
- Reubinoff, B.; Itsykson, P.; Turetsky, T.; Pera, M.; Reinhartz, E.; Itzik, A.; Ben-Hur, T. Neural progenitors from human embryonic stem cells. Nat. Biotechnol. 2001, 19, 1134–1140. [Google Scholar] [CrossRef]
- Pera, M.F.; Andrade, J.; Houssami, S.; Reubinoff, B.; Trounson, A.; Stanley, E.G.; Oostwaard, D.W.-V.; Mummery, C. Regulation of human embryonic stem cell differentiation by BMP-2 and its antagonist noggin. J. Cell Sci. 2004, 117, 1269–1280. [Google Scholar] [CrossRef] [Green Version]
- Keirstead, H.S.; Nistor, G.; Bernal, G.; Totoiu, M.; Cloutier, F.; Sharp, K.; Steward, O. Human embryonic stem cell-derived oligodendrocyte progenitor cell transplants remyelinate and restore locomotion after spinal cord injury. J. Neurosci. 2005, 25, 4694–4705. [Google Scholar] [CrossRef]
- Ben-Hur, T.; Idelson, M.; Khaner, H.; Pera, M.; Reinhartz, E.; Itzik, A.; Reubinoff, B.E. Transplantation of Human Embryonic Stem Cell-Derived Neural Progenitors Improves Behavioral Deficit in Parkinsonian Rats. Stem Cells 2004, 22, 1246–1255. [Google Scholar] [CrossRef]
- Chambers, S.M.; Fasano, C.A.; Papapetrou, E.P.; Tomishima, M.; Sadelain, M.; Studer, L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 2009, 27, 275–280. [Google Scholar] [CrossRef] [Green Version]
- Wattanapanitch, M.; Klincumhom, N.; Potirat, P.; Amornpisutt, R.; Lorthongpanich, C.; U-pratya, Y.; Laowtammathron, C.; Kheolamai, P.; Poungvarin, N.; Issaragrisil, S. Dual small-molecule targeting of SMAD signaling stimulates human induced pluripotent stem cells toward neural lineages. PLoS ONE 2014, 9, e106952. [Google Scholar]
- Takahashi, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, J.; Vodyanik, M.A.; Smuga-Otto, K.; Antosiewicz-Bourget, J.; Frane, J.L.; Tian, S.; Nie, J.; Jonsdottir, G.A.; Ruotti, V.; Stewart, R.; et al. Induced Pluripotent Stem Cell Lines Derived from Human Somatic Cells. Science 2007, 318, 1917–1920. [Google Scholar] [CrossRef] [PubMed]
- Simone, C.; Nizzardo, M.; Rizzo, F.; Ruggieri, M.; Riboldi, G.; Salani, S.; Bucchia, M.; Bresolin, N.; Comi, G.P.; Corti, S. iPSC-Derived neural stem cells act via kinase inhibition to exert neuroprotective effects in spinal muscular atrophy with respiratory distress type 1. Stem Cell Rep. 2014, 3, 297–311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meneghini, V.; Frati, G.; Sala, D.; De Cicco, S.; Luciani, M.; Cavazzin, C.; Paulis, M.; Mentzen, W.; Morena, F.; Giannelli, S.; et al. Generation of human induced pluripotent stem cell-derived bona fide neural stem cells for ex vivo gene therapy of metachromatic leukodystrophy. Stem Cells Transl. Med. 2017, 6, 352–368. [Google Scholar] [CrossRef]
- Hubert, C.G.; Bradley, R.; Ding, Y.; Toledo, C.M.; Herman, J.; Skutt-Kakaria, K.; Girard, E.; Davison, J.; Berndt, J.; Corrin, P.; et al. Genome-wide RNAi screens in human brain tumor isolates reveal a novel viability requirement for PHF5A. Genes Dev. 2013, 27, 1032–1045. [Google Scholar] [CrossRef] [Green Version]
- Cotney, J.; Muhle, R.A.; Sanders, S.; Liu, L.; Willsey, A.J.; Niu, W.; Liu, W.; Klei, L.; Lei, J.; Yin, J.; et al. The autism-associated chromatin modifier CHD8 regulates other autism risk genes during human neurodevelopment. Nat. Commun. 2015, 6, 6404. [Google Scholar] [CrossRef]
- Kumamaru, H.; Kadoya, K.; Adler, A.F.; Takashima, Y.; Graham, L.; Coppola, G.; Tuszynski, M.H. Generation and post-injury integration of human spinal cord neural stem cells. Nat. Methods 2018, 15, 723–731. [Google Scholar] [CrossRef]
- Poplawski, G.H.D.; Kawaguchi, R.; Van Niekerk, E.; Lu, P.; Mehta, N.; Canete, P.; Lie, R.; Dragatsis, I.; Meves, J.M.; Zheng, B.; et al. Injured adult neurons regress to an embryonic transcriptional growth state. Nat. Cell Biol. 2020, 581, 77–82. [Google Scholar] [CrossRef]
- Sato, N.; Meijer, L.; Skaltsounis, L.; Greengard, P.; Brivanlou, A.H. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat. Med. 2004, 10, 55–63. [Google Scholar] [CrossRef]
- Pera, M.F.; Tam, P.P.L. Extrinsic regulation of pluripotent stem cells. Nat. Cell Biol. 2010, 465, 713–720. [Google Scholar] [CrossRef]
- Ye, S.; Tan, L.; Yang, R.; Fang, B.; Qu, S.; Schulze, E.N.; Song, H.; Ying, Q.; Li, P. Pleiotropy of glycogen synthase kinase-3 inhibition by CHIR99021 promotes self-renewal of embryonic stem cells from refractory mouse strains. PLoS ONE 2012, 7, e35892. [Google Scholar]
- Ai, Z.; Niu, B.; Duan, K.; Si, C.; Wang, S.; Xiang, L.; Zhu, X.; Zhu, Q.; Feng, C.; Yin, Y.; et al. Modulation of Wnt and Activin/Nodal supports efficient derivation, cloning and suspension expansion of human pluripotent stem cells. Biomaterials 2020, 249, 120015. [Google Scholar] [CrossRef]
- Moris, N.; Anlas, K.; Brink, S.C.V.D.; Alemany, A.; Schröder, J.; Ghimire, S.; Balayo, T.; van Oudenaarden, A.; Arias, A.M. An in vitro model of early anteroposterior organization during human development. Nat. Cell Biol. 2020, 582, 410–415. [Google Scholar] [CrossRef]
- Zechner, D.; Fujita, Y.; Hulsken, J.; Müller, T.; Walther, I.; Taketo, M.M.; Crenshaw, E.B., 3rd; Birchmeier, W.; Birchmeier, C. beta-Catenin signals regulate cell growth and the balance between progenitor cell expansion and differentiation in the nervous system. Dev. Biol. 2003, 258, 406–418. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Liao, Y.; Qiu, M.; Shen, W. Wnt/beta-Catenin Signaling in Neural Stem Cell Homeostasis and Neurological Diseases. Neuroscientist 2021, 27, 58–72. [Google Scholar] [CrossRef] [PubMed]
- Goldman, S.A. Stem and Progenitor Cell-Based Therapy of the Central Nervous System: Hopes, Hype, and Wishful Thinking. Cell Stem Cell 2016, 18, 174–188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.-C.; Wernig, M.; Duncan, I.D.; Brüstle, O.; Thomson, J.A. In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat. Biotechnol. 2001, 19, 1129–1133. [Google Scholar] [CrossRef] [PubMed]
- Elkabetz, Y.; Panagiotakos, G.; Al Shamy, G.; Socci, N.D.; Tabar, V.; Studer, L. Human ES cell-derived neural rosettes reveal a functionally distinct early neural stem cell stage. Genes Dev. 2008, 22, 152–165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, W.N.; Cheng, C.; Theriault, K.M.; Sheridan, S.D.; Tsai, L.; Haggarty, S.J. A high-throughput screen for Wnt/beta-catenin signaling pathway modulators in human iPSC-derived neural progenitors. J. Biomol. Screen 2012, 17, 1252–1263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chailangkarn, T.; Trujillo, C.A.; Freitas, B.C.; Hrvoj-Mihic, B.; Herai, R.H.; Yu, D.X.; Brown, T.T.; Marchetto, M.C.; Bardy, C.; McHenry, L.; et al. A human neurodevelopmental model for Williams syndrome. Nat. Cell Biol. 2016, 536, 338–343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacob, F.; Salinas, R.D.; Zhang, D.Y.; Nguyen, P.T.; Schnoll, J.G.; Wong, S.Z.H.; Thokala, R.; Sheikh, S.; Saxena, D.; Prokop, S.; et al. A Patient-Derived Glioblastoma Organoid Model and Biobank Recapitulates Inter- and Intra-tumoral Heterogeneity. Cell 2020, 180, 188–204.e22. [Google Scholar] [CrossRef]
- Yilmaz, A.; Braverman-Gross, C.; Bialer-Tsypin, A.; Peretz, M.; Benvenisty, N. Mapping Gene Circuits Essential for Germ Layer Differentiation via Loss-of-Function Screens in Haploid Human Embryonic Stem Cells. Cell Stem Cell 2020, 27, 679–691.e6. [Google Scholar] [CrossRef]
- Narayanan, G.; Poonepalli, A.; Chen, J.; Sankaran, S.; Hariharan, S.; Yu, Y.H.; Robson, P.; Yang, H.; Ahmed, S. Single-Cell mRNA Profiling Identifies Progenitor Subclasses in Neurospheres. Stem Cells Dev. 2012, 21, 3351–3362. [Google Scholar] [CrossRef] [Green Version]
- Ninkovic, J.; Steiner-Mezzadri, A.; Jawerka, M.; Akinci, U.; Masserdotti, G.; Petricca, S.; Fischer, J.; von Holst, A.; Beckers, J.; Lie, D.C.; et al. The BAF Complex Interacts with Pax6 in Adult Neural Progenitors to Establish a Neurogenic Cross-Regulatory Transcriptional Network. Cell Stem Cell 2013, 13, 403–418. [Google Scholar] [CrossRef] [Green Version]
- Josephson, R.; Muller, T.; Pickel, J.; Okabe, S.; Reynolds, K.; Turner, P.; Zimmer, A.; McKay, R. POU transcription factors control expression of CNS stem cell-specific genes. Development 1998, 125, 3087–3100. [Google Scholar] [CrossRef]
- Decker, B.; Liput, M.; Abdellatif, H.; Yergeau, D.; Bae, Y.; Jornet, J.M.; Stachowiak, E.K.; Stachowiak, M.K. Global Genome Conformational Programming during Neuronal Development Is Associated with CTCF and Nuclear FGFR1-The Genome Archipelago Model. Int. J. Mol. Sci. 2020, 22, 347. [Google Scholar] [CrossRef]
- Fuccillo, M.; Joyner, A.L.; Fishell, G. Morphogen to mitogen: The multiple roles of hedgehog signalling in vertebrate neural development. Nat. Rev. Neurosci. 2006, 7, 772–783. [Google Scholar] [CrossRef]
- Ren, H.; Chen, X.; Tian, M.; Zhou, J.; Ouyang, H.; Zhang, Z. Regulation of Inflammatory Cytokines for Spinal Cord Injury Repair Through Local Delivery of Therapeutic Agents. Adv. Sci. 2018, 5, 1800529. [Google Scholar] [CrossRef] [Green Version]
- Cruz, C.D.; Coelho, A.; Antunes-Lopes, T.; Cruz, F. Biomarkers of spinal cord injury and ensuing bladder dysfunction. Adv. Drug Deliv. Rev. 2015, 82–83, 153–159. [Google Scholar] [CrossRef]
- Rahimi-Movaghar, V.; Sayyah, M.K.; Akbari, H.; Khorramirouz, R.; Rasouli, M.R.; Moradi-Lakeh, M.; Shokraneh, F.; Vaccaro, A.R. Epidemiology of Traumatic Spinal Cord Injury in Developing Countries: A Systematic Review. Neuroepidemiology 2013, 41, 65–85. [Google Scholar] [CrossRef]
- McDonald, J.W.; Sadowsky, C. Spinal-cord injury. Lancet 2002, 359, 417–425. [Google Scholar] [CrossRef]
- Long, J.; Federico, P.; Perez, M.A. A novel cortical target to enhance hand motor output in humans with spinal cord injury. Brain 2017, 140, 1619–1632. [Google Scholar] [CrossRef] [Green Version]
- Burnside, E.R.; De Winter, F.; Didangelos, A.; James, N.D.; Andreica, E.-C.; Horsfall, H.L.; Muir, E.M.; Verhaagen, J.; Bradbury, E. Immune-evasive gene switch enables regulated delivery of chondroitinase after spinal cord injury. Brain 2018, 141, 2362–2381. [Google Scholar] [CrossRef]
- Cheng, Z.; Zhu, W.; Cao, K.; Wu, F.; Li, J.; Wang, G.; Li, H.; Lu, M.; Ren, Y.; He, X. Anti-Inflammatory Mechanism of Neural Stem Cell Transplantation in Spinal Cord Injury. Int. J. Mol. Sci. 2016, 17, 1380. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.; Wang, F.; Chen, H.; Liu, N.; Qu, T.; Li, X.; Xu, D.; Liu, F.; Yin, Z.; Chen, Y. Co-Transplantation of Human Umbilical Cord Mesenchymal Stem Cells and Human Neural Stem Cells Improves the Outcome in Rats with Spinal Cord Injury. Cell Transplant. 2019, 28, 893–906. [Google Scholar] [CrossRef] [Green Version]
- Dulin, J.N.; Adler, A.F.; Kumamaru, H.; Poplawski, G.; Lee-Kubli, C.; Strobl, H.; Gibbs, D.; Kadoya, K.; Fawcett, J.; Lu, P.; et al. Injured adult motor and sensory axons regenerate into appropriate organotypic domains of neural progenitor grafts. Nat. Commun. 2018, 9, 84. [Google Scholar] [CrossRef]
- Dell’Anno, M.T.; Wang, X.; Onorati, M.; Li, M.; Talpo, F.; Sekine, Y.; Ma, S.; Liu, F.; Cafferty, W.B.J.; Sestan, N.; et al. Human neuroepithelial stem cell regional specificity enables spinal cord repair through a relay circuit. Nat. Commun. 2018, 9, 3419. [Google Scholar] [CrossRef] [Green Version]
- Hu, B.-Y.; Weick, J.P.; Yu, J.; Ma, L.; Zhang, X.-Q.; Thomson, J.A.; Zhang, S.-C. Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc. Natl. Acad. Sci. USA 2010, 107, 4335–4340. [Google Scholar] [CrossRef] [Green Version]
- Wu, B.; Li, L.; Li, B.; Gao, J.; Chen, Y.; Wei, M.; Yang, Z.; Zhang, B.; Li, S.; Li, K.; et al. Activin A and BMP4 Signaling Expands Potency of Mouse Embryonic Stem Cells in Serum-Free Media. Stem Cell Rep. 2020, 14, 241–255. [Google Scholar] [CrossRef] [Green Version]
- Bao, S.; Tang, W.W.; Wu, B.; Kim, S.; Li, J.; Li, L.; Kobayashi, T.; Lee, C.; Chen, Y.; Wei, M.; et al. Derivation of hypermethylated pluripotent embryonic stem cells with high potency. Cell Res. 2017, 28, 22–34. [Google Scholar] [CrossRef] [Green Version]
- Wu, B.; Li, Y.; Li, B.; Zhang, B.; Wang, Y.; Li, L.; Gao, J.; Fu, Y.; Li, S.; Chen, C.; et al. DNMTs Play an Important Role in Maintaining the Pluripotency of Leukemia Inhibitory Factor-Dependent Embryonic Stem Cells. Stem Cell Rep. 2021, 16, 582–596. [Google Scholar] [CrossRef]
- Theunissen, T.W.; Powell, B.E.; Wang, H.; Mitalipova, M.; Faddah, D.A.; Reddy, J.; Fan, Z.P.; Maetzel, D.; Ganz, K.; Shi, L.; et al. Systematic Identification of Culture Conditions for Induction and Maintenance of Naive Human Pluripotency. Cell Stem Cell 2014, 15, 471–487. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Liu, B.; Xu, J.; Wang, J.; Wu, J.; Shi, C.; Xu, Y.; Dong, J.; Wang, C.; Lai, W.; et al. Derivation of Pluripotent Stem Cells with In Vivo Embryonic and Extraembryonic Potency. Cell 2017, 169, 243–257.e25. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Xie, G.; Singh, M.; Ghanbarian, A.T.; Raskó, T.; Szvetnik, A.; Cai, H.; Besser, D.; Prigione, A.; Fuchs, N.V.; et al. Primate-specific endogenous retrovirus-driven transcription defines naive-like stem cells. Nat. Cell Biol. 2014, 516, 405–409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Y.; Ai, Z.; Yao, K.; Cao, L.; Du, J.; Shi, X.; Guo, Z.; Zhang, Y. CHIR99021 promotes self-renewal of mouse embryonic stem cells by modulation of protein-encoding gene and long intergenic non-coding RNA expression. Exp. Cell Res. 2013, 319, 2684–2699. [Google Scholar] [CrossRef] [PubMed]
- Sato, A.; Yamamoto, H.; Sakane, H.; Koyama, H.; Kikuchi, A. Wnt5a regulates distinct signalling pathways by binding to Frizzled2. EMBO J. 2009, 29, 41–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernando, C.V.; Kele, J.; Bye, C.R.; Niclis, J.C.; Alsanie, W.; Blakely, B.D.; Stenman, J.; Turner, B.J.; Parish, C.L. Diverse roles for Wnt7a in ventral midbrain neurogenesis and dopaminergic axon morphogenesis. Stem Cells Dev. 2014, 23, 1991–2003. [Google Scholar] [CrossRef]
- Ciani, L.; Boyle, K.; Dickins, E.; Sahores, M.; Anane, D.; Lopes, D.; Gibb, A.; Salinas, P.C. Wnt7a signaling promotes dendritic spine growth and synaptic strength through Ca2+/Calmodulin-dependent protein kinase II. Proc. Natl. Acad. Sci. USA 2011, 108, 10732–10737. [Google Scholar] [CrossRef] [Green Version]
- Delaunay, D.; Cortay, V.; Patti, D.; Knoblauch, K.; Dehay, C. Mitotic Spindle Asymmetry: A Wnt/PCP-Regulated Mechanism Generating Asymmetrical Division in Cortical Precursors. Cell Rep. 2014, 6, 400–414. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.P.; Laufer, E.; Jessell, T.M. Assigning the positional identity of spinal motor neurons: Rostrocaudal patterning of Hox-c expression by FGFs, Gdf11, and retinoids. Neuron 2001, 32, 997–1012. [Google Scholar] [CrossRef] [Green Version]
- Peljto, M.; Dasen, J.S.; Mazzoni, E.O.; Jessell, T.M.; Wichterle, H. Functional Diversity of ESC-Derived Motor Neuron Subtypes Revealed through Intraspinal Transplantation. Cell Stem Cell 2010, 7, 355–366. [Google Scholar] [CrossRef] [Green Version]
- Bel-Vialar, S.; Itasaki, N.; Krumlauf, R. Initiating Hox gene expression: In the early chick neural tube differential sensitivity to FGF and RA signaling subdivides the HoxB genes in two distinct groups. Development 2002, 129, 5103–5115. [Google Scholar] [CrossRef]
- Mahony, S.; Mazzoni, E.O.; McCuine, S.; Young, R.A.; Wichterle, H.; Gifford, D.K. Ligand-dependent dynamics of retinoic acid receptor binding during early neurogenesis. Genome Biol. 2011, 12, R2. [Google Scholar] [CrossRef] [Green Version]
- Mazzoni, E.O.; Mahony, S.; Peljto, M.; Patel, T.; Thornton, S.R.; McCuine, S.; Reeder, C.C.; Boyer, L.A.; Young, R.A.; Gifford, D.K.; et al. Saltatory remodeling of Hox chromatin in response to rostrocaudal patterning signals. Nat. Neurosci. 2013, 16, 1191–1198. [Google Scholar] [CrossRef] [Green Version]
- Gard, C.; Gonzalez Curto, G.; Frarma, Y.E.; Chollet, E.; Duval, N.; Auzié, V.; Aurade, F.; Vigier, L.; Relaix, F.; Pierani, A.; et al. Pax3- and Pax7-mediated Dbx1 regulation orchestrates the patterning of intermediate spinal interneurons. Dev. Biol. 2017, 432, 24–33. [Google Scholar] [CrossRef]
- Gao, X.; Nowak-Imialek, M.; Chen, X.; Chen, D.; Herrmann, D.; Ruan, D.; Chen, A.C.H.; Eckersley-Maslin, M.A.; Ahmad, S.; Lee, Y.L.; et al. Establishment of porcine and human expanded potential stem cells. Nat. Cell Biol. 2019, 21, 687–699. [Google Scholar] [CrossRef] [Green Version]
- Gelati, M.; Profico, D.; Projetti-Pensi, M.; Muzi, G.; Sgaravizzi, G.; Vescovi, A.L. Culturing and Expansion of “Clinical Grade” Precursors Cells from the Fetal Human Central Nervous System. Adv. Struct. Saf. Stud. 2013, 1059, 65–77. [Google Scholar]
- Li, T.; Shi, Y.; Wang, P.; Guachalla, L.M.; Sun, B.; Joerss, T.; Chen, Y.; Groth, M.; Krueger, A.; Platzer, M.; et al. Smg6/Est1 licenses embryonic stem cell differentiation via nonsense-mediated mRNA decay. EMBO J. 2015, 34, 1630–1647. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, B.-C.; Liu, F.-Y.; Gao, X.-Y.; Chen, Y.-L.; Meng, Q.-Q.; Song, Y.-L.; Li, X.-H.; Bao, S.-Q. Global Transcriptional Analyses of the Wnt-Induced Development of Neural Stem Cells from Human Pluripotent Stem Cells. Int. J. Mol. Sci. 2021, 22, 7473. https://doi.org/10.3390/ijms22147473
Liu B-C, Liu F-Y, Gao X-Y, Chen Y-L, Meng Q-Q, Song Y-L, Li X-H, Bao S-Q. Global Transcriptional Analyses of the Wnt-Induced Development of Neural Stem Cells from Human Pluripotent Stem Cells. International Journal of Molecular Sciences. 2021; 22(14):7473. https://doi.org/10.3390/ijms22147473
Chicago/Turabian StyleLiu, Bing-Chun, Fang-Yuan Liu, Xin-Yue Gao, Yang-Lin Chen, Qiao-Qiao Meng, Yong-Li Song, Xi-He Li, and Si-Qin Bao. 2021. "Global Transcriptional Analyses of the Wnt-Induced Development of Neural Stem Cells from Human Pluripotent Stem Cells" International Journal of Molecular Sciences 22, no. 14: 7473. https://doi.org/10.3390/ijms22147473
APA StyleLiu, B.-C., Liu, F.-Y., Gao, X.-Y., Chen, Y.-L., Meng, Q.-Q., Song, Y.-L., Li, X.-H., & Bao, S.-Q. (2021). Global Transcriptional Analyses of the Wnt-Induced Development of Neural Stem Cells from Human Pluripotent Stem Cells. International Journal of Molecular Sciences, 22(14), 7473. https://doi.org/10.3390/ijms22147473




