Oncolytic Viruses in Combination Therapeutic Approaches with Epigenetic Modulators: Past, Present, and Future Perspectives
Abstract
Simple Summary
Abstract
1. Introduction
2. Viral Families Most Used in Virotherapy
2.1. Herpesviridae
2.2. Adenoviridae
2.3. Rhabdoviridae
2.4. Reoviridae
2.5. Parvoviridae
2.6. Paramyxoviridae
2.7. Poxviridae
2.8. Picornaviridae
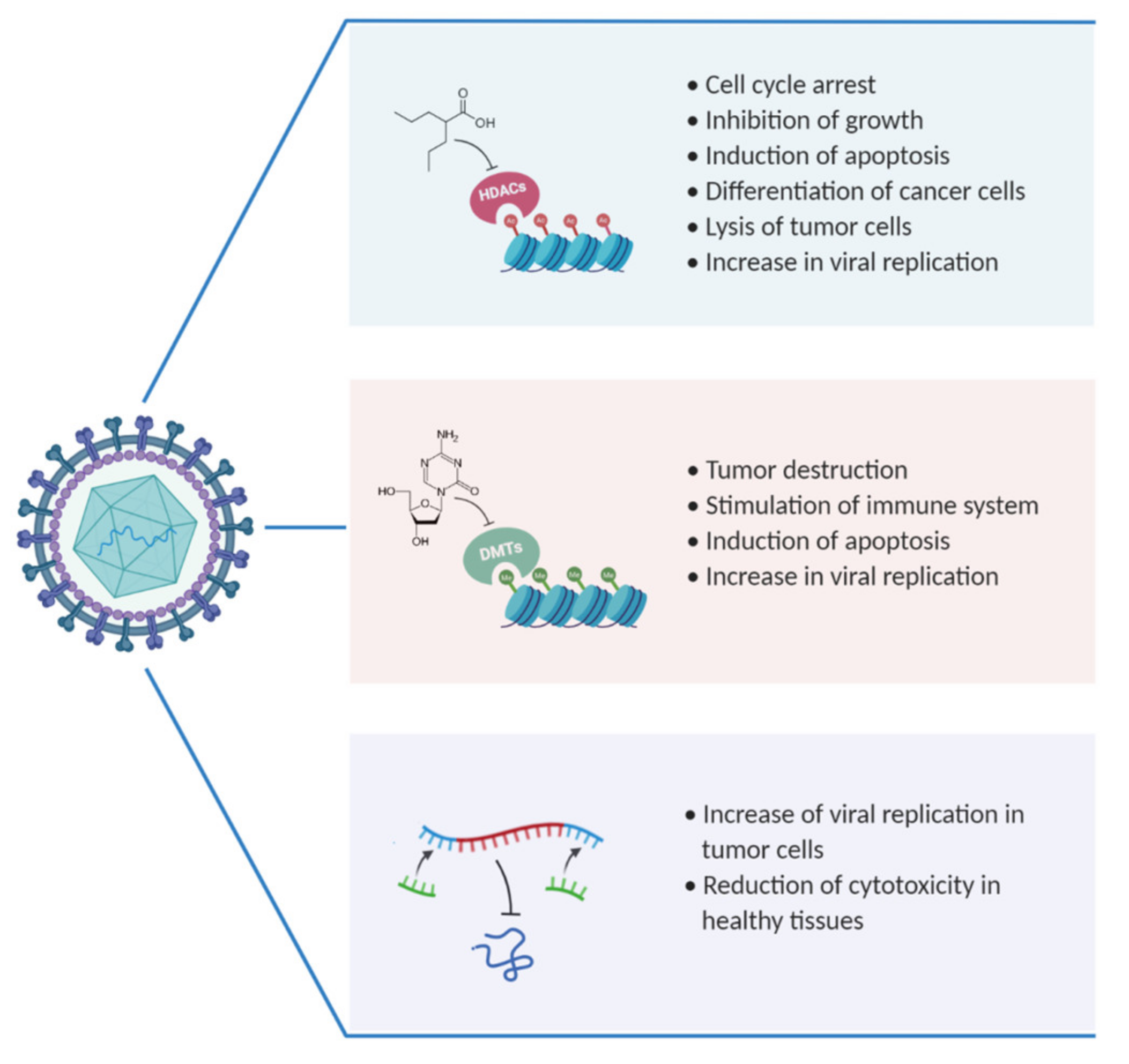
3. OVs and HDACs as Combinatorial Therapy
3.1. Herpesviridae
3.2. Adenoviridae
3.3. Rhabdoviridae
3.4. Reoviridae
3.5. Parvoviridae
3.6. Paramyxoviridae
3.7. Poxviridae
4. OVs and DNMTi as Combinatorial Therapy
4.1. Herpesviridae
4.2. Adenoviridae
4.3. Rhabdoviridae
5. OVs and miRNA: Promising Combinatorial Treatment
5.1. Herpesviridae
5.2. Adenoviridae
5.3. Picornaviridae
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nagai, H.; Kim, Y.H. Cancer prevention from the perspective of global cancer burden patterns. J. Thorac. Dis. 2017, 9, 448–451. [Google Scholar] [CrossRef] [PubMed]
- WHO. International Agency for Research Cancer: Lyon, France. 2018. Available online: https://www.who.int/cancer/PRGlobocanFinal.pdf (accessed on 31 May 2021).
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Rahib, L.; Smith, B.D.; Aizenberg, R.; Rosenzweig, A.B.; Fleshman, J.M.; Matrisian, L.M. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014, 74, 2913–2921. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef] [PubMed]
- Ascione, A.; Fontanella, L.; Imparato, M.; Rinaldi, L.; De Luca, M. Mortality from cirrhosis and hepatocellular carcinoma in Western Europe over the last 40 years. Liver Int. 2017, 37, 1193–1201. [Google Scholar] [CrossRef]
- Lenti, M.V.; Pasina, L.; Cococcia, S.; Cortesi, L.; Miceli, E.; Caccia Dominioni, C.; Pisati, M.; Mengoli, C.; Perticone, F.; Nobili, A.; et al. Mortality rate and risk factors for gastrointestinal bleeding in elderly patients. Eur. J. Intern. Med. 2019, 61, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.D.; Hainaut, P.; Gores, G.J.; Amadou, A.; Plymoth, A.; Roberts, L.R. A global view of hepatocellular carcinoma: Trends, risk, prevention and management. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 589–604. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, N.; Friedman, S.L.; Goossens, N.; Hoshida, Y. Risk factors and prevention of hepatocellular carcinoma in the era of precision medicine. J. Hepatol. 2018, 68, 526–549. [Google Scholar] [CrossRef]
- Anwanwan, D.; Singh, S.K.; Singh, S.; Saikam, V.; Singh, R. Challenges in liver cancer and possible treatment approaches. Biochim. Biophys. Acta Rev. Cancer 2020, 1873, 188314. [Google Scholar] [CrossRef] [PubMed]
- Di Francia, R.; Rinaldi, L.; Cillo, M.; Varriale, E.; Facchini, G.; D’Aniello, C.; Marotta, G.; Berretta, M. Antioxidant diet and genotyping as tools for the prevention of liver disease. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 5155–5163. [Google Scholar]
- Yu, J.; Wang, N.; Gong, Z.; Liu, L.; Yang, S.; Chen, G.G.; Lai, P.B.S. Cytochrome P450 1A2 overcomes nuclear factor kappa B-mediated sorafenib resistance in hepatocellular carcinoma. Oncogene 2021, 40, 492–507. [Google Scholar] [CrossRef]
- Yang, X.D.; Kong, F.E.; Qi, L.; Lin, J.X.; Yan, Q.; Loong, J.H.C.; Xi, S.Y.; Zhao, Y.; Zhang, Y.; Yuan, Y.F.; et al. PARP inhibitor Olaparib overcomes Sorafenib resistance through reshaping the pluripotent transcriptome in hepatocellular carcinoma. Mol. Cancer 2021, 20, 20. [Google Scholar] [CrossRef]
- Mendez-Blanco, C.; Fondevila, F.; Garcia-Palomo, A.; Gonzalez-Gallego, J.; Mauriz, J.L. Sorafenib resistance in hepatocarcinoma: Role of hypoxia-inducible factors. Exp. Mol. Med. 2018, 50, 1–9. [Google Scholar] [CrossRef]
- Zhu, Y.J.; Zheng, B.; Wang, H.Y.; Chen, L. New knowledge of the mechanisms of sorafenib resistance in liver cancer. Acta Pharmacol. Sin. 2017, 38, 614–622. [Google Scholar] [CrossRef] [PubMed]
- Rinaldi, L.; Valente, G.; Piai, G. Serial Liver Stiffness Measurements and Monitoring of Liver-Transplanted Patients in a Real-Life Clinical Practice. Hepat. Mon. 2016, 16, e41162. [Google Scholar] [CrossRef] [PubMed]
- Di Francia, R.; Rinaldi, L.; Troisi, A.; Di Benedetto, F.; Berretta, M. Effect of anti-oxidant agents in patients with hepatocellular diseases. Eur Rev. Med. Pharmacol. Sci. 2015, 19, 3993–3995. [Google Scholar] [PubMed]
- Narayan, V.; Vaughn, D. Pharmacokinetic and toxicity considerations in the use of neoadjuvant chemotherapy for bladder cancer. Expert Opin. Drug Metab. Toxicol. 2015, 11, 731–742. [Google Scholar] [CrossRef] [PubMed]
- Bahadoer, R.R.; Dijkstra, E.A.; van Etten, B.; Marijnen, C.A.M.; Putter, H.; Kranenbarg, E.M.; Roodvoets, A.G.H.; Nagtegaal, I.D.; Beets-Tan, R.G.H.; Blomqvist, L.K.; et al. Short-course radiotherapy followed by chemotherapy before total mesorectal excision (TME) versus preoperative chemoradiotherapy, TME, and optional adjuvant chemotherapy in locally advanced rectal cancer (RAPIDO): A randomised, open-label, phase 3 trial. Lancet Oncol. 2021, 22, 29–42. [Google Scholar] [CrossRef]
- Jindal, A.; Thadi, A.; Shailubhai, K. Hepatocellular Carcinoma: Etiology and Current and Future Drugs. J. Clin. Exp. Hepatol. 2019, 9, 221–232. [Google Scholar] [CrossRef]
- Bouattour, M.; Mehta, N.; He, A.R.; Cohen, E.I.; Nault, J.C. Systemic Treatment for Advanced Hepatocellular Carcinoma. Liver Cancer 2019, 8, 341–358. [Google Scholar] [CrossRef]
- Kelly, E.; Russell, S.J. History of oncolytic viruses: Genesis to genetic engineering. Mol. Ther. 2007, 15, 651–659. [Google Scholar] [CrossRef]
- Hoster, H.A.; Zanes, R.P., Jr.; Von Haam, E. Studies in Hodgkin’s syndrome; the association of viral hepatitis and Hodgkin’s disease; a preliminary report. Cancer Res. 1949, 9, 473–480. [Google Scholar]
- Lin, E.; Nemunaitis, J. Oncolytic viral therapies. Cancer Gene Ther. 2004, 11, 643–664. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.W.; Lee, J.S.; Kim, S.W.; Yun, C.O. Evolution of oncolytic adenovirus for cancer treatment. Adv. Drug Deliv. Rev. 2012, 64, 720–729. [Google Scholar] [CrossRef] [PubMed]
- Kalyanasundram, J.; Hamid, A.; Yusoff, K.; Chia, S.L. Newcastle disease virus strain AF2240 as an oncolytic virus: A review. Acta Trop. 2018, 183, 126–133. [Google Scholar] [CrossRef]
- Lin, C.Z.; Xiang, G.L.; Zhu, X.H.; Xiu, L.L.; Sun, J.X.; Zhang, X.Y. Advances in the mechanisms of action of cancer-targeting oncolytic viruses. Oncol. Lett. 2018, 15, 4053–4060. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; He, H.; Wang, H. Oncolytic herpes simplex virus and immunotherapy. BMC Immunol. 2018, 19, 40. [Google Scholar] [CrossRef]
- Yu, X.; He, S. The interplay between human herpes simplex virus infection and the apoptosis and necroptosis cell death pathways. Virol. J. 2016, 13, 77. [Google Scholar] [CrossRef]
- Suazo, P.A.; Ibanez, F.J.; Retamal-Diaz, A.R.; Paz-Fiblas, M.V.; Bueno, S.M.; Kalergis, A.M.; Gonzalez, P.A. Evasion of early antiviral responses by herpes simplex viruses. Mediat. Inflamm. 2015, 2015, 593757. [Google Scholar] [CrossRef] [PubMed]
- Alberts, P.; Tilgase, A.; Rasa, A.; Bandere, K.; Venskus, D. The advent of oncolytic virotherapy in oncology: The Rigvir(R) story. Eur. J. Pharmacol. 2018, 837, 117–126. [Google Scholar] [CrossRef]
- Fukuhara, H.; Ino, Y.; Todo, T. Oncolytic virus therapy: A new era of cancer treatment at dawn. Cancer Sci. 2016, 107, 1373–1379. [Google Scholar] [CrossRef] [PubMed]
- ClinicalTrials.gov. Available online: https://clinicaltrials.gov/ct2/resources/trends (accessed on 31 May 2021).
- Liang, M. Oncorine, the World First Oncolytic Virus Medicine and its Update in China. Curr. Cancer Drug Targets 2018, 18, 171–176. [Google Scholar] [CrossRef]
- Pol, J.; Kroemer, G.; Galluzzi, L. First oncolytic virus approved for melanoma immunotherapy. Oncoimmunology 2016, 5, e1115641. [Google Scholar] [CrossRef] [PubMed]
- Raja, J.; Ludwig, J.M.; Gettinger, S.N.; Schalper, K.A.; Kim, H.S. Oncolytic virus immunotherapy: Future prospects for oncology. J. Immunother. Cancer 2018, 6, 140. [Google Scholar] [CrossRef]
- Russell, S.J.; Peng, K.W.; Bell, J.C. Oncolytic virotherapy. Nat. Biotechnol. 2012, 30, 658–670. [Google Scholar] [CrossRef] [PubMed]
- Taguchi, S.; Fukuhara, H.; Todo, T. Oncolytic virus therapy in Japan: Progress in clinical trials and future perspectives. JPN J. Clin. Oncol. 2019, 49, 201–209. [Google Scholar] [CrossRef]
- Franco-Luzon, L.; Gonzalez-Murillo, A.; Alcantara-Sanchez, C.; Garcia-Garcia, L.; Tabasi, M.; Huertas, A.L.; Chesler, L.; Ramirez, M. Systemic oncolytic adenovirus delivered in mesenchymal carrier cells modulate tumor infiltrating immune cells and tumor microenvironment in mice with neuroblastoma. Oncotarget 2020, 11, 347–361. [Google Scholar] [CrossRef]
- Liu, G.B.; Zhao, L.; Zhang, L.; Zhao, K.N. Virus, Oncolytic Virus and Human Prostate Cancer. Curr. Cancer Drug Targets 2017, 17, 522–533. [Google Scholar] [CrossRef]
- Niemann, J.; Kuhnel, F. Oncolytic viruses: Adenoviruses. Virus Genes 2017, 53, 700–706. [Google Scholar] [CrossRef]
- Santos, J.M.; Heinio, C.; Cervera-Carrascon, V.; Quixabeira, D.C.A.; Siurala, M.; Havunen, R.; Butzow, R.; Zafar, S.; de Gruijl, T.; Lassus, H.; et al. Oncolytic adenovirus shapes the ovarian tumor microenvironment for potent tumor-infiltrating lymphocyte tumor reactivity. J. Immunother. Cancer 2020, 8, 34. [Google Scholar] [CrossRef]
- Wakabayashi, K.; Sakurai, F.; Ono, R.; Fujiwara, T.; Mizuguchi, H. Development of a Novel Oncolytic Adenovirus Expressing a Short-hairpin RNA Against Cullin 4A. Anticancer Res. 2020, 40, 161–168. [Google Scholar] [CrossRef]
- Passaro, C.; Alayo, Q.; DeLaura, I.; McNulty, J.; Grauwet, K.; Ito, H.; Bhaskaran, V.; Mineo, M.; Lawler, S.E.; Shah, K.; et al. Correction: Arming an Oncolytic Herpes Simplex Virus Type 1 with a Single-chain Fragment Variable Antibody against PD−1 for Experimental Glioblastoma Therapy. Clin. Cancer Res. 2020, 26, 758. [Google Scholar] [CrossRef] [PubMed]
- Sanchala, D.S.; Bhatt, L.K.; Prabhavalkar, K.S. Oncolytic Herpes Simplex Viral Therapy: A Stride toward Selective Targeting of Cancer Cells. Front. Pharmacol. 2017, 8, 270. [Google Scholar] [CrossRef] [PubMed]
- Sasso, E.; Froechlich, G.; Cotugno, G.; D’Alise, A.M.; Gentile, C.; Bignone, V.; De Lucia, M.; Petrovic, B.; Campadelli-Fiume, G.; Scarselli, E.; et al. Replicative conditioning of Herpes simplex type 1 virus by Survivin promoter, combined to ERBB2 retargeting, improves tumour cell-restricted oncolysis. Sci. Rep. 2020, 10, 4307. [Google Scholar] [CrossRef]
- Ye, Z.Q.; Zou, C.L.; Chen, H.B.; Lv, Q.Y.; Wu, R.Q.; Gu, D.N. Folate-conjugated herpes simplex virus for retargeting to tumor cells. J. Gene Med. 2020, 22, e3177. [Google Scholar] [CrossRef]
- Marichal, T.; Tsai, M.; Galli, S.J. Mast cells: Potential positive and negative roles in tumor biology. Cancer Immunol. Res. 2013, 1, 269–279. [Google Scholar] [CrossRef]
- Angelova, A.L.; Geletneky, K.; Nuesch, J.P.; Rommelaere, J. Tumor Selectivity of Oncolytic Parvoviruses: From in vitro and Animal Models to Cancer Patients. Front. Bioeng. Biotechnol. 2015, 3, 55. [Google Scholar] [CrossRef]
- Annels, N.E.; Arif, M.; Simpson, G.R.; Denyer, M.; Moller-Levet, C.; Mansfield, D.; Butler, R.; Shafren, D.; Au, G.; Knowles, M.; et al. Oncolytic Immunotherapy for Bladder Cancer Using Coxsackie A21 Virus. Mol. Ther. Oncolytics 2018, 9, 1–12. [Google Scholar] [CrossRef]
- Relph, K.; Annels, N.; Smith, C.; Kostalas, M.; Pandha, H. Oncolytic Immunotherapy for Bladder Cancer Using Coxsackie A21 Virus: Using a Bladder Tumor Precision-Cut Slice Model System to Assess Viral Efficacy. Methods Mol. Biol. 2020, 2058, 249–259. [Google Scholar] [CrossRef]
- Mahalingam, D.; Wilkinson, G.A.; Eng, K.H.; Fields, P.; Raber, P.; Moseley, J.L.; Cheetham, K.; Coffey, M.; Nuovo, G.; Kalinski, P.; et al. Pembrolizumab in Combination with the Oncolytic Virus Pelareorep and Chemotherapy in Patients with Advanced Pancreatic Adenocarcinoma: A Phase Ib Study. Clin. Cancer Res. 2020, 26, 71–81. [Google Scholar] [CrossRef]
- McLaughlin, M.; Pedersen, M.; Roulstone, V.; Bergerhoff, K.F.; Smith, H.G.; Whittock, H.; Kyula, J.N.; Dillon, M.T.; Pandha, H.S.; Vile, R.; et al. The PERK Inhibitor GSK2606414 Enhances Reovirus Infection in Head and Neck Squamous Cell Carcinoma via an ATF4-Dependent Mechanism. Mol. Ther. Oncolytics 2020, 16, 238–249. [Google Scholar] [CrossRef]
- Mohamed, A.; Smiley, J.R.; Shmulevitz, M. Polymorphisms in the Most Oncolytic Reovirus Strain Confer Enhanced Cell Attachment, Transcription, and Single-Step Replication Kinetics. J. Virol. 2020, 94, 45. [Google Scholar] [CrossRef]
- Samson, A.; Bentham, M.J.; Scott, K.; Nuovo, G.; Bloy, A.; Appleton, E.; Adair, R.A.; Dave, R.; Peckham-Cooper, A.; Toogood, G.; et al. Oncolytic reovirus as a combined antiviral and anti-tumour agent for the treatment of liver cancer. Gut 2018, 67, 562–573. [Google Scholar] [CrossRef] [PubMed]
- Aref, S.; Bailey, K.; Fielding, A. Measles to the Rescue: A Review of Oncolytic Measles Virus. Viruses 2016, 8, 294. [Google Scholar] [CrossRef] [PubMed]
- Aref, S.; Castleton, A.Z.; Bailey, K.; Burt, R.; Dey, A.; Leongamornlert, D.; Mitchell, R.J.; Okasha, D.; Fielding, A.K. Type 1 Interferon Responses Underlie Tumor-Selective Replication of Oncolytic Measles Virus. Mol. Ther. 2020, 28, 1043–1055. [Google Scholar] [CrossRef]
- Muhlebach, M.D.; Cattaneo, R. Development of Entry-Targeted Oncolytic Measles Viruses. Methods Mol. Biol 2020, 2058, 51–75. [Google Scholar] [CrossRef]
- Robinson, S.; Galanis, E. Potential and clinical translation of oncolytic measles viruses. Expert Opin. Biol. Ther. 2017, 17, 353–363. [Google Scholar] [CrossRef]
- Geletneky, K.; Hajda, J.; Angelova, A.L.; Leuchs, B.; Capper, D.; Bartsch, A.J.; Neumann, J.O.; Schoning, T.; Husing, J.; Beelte, B.; et al. Oncolytic H−1 Parvovirus Shows Safety and Signs of Immunogenic Activity in a First Phase I/IIa Glioblastoma Trial. Mol. Ther. 2017, 25, 2620–2634. [Google Scholar] [CrossRef]
- Lacroix, J.; Kis, Z.; Josupeit, R.; Schlund, F.; Stroh-Dege, A.; Frank-Stohr, M.; Leuchs, B.; Schlehofer, J.R.; Rommelaere, J.; Dinsart, C. Preclinical Testing of an Oncolytic Parvovirus in Ewing Sarcoma: Protoparvovirus H−1 Induces Apoptosis and Lytic Infection In Vitro but Fails to Improve Survival In Vivo. Viruses 2018, 10, 302. [Google Scholar] [CrossRef]
- Bishnoi, S.; Tiwari, R.; Gupta, S.; Byrareddy, S.N.; Nayak, D. Oncotargeting by Vesicular Stomatitis Virus (VSV): Advances in Cancer Therapy. Viruses 2018, 10, 90. [Google Scholar] [CrossRef]
- Seegers, S.L.; Frasier, C.; Greene, S.; Nesmelova, I.V.; Grdzelishvili, V.Z. Experimental Evolution Generates Novel Oncolytic Vesicular Stomatitis Viruses with Improved Replication in Virus-Resistant Pancreatic Cancer Cells. J. Virol. 2020, 94, 55. [Google Scholar] [CrossRef]
- Velazquez-Salinas, L.; Naik, S.; Pauszek, S.J.; Peng, K.W.; Russell, S.J.; Rodriguez, L.L. Oncolytic Recombinant Vesicular Stomatitis Virus (VSV) Is Nonpathogenic and Nontransmissible in Pigs, a Natural Host of VSV. Hum. Gene Ther. Clin. Dev. 2017, 28, 108–115. [Google Scholar] [CrossRef] [PubMed]
- Burke, S.; Shergold, A.; Elder, M.J.; Whitworth, J.; Cheng, X.; Jin, H.; Wilkinson, R.W.; Harper, J.; Carroll, D.K. Oncolytic Newcastle disease virus activation of the innate immune response and priming of antitumor adaptive responses in vitro. Cancer Immunol. Immunother. 2020, 69, 1015–1027. [Google Scholar] [CrossRef]
- Jiang, K.; Song, C.; Kong, L.; Hu, L.; Lin, G.; Ye, T.; Yao, G.; Wang, Y.; Chen, H.; Cheng, W.; et al. Recombinant oncolytic Newcastle disease virus displays antitumor activities in anaplastic thyroid cancer cells. BMC Cancer 2018, 18, 746. [Google Scholar] [CrossRef]
- Tayeb, S.; Zakay-Rones, Z.; Panet, A. Therapeutic potential of oncolytic Newcastle disease virus: A critical review. Oncolytic Virother. 2015, 4, 49–62. [Google Scholar] [CrossRef] [PubMed]
- ClinicalTrials.gov Identifier: NCT00769704. Available online: https://clinicaltrials.gov/ct2/show/results/NCT00769704?view=results (accessed on 31 May 2021).
- Available online: https://www.clinicaltrials.gov/ct2/show/NCT00402025 (accessed on 31 May 2021).
- ClinicalTrials.gov Identifier: NCT01161498. Available online: https://clinicaltrials.gov/ct2/show/NCT01161498 (accessed on 31 May 2021).
- ClinicalTrials.gov Identifier: NCT03152318. Available online: https://clinicaltrials.gov/ct2/show/NCT03152318 (accessed on 31 May 2021).
- Chiocca, E.A.; Nakashima, H.; Kasai, K.; Fernandez, S.A.; Oglesbee, M. Preclinical Toxicology of rQNestin34.5v.2: An Oncolytic Herpes Virus with Transcriptional Regulation of the ICP34.5 Neurovirulence Gene. Mol. Ther. Methods Clin. Dev. 2020, 17, 871–893. [Google Scholar] [CrossRef]
- ClinicalTrials.gov Identifier: NCT00028158. Available online: https://clinicaltrials.gov/ct2/show/NCT00028158 (accessed on 31 May 2021).
- Markert, J.M.; Razdan, S.N.; Kuo, H.C.; Cantor, A.; Knoll, A.; Karrasch, M.; Nabors, L.B.; Markiewicz, M.; Agee, B.S.; Coleman, J.M.; et al. A phase 1 trial of oncolytic HSV−1, G207, given in combination with radiation for recurrent GBM demonstrates safety and radiographic responses. Mol. Ther. 2014, 22, 1048–1055. [Google Scholar] [CrossRef]
- ClinicalTrials.gov Identifier: NCT03780049. Available online: https://clinicaltrials.gov/ct2/show/NCT03780049 (accessed on 31 May 2021).
- ClinicalTrials.gov Identifier: NCT04771676. Available online: https://clinicaltrials.gov/ct2/show/NCT04771676 (accessed on 31 May 2021).
- ClinicalTrials.gov Identifier: NCT04714983. Available online: https://clinicaltrials.gov/ct2/show/NCT04714983 (accessed on 31 May 2021).
- ClinicalTrials.gov Identifier: NCT03714334. Available online: https://clinicaltrials.gov/ct2/show/NCT03714334 (accessed on 31 May 2021).
- ClinicalTrials.gov Identifier: NCT01956734. Available online: https://clinicaltrials.gov/ct2/show/NCT01956734 (accessed on 31 May 2021).
- ClinicalTrials.gov Identifier: NCT02197169. Available online: https://clinicaltrials.gov/ct2/show/NCT02197169 (accessed on 31 May 2021).
- ClinicalTrials.gov Identifier: NCT01582516. Available online: https://clinicaltrials.gov/ct2/show/NCT01582516 (accessed on 31 May 2021).
- ClinicalTrials.gov Identifier: NCT04695327. Available online: https://clinicaltrials.gov/ct2/show/NCT04695327 (accessed on 31 May 2021).
- ClinicalTrials.gov Identifier: NCT04217473. Available online: https://clinicaltrials.gov/ct2/show/NCT04217473 (accessed on 31 May 2021).
- ClinicalTrials.gov Identifier: NCT02045602. Available online: https://www.clinicaltrials.gov/ct2/show/NCT02045602 (accessed on 31 May 2021).
- ClinicalTrials.gov Identifier: NCT02045589. Available online: https://clinicaltrials.gov/ct2/show/NCT02045589 (accessed on 31 May 2021).
- ClinicalTrials.gov Identifier: NCT03799744. Available online: https://clinicaltrials.gov/ct2/show/NCT03799744 (accessed on 31 May 2021).
- ClinicalTrials.gov Identifier: NCT02705196. Available online: https://www.clinicaltrials.gov/ct2/show/NCT02705196 (accessed on 31 May 2021).
- ClinicalTrials.gov Identifier: NCT04123470. Available online: https://clinicaltrials.gov/ct2/show/NCT04123470 (accessed on 31 May 2021).
- ClinicalTrials.gov Identifier: NCT03225989. Available online: https://clinicaltrials.gov/ct2/show/NCT03225989 (accessed on 31 May 2021).
- ClinicalTrials.gov Identifier: NCT01864759. Available online: https://clinicaltrials.gov/ct2/show/NCT01864759 (accessed on 31 May 2021).
- ClinicalTrials.gov Identifier: NCT04387461. Available online: https://clinicaltrials.gov/ct2/show/NCT04387461 (accessed on 31 May 2021).
- ClinicalTrials.gov Identifier: NCT04452591. Available online: https://clinicaltrials.gov/ct2/show/NCT04452591 (accessed on 31 May 2021).
- ClinicalTrials.gov Identifier: NCT02365818. Available online: https://clinicaltrials.gov/ct2/show/NCT02365818 (accessed on 31 May 2021).
- ClinicalTrials.gov Identifier: NCT03916510. Available online: https://clinicaltrials.gov/ct2/show/NCT03916510 (accessed on 31 May 2021).
- ClinicalTrials.gov Identifier: NCT03740256. Available online: https://clinicaltrials.gov/ct2/show/NCT03740256 (accessed on 31 May 2021).
- ClinicalTrials.gov Identifier: NCT04097002. Available online: https://clinicaltrials.gov/ct2/show/NCT04097002 (accessed on 31 May 2021).
- ClinicalTrials.gov Identifier: NCT04053283. Available online: https://clinicaltrials.gov/ct2/show/NCT04053283 (accessed on 31 May 2021).
- ClinicalTrials.gov Identifier: NCT00528684. Available online: https://clinicaltrials.gov/ct2/show/NCT00528684 (accessed on 31 May 2021).
- ClinicalTrials.gov Identifier: NCT02620423. Available online: https://clinicaltrials.gov/ct2/show/NCT02620423 (accessed on 31 May 2021).
- ClinicalTrials.gov Identifier: NCT03605719. Available online: https://clinicaltrials.gov/ct2/show/NCT03605719 (accessed on 31 May 2021).
- ClinicalTrials.gov Identifier: NCT00998192. Available online: https://clinicaltrials.gov/ct2/show/NCT00998192 (accessed on 31 May 2021).
- ClinicalTrials.gov Identifier: NCT00753038. Available online: https://clinicaltrials.gov/ct2/show/NCT00753038 (accessed on 31 May 2021).
- ClinicalTrials.gov Identifier: NCT00984464. Available online: https://clinicaltrials.gov/ct2/show/NCT00984464 (accessed on 31 May 2021).
- ClinicalTrials.gov Identifier: NCT04445844. Available online: https://clinicaltrials.gov/ct2/show/NCT04445844 (accessed on 31 May 2021).
- ClinicalTrials.gov Identifier: NCT01166542. Available online: https://clinicaltrials.gov/ct2/show/NCT01166542 (accessed on 31 May 2021).
- ClinicalTrials.gov Identifier: NCT01199263. Available online: https://clinicaltrials.gov/ct2/show/NCT01199263 (accessed on 31 May 2021).
- Geletneky, K.; Huesing, J.; Rommelaere, J.; Schlehofer, J.R.; Leuchs, B.; Dahm, M.; Krebs, O.; von Knebel Doeberitz, M.; Huber, B.; Hajda, J. Phase I/IIa study of intratumoral/intracerebral or intravenous/intracerebral administration of Parvovirus H−1 (ParvOryx) in patients with progressive primary or recurrent glioblastoma multiforme: ParvOryx01 protocol. BMC Cancer 2012, 12, 99. [Google Scholar] [CrossRef]
- ClinicalTrials.gov Identifier: NCT01301430. Available online: https://www.clinicaltrials.gov/ct2/show/NCT01301430 (accessed on 31 May 2021).
- ClinicalTrials.gov Identifier: NCT02653313. Available online: https://clinicaltrials.gov/ct2/show/NCT02653313 (accessed on 31 May 2021).
- ClinicalTrials.gov Identifier: NCT00390299. Available online: https://www.clinicaltrials.gov/ct2/show/NCT00390299 (accessed on 31 May 2021).
- ClinicalTrials.gov Identifier: NCT00408590. Available online: https://clinicaltrials.gov/ct2/show/NCT00408590 (accessed on 31 May 2021).
- ClinicalTrials.gov Identifier: NCT02192775. Available online: https://clinicaltrials.gov/ct2/show/NCT02192775 (accessed on 31 May 2021).
- ClinicalTrials.gov Identifier: NCT04521764. Available online: https://clinicaltrials.gov/ct2/show/NCT04521764 (accessed on 31 May 2021).
- ClinicalTrials.gov Identifier: NCT02068794. Available online: https://clinicaltrials.gov/ct2/show/NCT02068794 (accessed on 31 May 2021).
- ClinicalTrials.gov Identifier: NCT01503177. Available online: https://clinicaltrials.gov/ct2/show/NCT01503177 (accessed on 31 May 2021).
- ClinicalTrials.gov Identifier: NCT00450814. Available online: https://www.clinicaltrials.gov/ct2/show/NCT00450814 (accessed on 31 May 2021).
- ClinicalTrials.gov Identifier: NCT02700230. Available online: https://clinicaltrials.gov/ct2/show/NCT02700230 (accessed on 31 May 2021).
- ClinicalTrials.gov Identifier: NCT03294486. Available online: https://clinicaltrials.gov/ct2/show/NCT03294486 (accessed on 31 May 2021).
- ClinicalTrials.gov Identifier: NCT00554372. Available online: https://clinicaltrials.gov/ct2/show/NCT00554372 (accessed on 31 May 2021).
- ClinicalTrials.gov Identifier: NCT01636284. Available online: https://clinicaltrials.gov/ct2/show/NCT01636284 (accessed on 31 May 2021).
- ClinicalTrials.gov Identifier: NCT00429312. Available online: https://clinicaltrials.gov/ct2/show/NCT00429312 (accessed on 31 May 2021).
- ClinicalTrials.gov Identifier: NCT03294083. Available online: https://clinicaltrials.gov/ct2/show/NCT03294083 (accessed on 31 May 2021).
- ClinicalTrials.gov Identifier: NCT00629759. Available online: https://clinicaltrials.gov/ct2/show/NCT00629759 (accessed on 31 May 2021).
- Choi, A.H.; O’Leary, M.P.; Fong, Y.; Chen, N.G. From Benchtop to Bedside: A Review of Oncolytic Virotherapy. Biomedicines 2016, 4, 18. [Google Scholar] [CrossRef]
- Mohammad, R.M.; Muqbil, I.; Lowe, L.; Yedjou, C.; Hsu, H.Y.; Lin, L.T.; Siegelin, M.D.; Fimognari, C.; Kumar, N.B.; Dou, Q.P.; et al. Broad targeting of resistance to apoptosis in cancer. Semin. Cancer Biol. 2015, 35, S78–S103. [Google Scholar] [CrossRef]
- Gonzalez-Cao, M.; Karachaliou, N.; Santarpia, M.; Viteri, S.; Meyerhans, A.; Rosell, R. Activation of viral defense signaling in cancer. Ther. Adv. Med. Oncol. 2018, 10, 1758835918793105. [Google Scholar] [CrossRef]
- Guo, Z.S.; Thorne, S.H.; Bartlett, D.L. Oncolytic virotherapy: Molecular targets in tumor-selective replication and carrier cell-mediated delivery of oncolytic viruses. Biochim. Biophys. Acta 2008, 1785, 217–231. [Google Scholar] [CrossRef]
- Post, D.E.; Sandberg, E.M.; Kyle, M.M.; Devi, N.S.; Brat, D.J.; Xu, Z.; Tighiouart, M.; Van Meir, E.G. Targeted cancer gene therapy using a hypoxia inducible factor dependent oncolytic adenovirus armed with interleukin−4. Cancer Res. 2007, 67, 6872–6881. [Google Scholar] [CrossRef] [PubMed]
- Reinblatt, M.; Pin, R.H.; Federoff, H.J.; Fong, Y. Utilizing tumor hypoxia to enhance oncolytic viral therapy in colorectal metastases. Ann. Surg 2004, 239, 892–899. [Google Scholar] [CrossRef] [PubMed]
- Oh, E.; Hong, J.; Kwon, O.-J.; Yun, C.-O. A hypoxia-and telomerase-responsive oncolytic adenovirus expressing secretable trimeric TRAIL triggers tumour-specific apoptosis and promotes viral dispersion in TRAIL-resistant glioblastoma. Sci. Rep. 2018, 8, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Vojtechova, Z.; Tachezy, R. The Role of miRNAs in Virus-Mediated Oncogenesis. Int. J. Mol. Sci. 2018, 19, 1217. [Google Scholar] [CrossRef] [PubMed]
- Davola, M.E.; Mossman, K.L. Oncolytic viruses: How “lytic” must they be for therapeutic efficacy? Oncoimmunology 2019, 8, e1581528. [Google Scholar] [CrossRef] [PubMed]
- Kaufman, H.L.; Kohlhapp, F.J.; Zloza, A. Oncolytic viruses: A new class of immunotherapy drugs. Nat. Rev. Drug Discov. 2015, 14, 642–662. [Google Scholar] [CrossRef]
- Sokolowski, N.A.; Rizos, H.; Diefenbach, R.J. Oncolytic virotherapy using herpes simplex virus: How far have we come? Oncolytic Virother. 2015, 4, 207–219. [Google Scholar] [CrossRef]
- Shen, B.H.; Hermiston, T.W. Effect of hypoxia on Ad5 infection, transgene expression and replication. Gene Ther. 2005, 12, 902–910. [Google Scholar] [CrossRef]
- Maroun, J.; Munoz-Alia, M.; Ammayappan, A.; Schulze, A.; Peng, K.W.; Russell, S. Designing and building oncolytic viruses. Future Virol. 2017, 12, 193–213. [Google Scholar] [CrossRef]
- Badrinath, N.; Heo, J.; Yoo, S.Y. Viruses as nanomedicine for cancer. Int. J. Nanomed. 2016, 11, 4835–4847. [Google Scholar] [CrossRef]
- Marchini, A.; Scott, E.M.; Rommelaere, J. Overcoming Barriers in Oncolytic Virotherapy with HDAC Inhibitors and Immune Checkpoint Blockade. Viruses 2016, 8, 9. [Google Scholar] [CrossRef]
- Rinaldi, L.; Nevola, R.; Franci, G.; Perrella, A.; Corvino, G.; Marrone, A.; Berretta, M.; Morone, M.V.; Galdiero, M.; Giordano, M.; et al. Risk of Hepatocellular Carcinoma after HCV Clearance by Direct-Acting Antivirals Treatment Predictive Factors and Role of Epigenetics. Cancers 2020, 12, 1351. [Google Scholar] [CrossRef] [PubMed]
- Tischer, B.K.; Osterrieder, N. Herpesviruses—A zoonotic threat? Vet. Microbiol. 2010, 140, 266–270. [Google Scholar] [CrossRef] [PubMed]
- Franci, G.; Crudele, V.; Della Rocca, M.T.; Melardo, C.; Chianese, A.; Finamore, E.; Bencivenga, F.; Astorri, R.; Vitiello, M.; Galdiero, E.; et al. Epstein-Barr Virus Seroprevalence and Primary Infection at the University Hospital Luigi Vanvitelli of Naples from 2007 to 2017. Inter. Virol. 2019, 62, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Haarr, L.; Skulstad, S. The herpes simplex virus type 1 particle: Structure and molecular functions. Review article. APMIS 1994, 102, 321–346. [Google Scholar] [CrossRef] [PubMed]
- Stelitano, D.; Franci, G.; Chianese, A.; Galdiero, S.; Morelli, G.; Galdiero, M. HSV membrane glycoproteins, their function in viral entry and their use in vaccine studies. In Amino Acids, Peptides and Proteins; Royal Society of Chemistry: London, UK, 2019; pp. 14–43. [Google Scholar]
- Singh, M.; Zannella, C.; Folliero, V.; Di Girolamo, R.; Bajardi, F.; Chianese, A.; Altucci, L.; Damasco, A.; Del Sorbo, M.R.; Imperatore, C.; et al. Combating Actions of Green 2D-Materials on Gram Positive and Negative Bacteria and Enveloped Viruses. Front. Bioeng. Biotechnol. 2020, 8, 569967. [Google Scholar] [CrossRef]
- Coffin, R. Interview with Robert Coffin, inventor of T-VEC: The first oncolytic immunotherapy approved for the treatment of cancer. Immunotherapy 2016, 8, 103–106. [Google Scholar] [CrossRef]
- Liu, B.L.; Robinson, M.; Han, Z.Q.; Branston, R.H.; English, C.; Reay, P.; McGrath, Y.; Thomas, S.K.; Thornton, M.; Bullock, P.; et al. ICP34.5 deleted herpes simplex virus with enhanced oncolytic, immune stimulating, and anti-tumour properties. Gene Ther. 2003, 10, 292–303. [Google Scholar] [CrossRef]
- Andtbacka, R.H.; Ross, M.; Puzanov, I.; Milhem, M.; Collichio, F.; Delman, K.A.; Amatruda, T.; Zager, J.S.; Cranmer, L.; Hsueh, E.; et al. Patterns of Clinical Response with Talimogene Laherparepvec (T-VEC) in Patients with Melanoma Treated in the OPTiM Phase III Clinical Trial. Ann. Surg. Oncol. 2016, 23, 4169–4177. [Google Scholar] [CrossRef]
- Eissa, I.R.; Bustos-Villalobos, I.; Ichinose, T.; Matsumura, S.; Naoe, Y.; Miyajima, N.; Morimoto, D.; Mukoyama, N.; Zhiwen, W.; Tanaka, M.; et al. The Current Status and Future Prospects of Oncolytic Viruses in Clinical Trials against Melanoma, Glioma, Pancreatic, and Breast Cancers. Cancers 2018, 10, 356. [Google Scholar] [CrossRef]
- Johnson, D.B.; Puzanov, I.; Kelley, M.C. Talimogene laherparepvec (T-VEC) for the treatment of advanced melanoma. Immunotherapy 2015, 7, 611–619. [Google Scholar] [CrossRef] [PubMed]
- Masoud, S.J.; Hu, J.B.; Beasley, G.M.; Stewart, J.H.t.; Mosca, P.J. Efficacy of Talimogene Laherparepvec (T-VEC) Therapy in Patients with In-Transit Melanoma Metastasis Decreases with Increasing Lesion Size. Ann. Surg. Oncol. 2019, 26, 4633–4641. [Google Scholar] [CrossRef]
- Russell, L.; Peng, K.W. The emerging role of oncolytic virus therapy against cancer. Chin. Clin. Oncol. 2018, 7, 16. [Google Scholar] [CrossRef]
- Ahn, D.H.; Bekaii-Saab, T. The Continued Promise and Many Disappointments of Oncolytic Virotherapy in Gastrointestinal Malignancies. Biomedicines 2017, 5, 10. [Google Scholar] [CrossRef] [PubMed]
- Thomas, S.; Kuncheria, L.; Roulstone, V.; Kyula, J.N.; Mansfield, D.; Bommareddy, P.K.; Smith, H.; Kaufman, H.L.; Harrington, K.J.; Coffin, R.S. Development of a new fusion-enhanced oncolytic immunotherapy platform based on herpes simplex virus type 1. J. Immunother. Cancer 2019, 7, 1–17. [Google Scholar] [CrossRef]
- Hu, J.C.; Coffin, R.S.; Davis, C.J.; Graham, N.J.; Groves, N.; Guest, P.J.; Harrington, K.J.; James, N.D.; Love, C.A.; McNeish, I.; et al. A phase I study of OncoVEXGM-CSF, a second-generation oncolytic herpes simplex virus expressing granulocyte macrophage colony-stimulating factor. Clin. Cancer Res. 2006, 12, 6737–6747. [Google Scholar] [CrossRef]
- Senzer, N.N.; Kaufman, H.L.; Amatruda, T.; Nemunaitis, M.; Reid, T.; Daniels, G.; Gonzalez, R.; Glaspy, J.; Whitman, E.; Harrington, K.; et al. Phase II clinical trial of a granulocyte-macrophage colony-stimulating factor-encoding, second-generation oncolytic herpesvirus in patients with unresectable metastatic melanoma. J. Clin. Oncol. 2009, 27, 5763–5771. [Google Scholar] [CrossRef] [PubMed]
- Andtbacka, R.H.; Kaufman, H.L.; Collichio, F.; Amatruda, T.; Senzer, N.; Chesney, J.; Delman, K.A.; Spitler, L.E.; Puzanov, I.; Agarwala, S.S.; et al. Talimogene Laherparepvec Improves Durable Response Rate in Patients with Advanced Melanoma. J. Clin. Oncol. 2015, 33, 2780–2788. [Google Scholar] [CrossRef]
- Fukuhara, H.; Martuza, R.L.; Rabkin, S.D.; Ito, Y.; Todo, T. Oncolytic herpes simplex virus vector g47delta in combination with androgen ablation for the treatment of human prostate adenocarcinoma. Clin. Cancer Res. 2005, 11, 7886–7890. [Google Scholar] [CrossRef]
- Mineta, T.; Rabkin, S.D.; Yazaki, T.; Hunter, W.D.; Martuza, R.L. Attenuated multi-mutated herpes simplex virus−1 for the treatment of malignant gliomas. Nat. Med. 1995, 1, 938–943. [Google Scholar] [CrossRef]
- Foreman, P.M.; Friedman, G.K.; Cassady, K.A.; Markert, J.M. Oncolytic Virotherapy for the Treatment of Malignant Glioma. Neurotherapeutics 2017, 14, 333–344. [Google Scholar] [CrossRef]
- Kambara, H.; Okano, H.; Chiocca, E.A.; Saeki, Y. An oncolytic HSV−1 mutant expressing ICP34.5 under control of a nestin promoter increases survival of animals even when symptomatic from a brain tumor. Cancer Res. 2005, 65, 2832–2839. [Google Scholar] [CrossRef] [PubMed]
- Saha, B.; Wong, C.M.; Parks, R.J. The adenovirus genome contributes to the structural stability of the virion. Viruses 2014, 6, 3563–3583. [Google Scholar] [CrossRef] [PubMed]
- Mangel, W.F.; San Martin, C. Structure, function and dynamics in adenovirus maturation. Viruses 2014, 6, 116–4536. [Google Scholar] [CrossRef] [PubMed]
- Harada, J.N.; Berk, A.J. p53-Independent and -dependent requirements for E1B−55K in adenovirus type 5 replication. J. Virol. 1999, 73, 5333–5344. [Google Scholar] [CrossRef]
- Ries, S.; Korn, W.M. ONYX−015: Mechanisms of action and clinical potential of a replication-selective adenovirus. Br. J. Cancer 2002, 86, 5–11. [Google Scholar] [CrossRef]
- Garber, K. China approves world’s first oncolytic virus therapy for cancer treatment. J. Nat. Cancer Inst. 2006, 98, 298–300. [Google Scholar] [CrossRef]
- Ganly, I.; Kirn, D.; Eckhardt, G.; Rodriguez, G.I.; Soutar, D.S.; Otto, R.; Robertson, A.G.; Park, O.; Gulley, M.L.; Heise, C.; et al. A phase I study of Onyx−015, an E1B attenuated adenovirus, administered intratumorally to patients with recurrent head and neck cancer. Clin. Cancer Res. 2000, 6, 798–806. [Google Scholar] [PubMed]
- Nemunaitis, J.; Ganly, I.; Khuri, F.; Arseneau, J.; Kuhn, J.; McCarty, T.; Landers, S.; Maples, P.; Romel, L.; Randlev, B.; et al. Selective replication and oncolysis in p53 mutant tumors with ONYX−015, an E1B−55kD gene-deleted adenovirus, in patients with advanced head and neck cancer: A phase II trial. Cancer Res. 2000, 60, 6359–6366. [Google Scholar]
- Makower, D.; Rozenblit, A.; Kaufman, H.; Edelman, M.; Lane, M.E.; Zwiebel, J.; Haynes, H.; Wadler, S. Phase II clinical trial of intralesional administration of the oncolytic adenovirus ONYX−015 in patients with hepatobiliary tumors with correlative p53 studies. Clin. Cancer Res. 2003, 9, 693–702. [Google Scholar]
- Nemunaitis, J.; Khuri, F.; Ganly, I.; Arseneau, J.; Posner, M.; Vokes, E.; Kuhn, J.; McCarty, T.; Landers, S.; Blackburn, A.; et al. Phase II trial of intratumoral administration of ONYX−015, a replication-selective adenovirus, in patients with refractory head and neck cancer. J. Clin. Oncol. 2001, 19, 289–298. [Google Scholar] [CrossRef] [PubMed]
- Khuri, F.R.; Nemunaitis, J.; Ganly, I.; Arseneau, J.; Tannock, I.F.; Romel, L.; Gore, M.; Ironside, J.; MacDougall, R.H.; Heise, C.; et al. A controlled trial of intratumoral ONYX−015, a selectively-replicating adenovirus, in combination with cisplatin and 5-fluorouracil in patients with recurrent head and neck cancer. Nat. Med. 2000, 6, 879–885. [Google Scholar] [CrossRef] [PubMed]
- Nwanegbo, E.; Vardas, E.; Gao, W.; Whittle, H.; Sun, H.; Rowe, D.; Robbins, P.D.; Gambotto, A. Prevalence of neutralizing antibodies to adenoviral serotypes 5 and 35 in the adult populations of The Gambia, South Africa, and the United States. Clin. Diagn Lab. Immunol. 2004, 11, 351–357. [Google Scholar] [CrossRef] [PubMed]
- Peter, M.; Kühnel, F. Oncolytic Adenovirus in Cancer Immunotherapy. Cancers 2020, 12, 3354. [Google Scholar] [CrossRef]
- Philbrick, B.; Adamson, D.C. DNX−2401: An investigational drug for the treatment of recurrent glioblastoma. Expert Opin. Investig. Drugs 2019, 28, 1041–1049. [Google Scholar] [CrossRef]
- Lang, F.F.; Conrad, C.; Gomez-Manzano, C.; Yung, W.K.A.; Sawaya, R.; Weinberg, J.S.; Prabhu, S.S.; Rao, G.; Fuller, G.N.; Aldape, K.D.; et al. Phase I Study of DNX−2401 (Delta−24-RGD) Oncolytic Adenovirus: Replication and Immunotherapeutic Effects in Recurrent Malignant Glioma. J. Clin. Oncol. 2018, 36, 1419–1427. [Google Scholar] [CrossRef]
- Green, T.J.; Zhang, X.; Wertz, G.W.; Luo, M. Structure of the vesicular stomatitis virus nucleoprotein-RNA complex. Science 2006, 313, 357–360. [Google Scholar] [CrossRef]
- Stojdl, D.F.; Lichty, B.D.; tenOever, B.R.; Paterson, J.M.; Power, A.T.; Knowles, S.; Marius, R.; Reynard, J.; Poliquin, L.; Atkins, H.; et al. VSV strains with defects in their ability to shutdown innate immunity are potent systemic anti-cancer agents. Cancer Cell 2003, 4, 263–275. [Google Scholar] [CrossRef]
- Sahin, E.; Egger, M.E.; McMasters, K.M.; Zhou, H.S. Development of oncolytic reovirus for cancer therapy. J. Cancer Ther. 2013, 4, 16. [Google Scholar] [CrossRef]
- Gong, J.; Mita, M.M. Activated ras signaling pathways and reovirus oncolysis: An update on the mechanism of preferential reovirus replication in cancer cells. Front. Oncol. 2014, 4, 167. [Google Scholar] [CrossRef]
- Errington, F.; Steele, L.; Prestwich, R.; Harrington, K.J.; Pandha, H.S.; Vidal, L.; de Bono, J.; Selby, P.; Coffey, M.; Vile, R.; et al. Reovirus activates human dendritic cells to promote innate antitumor immunity. J. Immunol. 2008, 180, 6018–6026. [Google Scholar] [CrossRef] [PubMed]
- Mahalingam, D.; Goel, S.; Aparo, S.; Patel Arora, S.; Noronha, N.; Tran, H.; Chakrabarty, R.; Selvaggi, G.; Gutierrez, A.; Coffey, M.; et al. A Phase II Study of Pelareorep (REOLYSIN®) in Combination with Gemcitabine for Patients with Advanced Pancreatic Adenocarcinoma. Cancers 2018, 10, 160. [Google Scholar] [CrossRef] [PubMed]
- ClinicalTrials.gov Identifier: NCT01656538. Available online: https://clinicaltrials.gov/ct2/show/NCT01656538 (accessed on 31 May 2021).
- ClinicalTrials.gov Identifier: NCT01708993. Available online: https://clinicaltrials.gov/ct2/show/NCT01708993 (accessed on 31 May 2021).
- ClinicalTrials.gov Identifier: NCT01619813. Available online: https://clinicaltrials.gov/ct2/show/NCT01619813 (accessed on 31 May 2021).
- Kaufmann, B.; Simpson, A.A.; Rossmann, M.G. The structure of human parvovirus B19. Proc. Nat. Acad. Sci. USA 2004, 101, 11628–11633. [Google Scholar] [CrossRef]
- Li, J.; Bonifati, S.; Hristov, G.; Marttila, T.; Valmary-Degano, S.; Stanzel, S.; Schnolzer, M.; Mougin, C.; Aprahamian, M.; Grekova, S.P.; et al. Synergistic combination of valproic acid and oncolytic parvovirus H−1PV as a potential therapy against cervical and pancreatic carcinomas. EMBO Mol. Med. 2013, 5, 1537–1555. [Google Scholar] [CrossRef]
- Nuesch, J.P.; Lacroix, J.; Marchini, A.; Rommelaere, J. Molecular pathways: Rodent parvoviruses--mechanisms of oncolysis and prospects for clinical cancer treatment. Clin. Cancer Res. 2012, 18, 3516–3523. [Google Scholar] [CrossRef] [PubMed]
- Hristov, G.; Kramer, M.; Li, J.; El-Andaloussi, N.; Mora, R.; Daeffler, L.; Zentgraf, H.; Rommelaere, J.; Marchini, A. Through its nonstructural protein NS1, parvovirus H−1 induces apoptosis via accumulation of reactive oxygen species. J. Virol. 2010, 84, 5909–5922. [Google Scholar] [CrossRef] [PubMed]
- Josupeit, R.; Bender, S.; Kern, S.; Leuchs, B.; Hielscher, T.; Herold-Mende, C.; Schlehofer, J.R.; Dinsart, C.; Witt, O.; Rommelaere, J.; et al. Pediatric and Adult High-Grade Glioma Stem Cell Culture Models Are Permissive to Lytic Infection with Parvovirus H−1. Viruses 2016, 8, 138. [Google Scholar] [CrossRef] [PubMed]
- Palgen, J.L.; Jurgens, E.M.; Moscona, A.; Porotto, M.; Palermo, L.M. Unity in diversity: Shared mechanism of entry among paramyxoviruses. Prog. Mol. Biol. Transl. Sci. 2015, 129, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, H.C.; Henderson, B.A.; Zamora, J.L.; Johnston, G.P. Paramyxovirus Glycoproteins and the Membrane Fusion Process. Curr. Clin. Microbiol. Rep. 2016, 3, 142–154. [Google Scholar] [CrossRef] [PubMed]
- Chatziandreou, N.; Stock, N.; Young, D.; Andrejeva, J.; Hagmaier, K.; McGeoch, D.J.; Randall, R.E. Relationships and host range of human, canine, simian and porcine isolates of simian virus 5 (parainfluenza virus 5). J. Gen. Virol. 2004, 85, 3007–3016. [Google Scholar] [CrossRef] [PubMed]
- Fox, C.R.; Parks, G.D. Histone Deacetylase Inhibitors Enhance Cell Killing and Block Interferon-Beta Synthesis Elicited by Infection with an Oncolytic Parainfluenza Virus. Viruses 2019, 11, 431. [Google Scholar] [CrossRef]
- Wansley, E.K.; Dillon, P.J.; Gainey, M.D.; Tam, J.; Cramer, S.D.; Parks, G.D. Growth sensitivity of a recombinant simian virus 5 P/V mutant to type I interferon differs between tumor cell lines and normal primary cells. Virology 2005, 335, 131–144. [Google Scholar] [CrossRef]
- Foloppe, J.; Kempf, J.; Futin, N.; Kintz, J.; Cordier, P.; Pichon, C.; Findeli, A.; Vorburger, F.; Quemeneur, E.; Erbs, P. The Enhanced Tumor Specificity of TG6002, an Armed Oncolytic Vaccinia Virus Deleted in Two Genes Involved in Nucleotide Metabolism. Mol. Ther. Oncolytics 2019, 14, 1–14. [Google Scholar] [CrossRef]
- Heinrich, B.; Klein, J.; Delic, M.; Goepfert, K.; Engel, V.; Geberzahn, L.; Lusky, M.; Erbs, P.; Preville, X.; Moehler, M. Immunogenicity of oncolytic vaccinia viruses JX-GFP and TG6002 in a human melanoma in vitro model: Studying immunogenic cell death, dendritic cell maturation and interaction with cytotoxic T lymphocytes. Onco Targets Ther. 2017, 10, 2389–2401. [Google Scholar] [CrossRef]
- Lauer, U.M.; Schell, M.; Beil, J.; Berchtold, S.; Koppenhöfer, U.; Glatzle, J.; Königsrainer, A.; Möhle, R.; Nann, D.; Fend, F. Phase I study of oncolytic vaccinia virus GL-ONC1 in patients with peritoneal carcinomatosis. Clin. Cancer Res. 2018, 24, 4388–4398. [Google Scholar] [CrossRef]
- Minev, B.R.; Lander, E.; Feller, J.F.; Berman, M.; Greenwood, B.M.; Minev, I.; Santidrian, A.F.; Nguyen, D.; Draganov, D.; Killinc, M.O. First-in-human study of TK-positive oncolytic vaccinia virus delivered by adipose stromal vascular fraction cells. J. Transl. Med. 2019, 17, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Donina, S.; Strele, I.; Proboka, G.; Auzins, J.; Alberts, P.; Jonsson, B.; Venskus, D.; Muceniece, A. Adapted ECHO−7 virus Rigvir immunotherapy (oncolytic virotherapy) prolongs survival in melanoma patients after surgical excision of the tumour in a retrospective study. Melanoma Res. 2015, 25, 421–426. [Google Scholar] [CrossRef]
- Alberts, P.; Olmane, E.; Brokane, L.; Krastina, Z.; Romanovska, M.; Kupcs, K.; Isajevs, S.; Proboka, G.; Erdmanis, R.; Nazarovs, J.; et al. Long-term treatment with the oncolytic ECHO−7 virus Rigvir of a melanoma stage IV M1c patient, a small cell lung cancer stage IIIA patient, and a histiocytic sarcoma stage IV patient-three case reports. APMIS 2016, 124, 896–904. [Google Scholar] [CrossRef]
- Ismailov, Z.; Rasa, A.; Bandere, K.; Brokane, L.; Tilgase, A.; Olmane, E.; Nazarovs, J.; Alberts, P. A Case of Stage IV Chromophobe Renal Cell Carcinoma Treated with the Oncolytic ECHO−7 Virus, Rigvir(R). Am. J. Case Rep. 2019, 20, 48–52. [Google Scholar] [CrossRef]
- Li, G.; Tian, Y.; Zhu, W.G. The Roles of Histone Deacetylases and Their Inhibitors in Cancer Therapy. Front. Cell Dev. Biol. 2020, 8, 576946. [Google Scholar] [CrossRef] [PubMed]
- Verza, F.A.; Das, U.; Fachin, A.L.; Dimmock, J.R.; Marins, M. Roles of Histone Deacetylases and Inhibitors in Anticancer Therapy. Cancers 2020, 12, 1664. [Google Scholar] [CrossRef] [PubMed]
- Milazzo, G.; Mercatelli, D.; Di Muzio, G.; Triboli, L.; De Rosa, P.; Perini, G.; Giorgi, F.M. Histone Deacetylases (HDACs): Evolution, Specificity, Role in Transcriptional Complexes, and Pharmacological Actionability. Genes 2020, 11, 556. [Google Scholar] [CrossRef]
- Johnstone, R.W. Histone-deacetylase inhibitors: Novel drugs for the treatment of cancer. Nat. Rev. Drug Discov. 2002, 1, 287–299. [Google Scholar] [CrossRef]
- Mariadason, J.M. HDACs and HDAC inhibitors in colon cancer. Epigenetics 2008, 3, 28–37. [Google Scholar] [CrossRef]
- Park, J.; Thomas, S.; Munster, P.N. Epigenetic modulation with histone deacetylase inhibitors in combination with immunotherapy. Epigenomics 2015, 7, 641–652. [Google Scholar] [CrossRef] [PubMed]
- Secrist, J.P.; Zhou, X.; Richon, V.M. HDAC inhibitors for the treatment of cancer. Curr. Opin. Investig. Drugs 2003, 4, 1422–1427. [Google Scholar] [PubMed]
- Nehme, Z.; Pasquereau, S.; Herbein, G. Control of viral infections by epigenetic-targeted therapy. Clin. Epigenetics 2019, 11, 55. [Google Scholar] [CrossRef]
- Zhao, C.; Dong, H.; Xu, Q.; Zhang, Y. Histone deacetylase (HDAC) inhibitors in cancer: A patent review (2017-present). Expert Opin. Ther. Pat. 2020, 30, 263–274. [Google Scholar] [CrossRef]
- Sanaei, M.; Kavoosi, F. Histone Deacetylases and Histone Deacetylase Inhibitors: Molecular Mechanisms of Action in Various Cancers. Adv. Biomed. Res. 2019, 8, 63. [Google Scholar] [CrossRef]
- Hassell, K.N. Histone Deacetylases and their Inhibitors in Cancer Epigenetics. Diseases 2019, 7, 57. [Google Scholar] [CrossRef] [PubMed]
- Abbas, A.; Gupta, S. The role of histone deacetylases in prostate cancer. Epigenetics 2008, 3, 300–309. [Google Scholar] [CrossRef] [PubMed]
- Bubna, A.K. Vorinostat-An Overview. Indian J. Dermatol. 2015, 60, 419. [Google Scholar] [CrossRef]
- Bali, P.; Pranpat, M.; Bradner, J.; Balasis, M.; Fiskus, W.; Guo, F.; Rocha, K.; Kumaraswamy, S.; Boyapalle, S.; Atadja, P.; et al. Inhibition of histone deacetylase 6 acetylates and disrupts the chaperone function of heat shock protein 90: A novel basis for antileukemia activity of histone deacetylase inhibitors. J. Biol. Chem. 2005, 280, 26729–26734. [Google Scholar] [CrossRef] [PubMed]
- Rivero-Cruz, J.F.; Lezutekong, R.; Lobo-Echeverri, T.; Ito, A.; Mi, Q.; Chai, H.B.; Soejarto, D.D.; Cordell, G.A.; Pezzuto, J.M.; Swanson, S.M. Cytotoxic constituents of the twigs of Simarouba glauca collected from a plot in Southern Florida. Phytother. Res. 2005, 19, 136–140. [Google Scholar] [CrossRef]
- Rikiishi, H. Autophagic and apoptotic effects of HDAC inhibitors on cancer cells. J. Biomed. Biotechnol. 2011, 2011, 830260. [Google Scholar] [CrossRef]
- Chinnaiyan, P.; Vallabhaneni, G.; Armstrong, E.; Huang, S.M.; Harari, P.M. Modulation of radiation response by histone deacetylase inhibition. Int. J. Radiat. Oncol. Biol. Phys. 2005, 62, 223–229. [Google Scholar] [CrossRef]
- Munster, P.N.; Troso-Sandoval, T.; Rosen, N.; Rifkind, R.; Marks, P.A.; Richon, V.M. The histone deacetylase inhibitor suberoylanilide hydroxamic acid induces differentiation of human breast cancer cells. Cancer Res. 2001, 61, 8492–8497. [Google Scholar]
- Kim, H.J.; Bae, S.C. Histone deacetylase inhibitors: Molecular mechanisms of action and clinical trials as anti-cancer drugs. Am. J. Transl. Res. 2011, 3, 166–179. [Google Scholar]
- Vanhaecke, T.; Papeleu, P.; Elaut, G.; Rogiers, V. Trichostatin A-like hydroxamate histone deacetylase inhibitors as therapeutic agents: Toxicological point of view. Curr. Med. Chem. 2004, 11, 1629–1643. [Google Scholar] [CrossRef]
- Damaskos, C.; Garmpis, N.; Valsami, S.; Kontos, M.; Spartalis, E.; Kalampokas, T.; Kalampokas, E.; Athanasiou, A.; Moris, D.; Daskalopoulou, A.; et al. Histone Deacetylase Inhibitors: An Attractive Therapeutic Strategy Against Breast Cancer. Anticancer Res. 2017, 37, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Jiang, X.; Pu, H.; Zhang, W.; An, C.; Hu, X.; Liou, A.K.; Leak, R.K.; Gao, Y.; Chen, J. Scriptaid, a novel histone deacetylase inhibitor, protects against traumatic brain injury via modulation of PTEN and AKT pathway: Scriptaid protects against TBI via AKT. Neurotherapeutics 2013, 10, 124–142. [Google Scholar] [CrossRef]
- Kuefer, R.; Hofer, M.D.; Altug, V.; Zorn, C.; Genze, F.; Kunzi-Rapp, K.; Hautmann, R.E.; Gschwend, J.E. Sodium butyrate and tributyrin induce in vivo growth inhibition and apoptosis in human prostate cancer. Br. J. Cancer 2004, 90, 535–541. [Google Scholar] [CrossRef] [PubMed]
- Xiong, F.; Mou, Y.Z.; Xiang, X.Y. Inhibition of mouse B16 melanoma by sodium butyrate correlated to tumor associated macrophages differentiation suppression. Int. J. Clin. Exp. Med. 2015, 8, 4170–4174. [Google Scholar] [PubMed]
- Fortunati, N.; Bertino, S.; Costantino, L.; Bosco, O.; Vercellinatto, I.; Catalano, M.G.; Boccuzzi, G. Valproic acid is a selective antiproliferative agent in estrogen-sensitive breast cancer cells. Cancer Lett. 2008, 259, 156–164. [Google Scholar] [CrossRef]
- Jennings, V.A.; Scott, G.B.; Rose, A.M.S.; Scott, K.J.; Migneco, G.; Keller, B.; Reilly, K.; Donnelly, O.; Peach, H.; Dewar, D.; et al. Potentiating Oncolytic Virus-Induced Immune-Mediated Tumor Cell Killing Using Histone Deacetylase Inhibition. Mol. Ther. 2019, 27, 1139–1152. [Google Scholar] [CrossRef]
- Nakashima, H.; Nguyen, T.; Chiocca, E.A. Combining HDAC inhibitors with oncolytic virotherapy for cancer therapy. Oncolytic Virother. 2015, 4, 183–191. [Google Scholar] [CrossRef]
- Otsuki, A.; Patel, A.; Kasai, K.; Suzuki, M.; Kurozumi, K.; Chiocca, E.A.; Saeki, Y. Histone deacetylase inhibitors augment antitumor efficacy of herpes-based oncolytic viruses. Mol. Ther. 2008, 16, 1546–1555. [Google Scholar] [CrossRef]
- Connolly, R.M.; Rudek, M.A.; Piekarz, R. Entinostat: A promising treatment option for patients with advanced breast cancer. Future Oncol. 2017, 13, 1137–1148. [Google Scholar] [CrossRef]
- Schech, A.; Kazi, A.; Yu, S.; Shah, P.; Sabnis, G. Histone Deacetylase Inhibitor Entinostat Inhibits Tumor-Initiating Cells in Triple-Negative Breast Cancer Cells. Mol. Cancer Ther. 2015, 14, 1848–1857. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.S.; Parmigiani, R.B.; Marks, P.A. Histone deacetylase inhibitors: Molecular mechanisms of action. Oncogene 2007, 26, 5541–5552. [Google Scholar] [CrossRef] [PubMed]
- Barbarotta, L.; Hurley, K. Romidepsin for the Treatment of Peripheral T-Cell Lymphoma. J. Adv. Pract. Oncol. 2015, 6, 22–36. [Google Scholar]
- Suraweera, A.; O’Byrne, K.J.; Richard, D.J. Combination Therapy with Histone Deacetylase Inhibitors (HDACi) for the Treatment of Cancer: Achieving the Full Therapeutic Potential of HDACi. Front. Oncol. 2018, 8, 92. [Google Scholar] [CrossRef]
- Furumai, R.; Matsuyama, A.; Kobashi, N.; Lee, K.H.; Nishiyama, M.; Nakajima, H.; Tanaka, A.; Komatsu, Y.; Nishino, N.; Yoshida, M.; et al. FK228 (depsipeptide) as a natural prodrug that inhibits class I histone deacetylases. Cancer Res. 2002, 62, 4916–4921. [Google Scholar] [PubMed]
- ClinicalTrials.gov Identifier: NCT00943449. Available online: https://clinicaltrials.gov/ct2/show/NCT00943449 (accessed on 31 May 2021).
- Marrone, A.; Franci, G.; Perrella, A.; Nevola, R.; Chianese, A.; Adinolfi, L.E.; Sasso, F.C.; Rinaldi, L. Editorial—HCC in HCV patients and the direct acting antivirals: Is there really a link? Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 983–987. [Google Scholar] [CrossRef] [PubMed]
- Rinaldi, L.; Pafundi, P.C.; Galiero, R.; Caturano, A.; Morone, M.V.; Silvestri, C.; Giordano, M.; Salvatore, T.; Sasso, F.C. Mechanisms of Non-Alcoholic Fatty Liver Disease in the Metabolic Syndrome. A Narrative Review. Antioxidants 2021, 10, 270. [Google Scholar] [CrossRef]
- Ruf, B.; Berchtold, S.; Venturelli, S.; Burkard, M.; Smirnow, I.; Prenzel, T.; Henning, S.W.; Lauer, U.M. Combination of the oral histone deacetylase inhibitor resminostat with oncolytic measles vaccine virus as a new option for epi-virotherapeutic treatment of hepatocellular carcinoma. Mol. Ther. Oncolytics 2015, 2, 15019. [Google Scholar] [CrossRef]
- Soukupova, J.; Bertran, E.; Penuelas-Haro, I.; Urdiroz-Urricelqui, U.; Borgman, M.; Kohlhof, H.; Fabregat, I. Resminostat induces changes in epithelial plasticity of hepatocellular carcinoma cells and sensitizes them to sorafenib-induced apoptosis. Oncotarget 2017, 8, 110367–110379. [Google Scholar] [CrossRef]
- Katsura, T.; Iwai, S.; Ota, Y.; Shimizu, H.; Ikuta, K.; Yura, Y. The effects of trichostatin A on the oncolytic ability of herpes simplex virus for oral squamous cell carcinoma cells. Cancer Gene Ther. 2009, 16, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.L.; Wilson, M.G.; Hiscott, J. Oncolytic viruses and histone deacetylase inhibitors--a multi-pronged strategy to target tumor cells. Cytokine Growth Factor Rev. 2010, 21, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Franci, G.; Falanga, A.; Zannella, C.; Folliero, V.; Martora, F.; Galdiero, M.; Galdiero, S.; Morelli, G.; Galdiero, M. Infectivity inhibition by overlapping synthetic peptides derived from the gH/gL heterodimer of herpes simplex virus type 1. J. Pept. Sci. 2017, 23, 311–319. [Google Scholar] [CrossRef]
- Liu, T.C.; Castelo-Branco, P.; Rabkin, S.D.; Martuza, R.L. Trichostatin A and oncolytic HSV combination therapy shows enhanced antitumoral and antiangiogenic effects. Mol. Ther. 2008, 16, 1041–1047. [Google Scholar] [CrossRef]
- Kim, D.R.; Park, M.Y.; Lim, H.J.; Park, J.S.; Cho, Y.J.; Lee, S.W.; Yoon, H.I.; Lee, J.H.; Kim, Y.S.; Lee, C.T. Combination therapy of conditionally replicating adenovirus and histone deacetylase inhibitors. Int. J. Mol. Med. 2012, 29, 218–224. [Google Scholar] [CrossRef]
- Ma, J.; Zhao, J.; Lu, J.; Jiang, Y.; Yang, H.; Li, P.; Zhao, M.; Liu, K.; Dong, Z. Coxsackievirus and adenovirus receptor promotes antitumor activity of oncolytic adenovirus H101 in esophageal cancer. Int. J. Mol. Med. 2012, 30, 1403–1409. [Google Scholar] [CrossRef]
- Ma, J.; Li, N.; Zhao, J.; Lu, J.; Ma, Y.; Zhu, Q.; Dong, Z.; Liu, K.; Ming, L. Histone deacetylase inhibitor trichostatin A enhances the antitumor effect of the oncolytic adenovirus H101 on esophageal squamous cell carcinoma in vitro and in vivo. Oncol. Lett. 2017, 13, 4868–4874. [Google Scholar] [CrossRef][Green Version]
- Kitazono, M.; Goldsmith, M.E.; Aikou, T.; Bates, S.; Fojo, T. Enhanced adenovirus transgene expression in malignant cells treated with the histone deacetylase inhibitor FR901228. Cancer Res. 2001, 61, 6328–6330. [Google Scholar] [PubMed]
- Han, X.; Wang, S.; Zhou, W.; Li, Y.; Lei, W.; Lv, W. Synergistic combination of histone deacetylase inhibitor suberoylanilide hydroxamic acid and oncolytic adenovirus ZD55-TRAIL as a therapy against cervical cancer. Mol. Med. Rep. 2015, 12, 435–441. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pei, Z.; Chu, L.; Zou, W.; Zhang, Z.; Qiu, S.; Qi, R.; Gu, J.; Qian, C.; Liu, X. An oncolytic adenoviral vector of Smac increases antitumor activity of TRAIL against HCC in human cells and in mice. Hepatology 2004, 39, 1371–1381. [Google Scholar] [CrossRef]
- Nguyen, T.L.; Abdelbary, H.; Arguello, M.; Breitbach, C.; Leveille, S.; Diallo, J.S.; Yasmeen, A.; Bismar, T.A.; Kirn, D.; Falls, T.; et al. Chemical targeting of the innate antiviral response by histone deacetylase inhibitors renders refractory cancers sensitive to viral oncolysis. Proc. Nat. Acad. Sci. USA 2008, 105, 14981–14986. [Google Scholar] [CrossRef]
- Shulak, L.; Beljanski, V.; Chiang, C.; Dutta, S.M.; Van Grevenynghe, J.; Belgnaoui, S.M.; Nguyen, T.L.; Di Lenardo, T.; Semmes, O.J.; Lin, R.; et al. Histone deacetylase inhibitors potentiate vesicular stomatitis virus oncolysis in prostate cancer cells by modulating NF-kappaB-dependent autophagy. J. Virol. 2014, 88, 2927–2940. [Google Scholar] [CrossRef] [PubMed]
- Eckschlager, T.; Plch, J.; Stiborova, M.; Hrabeta, J. Histone Deacetylase Inhibitors as Anticancer Drugs. Int. J. Mol. Sci 2017, 18, 1414. [Google Scholar] [CrossRef] [PubMed]
- Thurn, K.T.; Thomas, S.; Moore, A.; Munster, P.N. Rational therapeutic combinations with histone deacetylase inhibitors for the treatment of cancer. Future Oncol. 2011, 7, 263–283. [Google Scholar] [CrossRef] [PubMed]
- Michan, S.; Sinclair, D. Sirtuins in mammals: Insights into their biological function. Biochem. J. 2007, 404, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Muscolini, M.; Castiello, L.; Palermo, E.; Zevini, A.; Ferrari, M.; Olagnier, D.; Hiscott, J. SIRT1 Modulates the Sensitivity of Prostate Cancer Cells to Vesicular Stomatitis Virus Oncolysis. J. Virol. 2019, 93, 208. [Google Scholar] [CrossRef]
- Kapoor, P.; Kumar, S.; Fonseca, R.; Lacy, M.Q.; Witzig, T.E.; Hayman, S.R.; Dispenzieri, A.; Buadi, F.; Bergsagel, P.L.; Gertz, M.A.; et al. Impact of risk stratification on outcome among patients with multiple myeloma receiving initial therapy with lenalidomide and dexamethasone. Blood 2009, 114, 518–521. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.; Ma, J.; Zou, Z.; Jemal, A. Cancer statistics, 2014. CA Cancer J. Clin. 2014, 64, 9–29. [Google Scholar] [CrossRef]
- Cai, W.; Chen, G.; Luo, Q.; Liu, J.; Guo, X.; Zhang, T.; Ma, F.; Yuan, L.; Li, B.; Cai, J. PMP22 regulates self-renewal and chemoresistance of gastric cancer cells. Mol. Cancer Ther. 2017, 16, 1187–1198. [Google Scholar] [CrossRef] [PubMed]
- Jaime-Ramirez, A.C.; Yu, J.G.; Caserta, E.; Yoo, J.Y.; Zhang, J.; Lee, T.J.; Hofmeister, C.; Lee, J.H.; Kumar, B.; Pan, Q.; et al. Reolysin and Histone Deacetylase Inhibition in the Treatment of Head and Neck Squamous Cell Carcinoma. Mol. Ther. Oncolytics 2017, 5, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Bretscher, C.; Marchini, A. H−1 Parvovirus as a Cancer-Killing Agent: Past, Present, and Future. Viruses 2019, 11, 562. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Seto, E. HDACs and HDAC Inhibitors in Cancer Development and Therapy. Cold Spring Harb. Perspect. Med. 2016, 6, 218. [Google Scholar] [CrossRef] [PubMed]
- MacTavish, H.; Diallo, J.S.; Huang, B.; Stanford, M.; Le Boeuf, F.; De Silva, N.; Cox, J.; Simmons, J.G.; Guimond, T.; Falls, T.; et al. Enhancement of vaccinia virus based oncolysis with histone deacetylase inhibitors. PLoS ONE 2010, 5, e14462. [Google Scholar] [CrossRef] [PubMed]
- Lyko, F. The DNA methyltransferase family: A versatile toolkit for epigenetic regulation. Nat. Rev. Genet. 2018, 19, 81–92. [Google Scholar] [CrossRef]
- Barau, J.; Teissandier, A.; Zamudio, N.; Roy, S.; Nalesso, V.; Herault, Y.; Guillou, F.; Bourc’his, D. The DNA methyltransferase DNMT3C protects male germ cells from transposon activity. Science 2016, 354, 909–912. [Google Scholar] [CrossRef] [PubMed]
- Bestor, T.H. The DNA methyltransferases of mammals. Hum. Mol. Genet. 2000, 9, 2395–2402. [Google Scholar] [CrossRef] [PubMed]
- Turek-Plewa, J.; Jagodzinski, P.P. The role of mammalian DNA methyltransferases in the regulation of gene expression. Cell Mol. Biol. Lett. 2005, 10, 631–647. [Google Scholar] [PubMed]
- Goll, M.G.; Kirpekar, F.; Maggert, K.A.; Yoder, J.A.; Hsieh, C.L.; Zhang, X.; Golic, K.G.; Jacobsen, S.E.; Bestor, T.H. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science 2006, 311, 395–398. [Google Scholar] [CrossRef]
- Margot, J.B.; Ehrenhofer-Murray, A.E.; Leonhardt, H. Interactions within the mammalian DNA methyltransferase family. BMC Mol. Biol. 2003, 4, 7. [Google Scholar] [CrossRef]
- Gnyszka, A.; Jastrzebski, Z.; Flis, S. DNA methyltransferase inhibitors and their emerging role in epigenetic therapy of cancer. Anticancer Res. 2013, 33, 2989–2996. [Google Scholar]
- Cui, H.; Cruz-Correa, M.; Giardiello, F.M.; Hutcheon, D.F.; Kafonek, D.R.; Brandenburg, S.; Wu, Y.; He, X.; Powe, N.R.; Feinberg, A.P. Loss of IGF2 imprinting: A potential marker of colorectal cancer risk. Science 2003, 299, 1753–1755. [Google Scholar] [CrossRef]
- Cui, H.; Onyango, P.; Brandenburg, S.; Wu, Y.; Hsieh, C.L.; Feinberg, A.P. Loss of imprinting in colorectal cancer linked to hypomethylation of H19 and IGF2. Cancer Res. 2002, 62, 6442–6446. [Google Scholar] [PubMed]
- Jelinic, P.; Shaw, P. Loss of imprinting and cancer. J. Pathol. 2007, 211, 261–268. [Google Scholar] [CrossRef]
- Murphy, S.K.; Huang, Z.; Wen, Y.; Spillman, M.A.; Whitaker, R.S.; Simel, L.R.; Nichols, T.D.; Marks, J.R.; Berchuck, A. Frequent IGF2/H19 domain epigenetic alterations and elevated IGF2 expression in epithelial ovarian cancer. Mol. Cancer Res. 2006, 4, 283–292. [Google Scholar] [CrossRef] [PubMed]
- Monda, V.; Valenzano, A.; Moscatelli, F.; Messina, A.; Piombino, L.; Zannella, C.; Viggiano, E.; Monda, G.; De Luca, V.; Chieffi, S. Modifications of activity of autonomic nervous system, and resting energy expenditure in women using hormone-replacement therapy. Biol. Med. 2016, 8, 1. [Google Scholar]
- Andrews, S.V.; Ellis, S.E.; Bakulski, K.M.; Sheppard, B.; Croen, L.A.; Hertz-Picciotto, I.; Newschaffer, C.J.; Feinberg, A.P.; Arking, D.E.; Ladd-Acosta, C.; et al. Cross-tissue integration of genetic and epigenetic data offers insight into autism spectrum disorder. Nat. Commun. 2017, 8, 1011. [Google Scholar] [CrossRef]
- Cuddapah, V.A.; Pillai, R.B.; Shekar, K.V.; Lane, J.B.; Motil, K.J.; Skinner, S.A.; Tarquinio, D.C.; Glaze, D.G.; McGwin, G.; Kaufmann, W.E.; et al. Methyl-CpG-binding protein 2 (MECP2) mutation type is associated with disease severity in Rett syndrome. J. Med. Genet. 2014, 51, 152–158. [Google Scholar] [CrossRef] [PubMed]
- Jin, Z.; Liu, Y. DNA methylation in human diseases. Genes Dis. 2018, 5, 1–8. [Google Scholar] [CrossRef]
- Liu, Y.; Aryee, M.J.; Padyukov, L.; Fallin, M.D.; Hesselberg, E.; Runarsson, A.; Reinius, L.; Acevedo, N.; Taub, M.; Ronninger, M.; et al. Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nat. Biotechnol. 2013, 31, 142–147. [Google Scholar] [CrossRef]
- Sun, Z.H.; Liu, Y.H.; Liu, J.D.; Xu, D.D.; Li, X.F.; Meng, X.M.; Ma, T.T.; Huang, C.; Li, J. MeCP2 Regulates PTCH1 Expression Through DNA Methylation in Rheumatoid Arthritis. Inflammation 2017, 40, 1497–1508. [Google Scholar] [CrossRef]
- Okemoto, K.; Kasai, K.; Wagner, B.; Haseley, A.; Meisen, H.; Bolyard, C.; Mo, X.; Wehr, A.; Lehman, A.; Fernandez, S.; et al. DNA demethylating agents synergize with oncolytic HSV1 against malignant gliomas. Clin. Cancer Res. 2013, 19, 5952–5959. [Google Scholar] [CrossRef]
- Cuddington, B.P.; Verschoor, M.; Ashkar, A.; Mossman, K.L. Enhanced efficacy with azacytidine and oncolytic BHV−1 in a tolerized cotton rat model of breast adenocarcinoma. Mol. Ther. Oncolytics 2015, 2, 15004. [Google Scholar] [CrossRef] [PubMed]
- Stiff, A.; Caserta, E.; Sborov, D.W.; Nuovo, G.J.; Mo, X.; Schlotter, S.Y.; Canella, A.; Smith, E.; Badway, J.; Old, M.; et al. Histone Deacetylase Inhibitors Enhance the Therapeutic Potential of Reovirus in Multiple Myeloma. Mol. Cancer Ther. 2016, 15, 830–841. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, R.; Cuddington, B.; Mossman, K. Bovine herpesvirus type 1 as a novel oncolytic virus. Cancer Gene Ther. 2010, 17, 344–355. [Google Scholar] [CrossRef]
- Chen, X.J.; Zhang, K.; Xin, Y.; Jiang, G. Oncolytic adenovirus-expressed RNA interference of O(6)-methylguanine DNA methyltransferase activity may enhance the antitumor effects of temozolomide. Oncol. Lett. 2014, 8, 2201–2202. [Google Scholar] [CrossRef][Green Version]
- Bleehen, N.M.; Newlands, E.S.; Lee, S.M.; Thatcher, N.; Selby, P.; Calvert, A.H.; Rustin, G.J.; Brampton, M.; Stevens, M.F. Cancer Research Campaign phase II trial of temozolomide in metastatic melanoma. J. Clin. Oncol. 1995, 13, 910–913. [Google Scholar] [CrossRef] [PubMed]
- O’Reilly, S.M.; Newlands, E.S.; Glaser, M.G.; Brampton, M.; Rice-Edwards, J.M.; Illingworth, R.D.; Richards, P.G.; Kennard, C.; Colquhoun, I.R.; Lewis, P.; et al. Temozolomide: A new oral cytotoxic chemotherapeutic agent with promising activity against primary brain tumours. Eur. J. Cancer 1993, 29A, 940–942. [Google Scholar] [CrossRef]
- Plummer, E.R.; Middleton, M.R.; Jones, C.; Olsen, A.; Hickson, I.; McHugh, P.; Margison, G.P.; McGown, G.; Thorncroft, M.; Watson, A.J.; et al. Temozolomide pharmacodynamics in patients with metastatic melanoma: Dna damage and activity of repair enzymes O6-alkylguanine alkyltransferase and poly(ADP-ribose) polymerase−1. Clin. Cancer Res. 2005, 11, 3402–3409. [Google Scholar] [CrossRef]
- Zheng, M.; Huang, J.; Tong, A.; Yang, H. Oncolytic Viruses for Cancer Therapy: Barriers and Recent Advances. Mol. Ther. Oncolytics 2019, 15, 234–247. [Google Scholar] [CrossRef]
- Shi, T.; Song, X.; Wang, Y.; Liu, F.; Wei, J. Combining Oncolytic Viruses with Cancer Immunotherapy: Establishing a New Generation of Cancer Treatment. Front. Immunol. 2020, 11, 2076. [Google Scholar] [CrossRef]
- Marzulli, M.; Mazzacurati, L.; Zhang, M.; Goins, W.F.; Hatley, M.E.; Glorioso, J.C.; Cohen, J.B. A Novel Oncolytic Herpes Simplex Virus Design based on the Common Overexpression of microRNA−21 in Tumors. J. Gene Ther. 2018, 3, 2060. [Google Scholar] [CrossRef]
- Mazzacurati, L.; Marzulli, M.; Reinhart, B.; Miyagawa, Y.; Uchida, H.; Goins, W.F.; Li, A.; Kaur, B.; Caligiuri, M.; Cripe, T.; et al. Use of miRNA response sequences to block off-target replication and increase the safety of an unattenuated, glioblastoma-targeted oncolytic HSV. Mol. Ther. 2015, 23, 99–107. [Google Scholar] [CrossRef]
- Li, J.M.; Kao, K.C.; Li, L.F.; Yang, T.M.; Wu, C.P.; Horng, Y.M.; Jia, W.W.; Yang, C.T. MicroRNA−145 regulates oncolytic herpes simplex virus−1 for selective killing of human non-small cell lung cancer cells. Virol. J. 2013, 10, 241. [Google Scholar] [CrossRef]
- Fu, X.; Rivera, A.; Tao, L.; De Geest, B.; Zhang, X. Construction of an oncolytic herpes simplex virus that precisely targets hepatocellular carcinoma cells. Mol. Ther. 2012, 20, 339–346. [Google Scholar] [CrossRef]
- Hodzic, J.; Sie, D.; Vermeulen, A.; van Beusechem, V.W. Functional Screening Identifies Human miRNAs that Modulate Adenovirus Propagation in Prostate Cancer Cells. Hum. Gene Ther. 2017, 28, 766–780. [Google Scholar] [CrossRef] [PubMed]
- Bofill-De Ros, X.; Villanueva, E.; Fillat, C. Late-phase miRNA-controlled oncolytic adenovirus for selective killing of cancer cells. Oncotarget 2015, 6, 6179–6190. [Google Scholar] [CrossRef] [PubMed]
- Rovira-Rigau, M.; Raimondi, G.; Marin, M.A.; Gironella, M.; Alemany, R.; Fillat, C. Bioselection Reveals miR−99b and miR−485 as Enhancers of Adenoviral Oncolysis in Pancreatic Cancer. Mol. Ther. 2019, 27, 230–243. [Google Scholar] [CrossRef]
- Lou, W.; Chen, Q.; Ma, L.; Liu, J.; Yang, Z.; Shen, J.; Cui, Y.; Bian, X.W.; Qian, C. Oncolytic adenovirus co-expressing miRNA−34a and IL−24 induces superior antitumor activity in experimental tumor model. J. Mol. Med. 2013, 91, 715–725. [Google Scholar] [CrossRef]
- Moshiri, F.; Callegari, E.; D’Abundo, L.; Corra, F.; Lupini, L.; Sabbioni, S.; Negrini, M. Inhibiting the oncogenic mir−221 by microRNA sponge: Toward microRNA-based therapeutics for hepatocellular carcinoma. Gastroenterol. Hepatol. Bed. Bench 2014, 7, 43–54. [Google Scholar]
- Callegari, E.; Elamin, B.K.; D’Abundo, L.; Falzoni, S.; Donvito, G.; Moshiri, F.; Milazzo, M.; Altavilla, G.; Giacomelli, L.; Fornari, F.; et al. Anti-tumor activity of a miR−199-dependent oncolytic adenovirus. PLoS ONE 2013, 8, e73964. [Google Scholar] [CrossRef] [PubMed]
- Leber, M.F.; Baertsch, M.A.; Anker, S.C.; Henkel, L.; Singh, H.M.; Bossow, S.; Engeland, C.E.; Barkley, R.; Hoyler, B.; Albert, J.; et al. Enhanced Control of Oncolytic Measles Virus Using MicroRNA Target Sites. Mol. Ther. Oncolytics 2018, 9, 30–40. [Google Scholar] [CrossRef]
- Geekiyanage, H.; Galanis, E. MiR−31 and miR−128 regulates poliovirus receptor-related 4 mediated measles virus infectivity in tumors. Mol. Oncol. 2016, 10, 1387–1403. [Google Scholar] [CrossRef]
- Lei, W.; Wang, S.; Yang, C.; Huang, X.; Chen, Z.; He, W.; Shen, J.; Liu, X.; Qian, W. Combined expression of miR−34a and Smac mediated by oncolytic vaccinia virus synergistically promote anti-tumor effects in Multiple Myeloma. Sci. Rep. 2016, 6, 1–11. [Google Scholar] [CrossRef]
- Jia, Y.; Miyamoto, S.; Soda, Y.; Takishima, Y.; Sagara, M.; Liao, J.; Hirose, L.; Hijikata, Y.; Miura, Y.; Hara, K.; et al. Extremely Low Organ Toxicity and Strong Antitumor Activity of miR−34-Regulated Oncolytic Coxsackievirus B3. Mol. Ther. Oncolytics 2019, 12, 246–258. [Google Scholar] [CrossRef]
- Ruiz, A.J.; Hadac, E.M.; Nace, R.A.; Russell, S.J. MicroRNA-Detargeted Mengovirus for Oncolytic Virotherapy. J. Virol. 2016, 90, 4078–4092. [Google Scholar] [CrossRef]
- Friedman, R.C.; Farh, K.K.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef] [PubMed]
- Ameres, S.L.; Zamore, P.D. Diversifying microRNA sequence and function. Nat. Rev. Mol. Cell Biol. 2013, 14, 475–488. [Google Scholar] [CrossRef]
- Amiel, J.; de Pontual, L.; Henrion-Caude, A. miRNA, development and disease. Adv. Genet. 2012, 80, 1–36. [Google Scholar] [CrossRef]
- Lujambio, A.; Lowe, S.W. The microcosmos of cancer. Nature 2012, 482, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Volinia, S.; Calin, G.A.; Liu, C.-G.; Ambs, S.; Cimmino, A.; Petrocca, F.; Visone, R.; Iorio, M.; Roldo, C.; Ferracin, M. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 2006, 103, 2257–2261. [Google Scholar] [CrossRef] [PubMed]
- Gu, S.; Jin, L.; Zhang, F.; Sarnow, P.; Kay, M.A. Biological basis for restriction of microRNA targets to the 3′ untranslated region in mammalian mRNAs. Nat. Struct. Mol. Biol. 2009, 16, 144–150. [Google Scholar] [CrossRef]
- Kelly, E.J.; Hadac, E.M.; Greiner, S.; Russell, S.J. Engineering microRNA responsiveness to decrease virus pathogenicity. Nat. Med. 2008, 14, 1278–1283. [Google Scholar] [CrossRef]
- Ylosmaki, E.; Hakkarainen, T.; Hemminki, A.; Visakorpi, T.; Andino, R.; Saksela, K. Generation of a conditionally replicating adenovirus based on targeted destruction of E1A mRNA by a cell type-specific MicroRNA. J. Virol. 2008, 82, 11009–11015. [Google Scholar] [CrossRef]
- Leber, M.F.; Bossow, S.; Leonard, V.H.; Zaoui, K.; Grossardt, C.; Frenzke, M.; Miest, T.; Sawall, S.; Cattaneo, R.; von Kalle, C.; et al. MicroRNA-sensitive oncolytic measles viruses for cancer-specific vector tropism. Mol. Ther. 2011, 19, 1097–1106. [Google Scholar] [CrossRef]
- Shayestehpour, M.; Moghim, S.; Salimi, V.; Jalilvand, S.; Yavarian, J.; Romani, B.; Mokhtari-Azad, T. Targeting human breast cancer cells by an oncolytic adenovirus using microRNA-targeting strategy. Virus Res. 2017, 240, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Santella, B.; Pignataro, D.; Lavano, M.A.; Rinaldi, M.; Galdiero, F. Comment on: Expressions of MiR−132 in patients with chronic hepatitis B, posthepatitic cirrhosis and hepatitis B virus-related hepatocellular carcinoma. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 1384–1385. [Google Scholar] [CrossRef]
- Santella, B.; Folliero, V.; Pirofalo, G.M.; Serretiello, E.; Zannella, C.; Moccia, G.; Santoro, E.; Sanna, G.; Motta, O.; De Caro, F.; et al. Sepsis-A Retrospective Cohort Study of Bloodstream Infections. Antibiotics 2020, 9, 851. [Google Scholar] [CrossRef] [PubMed]
- Anesti, A.-M.; Simpson, G.R.; Price, T.; Pandha, H.S.; Coffin, R.S. Expression of RNA interference triggers from an oncolytic herpes simplex virus results in specific silencing in tumour cells in vitro and tumours in vivo. BMC Cancer 2010, 10, 486. [Google Scholar] [CrossRef]
- Chen, Z.; Zeng, H.; Guo, Y.; Liu, P.; Pan, H.; Deng, A.; Hu, J. miRNA−145 inhibits non-small cell lung cancer cell proliferation by targeting c-Myc. J. Exp. Clin. Cancer Res. 2010, 29, 151. [Google Scholar] [CrossRef]
- Guan, P.; Yin, Z.; Li, X.; Wu, W.; Zhou, B. Meta-analysis of human lung cancer microRNA expression profiling studies comparing cancer tissues with normal tissues. J. Exp. Clin. Cancer Res. 2012, 31, 54. [Google Scholar] [CrossRef] [PubMed]
- Krichevsky, A.M.; Gabriely, G. miR−21: A small multi-faceted RNA. J. Cell Mol. Med. 2009, 13, 39–53. [Google Scholar] [CrossRef] [PubMed]
- Ylösmäki, E.; Lavilla-Alonso, S.; Jäämaa, S.; Vähä-Koskela, M.; af Hällström, T.; Hemminki, A.; Arola, J.; Mäkisalo, H.; Saksela, K. MicroRNA-Mediated Suppression of Oncolytic Adenovirus Replication in Human Liver. PLoS ONE 2013, 8, e54506. [Google Scholar] [CrossRef]
- Sugio, K.; Sakurai, F.; Katayama, K.; Tashiro, K.; Matsui, H.; Kawabata, K.; Kawase, A.; Iwaki, M.; Hayakawa, T.; Fujiwara, T.; et al. Enhanced Safety Profiles of the Telomerase-Specific Replication- Competent Adenovirus by Incorporation of Normal Cell-Specific microRNA-Targeted Sequences. Clin. Cancer Res. 2011, 17, 2807–2818. [Google Scholar] [CrossRef]
- Bofill-De Ros, X.; Gironella, M.; Fillat, C. miR−148a- and miR−216a-regulated oncolytic adenoviruses targeting pancreatic tumors attenuate tissue damage without perturbation of miRNA activity. Mol. Ther. 2014, 22, 1665–1677. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, S.; Inoue, H.; Nakamura, T.; Yamada, M.; Sakamoto, C.; Urata, Y.; Okazaki, T.; Marumoto, T.; Takahashi, A.; Takayama, K.; et al. Coxsackievirus B3 is an oncolytic virus with immunostimulatory properties that is active against lung adenocarcinoma. Cancer Res. 2012, 72, 2609–2621. [Google Scholar] [CrossRef]
- Shafren, D.R.; Williams, D.T.; Barry, R.D. A decay-accelerating factor-binding strain of coxsackievirus B3 requires the coxsackievirus-adenovirus receptor protein to mediate lytic infection of rhabdomyosarcoma cells. J. Virol. 1997, 71, 9844–9848. [Google Scholar] [CrossRef]
- Fulci, G.; Breymann, L.; Gianni, D.; Kurozomi, K.; Rhee, S.S.; Yu, J.; Kaur, B.; Louis, D.N.; Weissleder, R.; Caligiuri, M.A.; et al. Cyclophosphamide enhances glioma virotherapy by inhibiting innate immune responses. Proc. Natl. Acad. Sci. USA 2006, 103, 12873–12878. [Google Scholar] [CrossRef]
- Ruiz, A.J.; Russell, S.J. MicroRNAs and oncolytic viruses. Curr. Opin. Virol. 2015, 13, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Mell, L.K.; Brumund, K.T.; Daniels, G.A.; Advani, S.J.; Zakeri, K.; Wright, M.E.; Onyeama, S.J.; Weisman, R.A.; Sanghvi, P.R.; Martin, P.J.; et al. Phase I Trial of Intravenous Oncolytic Vaccinia Virus (GL-ONC1) with Cisplatin and Radiotherapy in Patients with Locoregionally Advanced Head and Neck Carcinoma. Clin. Cancer Res. 2017, 23, 5696–5702. [Google Scholar] [CrossRef]
| Virus Family | Virus | Institution | Tumor | Phase | Status | Trial N° | Source |
|---|---|---|---|---|---|---|---|
| Herpesviridae | Imlygic | BioVex Limited | Melanoma | III | Completed | NCT00769704 NCT01368276 | [67,68] |
| BioVex Limited | Pancreatic Cancer | I | Completed | NCT00402025 | [69] | ||
| BioVex Limited | Squamous Cell Carcinoma; Head and Neck\Cancer | III | Completed | NCT01161498 | [70] | ||
| G47∆ | The University of Tokyo Hospital | Prostate cancer | I | Completed | UMIN000010463 | [32] | |
| The IMSUT Hospital | Glioblastoma | II | Completed | UMIN000015995 | [71] | ||
| G47∆ | The University of Tokyo Hospital | Prostate cancer | I | Completed | UMIN000010463 | [32] | |
| The IMSUT Hospital | Glioblastoma | II | Completed | UMIN000015995 | [71] | ||
| rQNestin34.5 | National Institutes of Health (NIH); Candel Therapeutics, Inc.; Dana-Farber Cancer Institute | Glioma; Astrocytoma; Glioblastoma; | I | Recruiting | NCT03152318 | [72] | |
| G207 | MediGene | Glioma; Astrocytoma; Glioblastoma; | I, II | Completed | NCT00028158 | [73] | |
| MediGene | Malignant glioma | I | Completed | NCT00157703 | [74] | ||
| Adenoviridae | H101 (Oncorine) | Sun Yat-sen University | Hepatocellular Carcinoma | III | Recruiting | NCT03780049 | [75] |
| Fudan University | Malignant Ascites | II | Recruiting | NCT04771676 | [76] | ||
| DNX-2440 | DNAtrix, Inc., H. Lee Moffitt Cancer Center and Research Institute | Colon cancer; Colorectal Cancer; Breast Cancer; Melanoma; Renal Cell Cancer; Sarcoma; Squamous Cell Carcinoma | I | Recruiting | NCT04714983 | [77] | |
| Clinica Universidad de Navarra, Universidad de Navarra, DNAtrix, Inc. | Glioblastoma | I | Recruiting | NCT03714334 | [78] | ||
| DNX-2401 (Delta-24-RGD) | Clinica Universidad de Navarra, Universidad de Navarra, DNAtrix, Inc. | Glioblastoma | I | Completed | NCT01956734 NCT02197169 NCT01582516 | [79,80,81] | |
| TILT-123 | TILT Biotherapeutics Ltd. | Solid Tumor; Metastatic Melanoma | I | Recruiting | NCT04695327 NCT04217473 | [82,83] | |
| VCN-01 | VCN Biosciences, S.L. | Solid Tumors; Pancreatic Adenocarcinoma | I | Completed | NCT02045602 NCT02045589 | [84,85] | |
| Institut Català d’Oncologia, VCN Biosciences, S.L. BioClever 2005 S.L. AstraZeneca | Head and Neck Neoplasms Carcinoma | I | Recruiting | NCT03799744 | [86] | ||
| LOAd703 | Lokon Pharma AB | Pancreatic Cancer | I, II | Recruiting | NCT02705196 | [87] | |
| Lokon Pharma AB, Precision Oncology LLC | Malignant Melanoma | I, II | Recruiting | NCT04123470 | [88] | ||
| Lokon Pharma AB, Uppsala University | Pancreatic Adenocarcinoma; Ovarian Cancer; Biliary Carcinoma; Colorectal Cancer | I, II | Recruiting | NCT03225989 | [89] | ||
| ICOVIR-5 | Institut Català d’Oncologia | Melanoma | I | Completed | NCT01864759 | [90] | |
| CG0070 | CG Oncology, Inc., Merck Sharp & Dohme Corp. | Non Muscle Invasive Bladder Cancer | II, III | Recruiting | NCT04387461 NCT04452591 NCT02365818 | [91,92,93] | |
| Enadenotucirev | University of Oxford, PsiOxus Therapeutics Ltd. Cancer Research UK | Rectal Cancer | I | Recruiting | NCT03916510 | [94] | |
| CAdVEC | Baylor College of Medicine, The Methodist Hospital System, Daniel Wang, Baylor College of Medicine | Bladder Cancer; Head and Neck Squamous Cell Carcinoma; Breast cancer; Colorectal Cancer; Pancreatic Cancer | I | Recruiting | NCT03740256 | [95] | |
| ORCA-010 | Orca Therapeutics B.V., CMX Research | Prostate Cancer | I, II | Recruiting | NCT04097002 | [96] | |
| NG-641 | PsiOxus Therapeutics Ltd. | Epithelial Tumor | I | Recruiting | NCT04053283 | [97] | |
| Reoviridae | Reolysin | Oncolytics Biotech | Malignant Glioma | I | Completed | NCT00528684 | [98] |
| Oncolytics Biotech | Pancreatic Adenocarcinoma | I | Completed | NCT02620423 | [99] | ||
| Emory University, Bristol-Myers Squibb Oncolytics Biotech University of Utah City of Hope Medical Center Phylogeny | Recurrent Plasma Cell Myeloma | I | Recruiting | NCT03605719 | [100] | ||
| Oncolytics Biotech | Squamous Cell Carcinoma of the Lung | II | Completed | NCT00998192 | [101] | ||
| Oncolytics Biotech | Squamous Cell Carcinoma of the Head and Neck | II | Completed | NCT00753038 | [102] | ||
| Oncolytics Biotech | Melanoma | II | Completed | NCT00984464 | [103] | ||
| Incyte Corporation; Oncolytics Biotech; National Cancer Institute (NCI) | Breast Cancer | II | Recruiting | NCT04445844 | [104] | ||
| Oncolytics Biotech | Carcinoma, Squamous Cell of the Head and Neck | III | Completed | NCT01166542 | [105] | ||
| National Cancer Institute (NCI) | Ovarian Cancer | II | Completed | NCT01199263 | [106] | ||
| Parvoviridae | H1-PV | Oryx GmbH & Co. KG | Glioblastoma Multiforme | I, II | Completed | NCT01301430 | [107,108] |
| Oryx GmbH & Co. KG | Pancreatic Ductal Carcinoma | I, II | Completed | NCT02653313 | [109] | ||
| Paramyxoviridae | Measles virus | National Cancer Institute (NCI) | Anaplastic Astrocytoma; Anaplastic Oligodendroglioma Mixed Glioma; Recurrent Glioblastoma | I | Completed | NCT00390299 | [110] |
| National Cancer Institute (NCI) | Ovarian Cancer; Primary Peritoneal Cavity Cancer | I | Completed | NCT00408590 | [111] | ||
| University of Arkansas | Multiple Myeloma | II | Completed | NCT02192775 | [112] | ||
| National Cancer Institute (NCI) | Breast Cancer | I | Recruiting | NCT04521764 | [113] | ||
| Mayo Clinic, National Cancer Institute (NCI) | Ovarian Carcinoma, Peritoneal Carcinoma, Fallopian Tube Transitional Cell Carcinoma | I, II | Recruiting | NCT02068794 | [114] | ||
| Mayo Clinic, National Cancer Institute (NCI) | Malignant Mesothelioma | I | Completed | NCT01503177 | [115] | ||
| Mayo Clinic, National Cancer Institute (NCI) | Plasma Cell Myeloma | I, II | Completed | NCT00450814 | [116] | ||
| Mayo Clinic, National Cancer Institute (NCI) | Malignant peripheral nerve sheath tumor | I | Recruiting | NCT02700230 | [117] | ||
| Poxviridae | Vaccinia virus | Assistance Publique-Hopitaux de Paris | Glioblastoma; Brain Cancer; | I, II | Recruiting | NCT03294486 | [118] |
| Jennerex Biotherapeutics, SillaJen, Inc. | Hepatocellular Carcinoma | II, III | Completed | NCT00554372 NCT01636284 | [119,120] | ||
| Jennerex Biotherapeutics | Melanoma | I, II | Completed | NCT00429312 | [121] | ||
| SillaJen, Inc., Regeneron Pharmaceuticals | Renal Cell Carcinoma | I, II | Recruiting | NCT03294083 | [122] | ||
| Jennerex Biotherapeutics, Green Cross Corporation | Hepatic carcinoma | I | Completed | NCT00629759 | [123] |
| Virus Family | Oncolytic Virus | Epigenetic Modulators | Tumor Type | Effects | Source |
|---|---|---|---|---|---|
| Herpesviridae | G∆47 | TSA | Glioma; Colorectal cancer | Reduced VEGF and cyclin D1 | [138] |
| R849 | TSA | Oral squamous cell carcinoma (SCC) | Increased NF-kB activity Increased viral production Increased p21 interrupt cell cycle in G0/G1 phase | [240] | |
| rQNestin34.5 | VPA; TSA; NaB 5-AZA | Glioblastoma Glioma | Increased viral replication Increased IFN-I expression Increased tumor apoptosis | [228,280] | |
| BHV-1 | 5-AZA | Spontaneous breast fibrosarcomas | Increased tumor apoptosis | [281] | |
| H101 | TSA; romidepsin | Esophageal squamous cell carcinoma (ESCC) | Increased CAR and viral activity Increased viral entry | [247] | |
| Adenoviridae | ZD55-Trial | SAHA | Cervical cancer | Increased cell cycle block in phase G2 Increased apoptosis | [248,249] |
| Rhabdoviridae | VSV∆51 | vorinostat; MS275; SIRT1; decitabine | Prostate cancer T-cell lymphocytic leukemia | Increased apoptosis Increased viral oncolysis Increased remission tumor | [138,176,255] |
| Reoviridae | RV | entinostat | Multiple myeloma (MM); Head and neck squamous cell carcinomas (HNSCC) | Increased antiviral activity | [259,282] |
| Parvoviridae | H1PV | VPA; NaB | Cervical cancer; Pancreatic cancer Carcinoma | Increased apoptosis Increased viral oncotoxicity | [138,185] |
| P/V-CPI | scriptaid | Small cell lung cancer; Laryngeal carcinoma cells | Increased apoptosis | [192] | |
| Paramyxoviridae | MeV | Res | HCC | Increased apoptosis Increased viral replication | [238] |
| Poxviridae | VV | TSA | Colon carcinoma | Increased viral replication | [138,262] |
| Virus Family | Engineered Virus | miRNA | Target cell/Tissue | Results/Effects | Source |
|---|---|---|---|---|---|
| Herpesviridae | dnU L 9-T21 | miR-21 | Glioblastoma cell lines | Increased cell specificity | [290] |
| KG4: T-124 | miR-124 | Brain tissue | Increased safety | [291] | |
| AP27i145 | miR-145 | Non-small cell lung cancer cells | Increased selectivity | [292] | |
| LCSOV | miR-7;miR-122; miR-124 | HCC | Increased selectivity Increased safety | [293] | |
| Adenoviridae | AdΔ24.CMV-GFP | miR-26b | Prostate cancer | Increased propagation Reduced spread | [294] |
| Ad-L5-8miR148aT | miR-148a | Hepatocytes/liver | Increased hepatotoxicity Increased anticancer response | [295] | |
| ICOVIR15 | miR-99b; miR-485 | Pancreatic Cancer | Increased anticancer activity Increased oncolytic potential | [296] | |
| AdCN205-IL-24-miR-34a | miR-34a | HCC | Complete tumor regression | [297] | |
| rAd-199T-miR-221 | mir-221 | HCC | Reduced apoptosis Increased miR-221 levels | [298] | |
| Ad-199T | miR-199 | HCC | Reduced hepatotoxicity Increased cell specificity | [299] | |
| Paramyxoviridae | MV-EGFP-H miRTS7, MV-EGFP-H miRTS7rc, MV-EGFP-H miRTS122, MV-EGFP-H miRTS122rc, MV-EGFP-H miRTS124 | miR-124; miR-125b; miR-7 | Brain tissue | Increased replicative control | [300] |
| SLAM of Measles Virus | miR-31 miR-128 | Glioblastoma | Increased infectivity Increased anticancer effect | [301] | |
| Poxviridae | VV-miR-34a VV-Smac | miR-34a | Multiple Myeloma | Increased anticancer effect Increased apoptosis Reduced tumor growth | [302] |
| Picornaviridae | 53a-CVB | miR-34a/c | Human non-small cell Lung cancer cells | Reduced cytotoxicity | [303] |
| vMC24-NC | miR-124 miR-125 miR-133 miR-208 miR-142 | Nervous tissue Cardiac tissue Hematopoietic tissues | Reduced pathogenesis Increased safety | [304] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chianese, A.; Santella, B.; Ambrosino, A.; Stelitano, D.; Rinaldi, L.; Galdiero, M.; Zannella, C.; Franci, G. Oncolytic Viruses in Combination Therapeutic Approaches with Epigenetic Modulators: Past, Present, and Future Perspectives. Cancers 2021, 13, 2761. https://doi.org/10.3390/cancers13112761
Chianese A, Santella B, Ambrosino A, Stelitano D, Rinaldi L, Galdiero M, Zannella C, Franci G. Oncolytic Viruses in Combination Therapeutic Approaches with Epigenetic Modulators: Past, Present, and Future Perspectives. Cancers. 2021; 13(11):2761. https://doi.org/10.3390/cancers13112761
Chicago/Turabian StyleChianese, Annalisa, Biagio Santella, Annalisa Ambrosino, Debora Stelitano, Luca Rinaldi, Massimiliano Galdiero, Carla Zannella, and Gianluigi Franci. 2021. "Oncolytic Viruses in Combination Therapeutic Approaches with Epigenetic Modulators: Past, Present, and Future Perspectives" Cancers 13, no. 11: 2761. https://doi.org/10.3390/cancers13112761
APA StyleChianese, A., Santella, B., Ambrosino, A., Stelitano, D., Rinaldi, L., Galdiero, M., Zannella, C., & Franci, G. (2021). Oncolytic Viruses in Combination Therapeutic Approaches with Epigenetic Modulators: Past, Present, and Future Perspectives. Cancers, 13(11), 2761. https://doi.org/10.3390/cancers13112761








