Measuring Statistical Dependence via Characteristic Function IPM
Abstract
1. Introduction
- Theoretical and methodological contributions. We propose a new IPM-based statistical dependence measure (UFDM) and derive its properties. The main theoretical result of this paper is the structural characterisation of UFDM, which includes invariance under linear transformations and augmentation with independent noise, monotonicity under linear dimension reduction, vanishing under independence, and a concentration bound for its empirical estimator. We additionally propose a gradient-based estimation algorithm with an SVD warm-up to ensure numerical stability.
- Empirical analysis. We conduct an empirical study demonstrating the practical effectiveness of UFDM in permutation-based independence testing across diverse linear, nonlinear, and geometrically structured patterns, as well as in supervised feature-extraction tasks on real datasets.
1.1. IPM Framework
1.2. Characteristic Functions
2. Previous Work
- Distance correlation. DCOR [13] is defined as
- HSIC. For reproducing kernel Hilbert spaces (RKHS) and with kernels k and l, it is defined as
- MEF. Shannon mutual information is defined by [19]. The neural estimation of mutual information (MINE, [26]) uses its variational (Donsker–Varadhan) representation , since it allows avoiding density estimation (here is a neural network with parameters ). In this case, the optimisation is performed over the space of neural network parameters, which often leads to unstable training and biased estimates due to the unboundedness of the objective and the difficulty of balancing the exponential term. The matrix-based Rényi’s -order entropy functional (MEF) [20,21,27] provides a kernel version of mutual information that avoids both density estimation and neural optimization. For random variables X and Y with distributions , , and , it is defined as
Motivation
3. Proposed Measure
- .
- .
- if and only if (⊥ denotes statistical independence).
- For Gaussian random vectors , with cross-covariance matrix we have .
- Invariance under full-rank linear transformation: for any full-rank matrices , and vectors , .
- Linear dimension reduction does not increase .
- If , for any continuous function , , if has a density.
- If X and Y have densities, then , where is mutual information.
- Invariance to augmentation with independent noise: let be random vectors such that . Then .
- Interpretation of UFDM via canonical correlation analysis (CCA). In the Gaussian case, the UFDM objective reduces analytically to CCA via a closed-form expression (Theorem 1, Property 4): after whitening (setting and ), it becomes , where . By von Neumann’s inequality, the maximizers align with the leading singular vectors of K, corresponding to the top CCA pair. Note that since Gaussian independence is equivalent to the vanishing of the leading canonical correlation (as all remaining correlations , must also vanish), UFDM’s focus on the leading canonical correlation entails no loss of discriminatory power.
- Interpretation of UFDM via cumulants. Let us recall that , , . For general distributions, writing offers a cumulant-series factorization, with , where are cross-cumulants and are the -order tensors formed by the tensor product of p copies of and q copies of . The leading term, corresponding to , is (with for centered variables), which aligns with the CCA interpretation, while higher-order terms capture non-Gaussian deviations, interpreting UFDM as a frequency-domain approach that aligns with cross-cumulant directions under marginal damping by .
- Remark on the representations of CFs. Since , the UFDM objective naturally operates on the real two-dimensional vector formed by the real and imaginary parts of . This aligns with recent work on real-vector representations of characteristic functions [17] and shows that UFDM does not rely on any special algebraic role of the imaginary unit.
3.1. Estimation
- Empirical estimator. Let us define the empirical estimator of for a fixed :
3.2. Estimator Convergence
- This implies the convergence of the empirical estimator Equation (12):
3.3. Estimator Computation
| Algorithm 1 UFDM estimation |
| Require: Number of iterations N, batch size , initial . for to N do Sample batch . Standardise to zero mean and unit variance. . end for return , , |
| Algorithm 2 SVD warm-up |
| Require:
Batch size . Sample batch . Compute cross-covariance . Decompose: . . return , |
4. Experiments
4.1. Permutation Tests
- Permutation tests with UFDM. We compared , DCOR, HSIC, and MEF in permutation-based statistical independence testing ( versus the alternative ) using a set of multivariate distributions. We investigated scenarios with a sample size of and data dimensions (). To ensure valid finite-sample calibration, permutation p-values were computed with the Phipson–Smyth correction [31].
- Hyperparameters. We used 500 permutations per p-value. The number of iterations in UFDM estimation Algorithm 1 was set to 100. The batch size equaled the sample size (). We used a learning rate of . Due to the high computation time (permutation tests took days on five machines with Intel i7 CPU, 16GB of RAM, and Nvidia GeForce RTX 2060 12 GB GPU), we relied on 500 p-values for each test in the scenario and on 100 p-values for each test in the scenario.
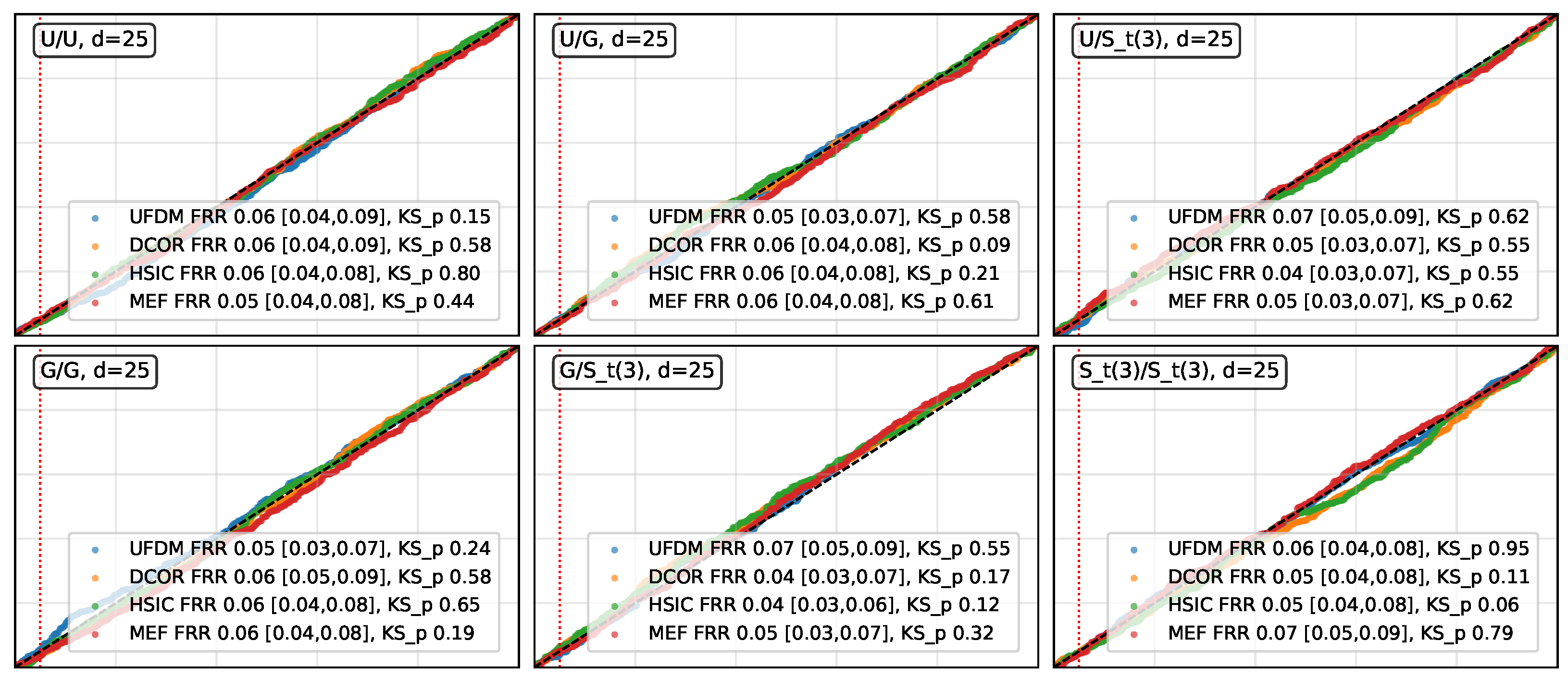
- Distributions analysed. In the case, X was sampled from multivariate uniform, Gaussian, and Student distributions (corresponding to no-tail, light-tail, and heavy-tail scenarios, respectively), and Y was independently sampled from the same set of distributions. Afterwards, we examined the uniformity of the p-values obtained from permutation tests using different statistical measures, through QQ-plots and Kolmogorov–Smirnov (KS) tests.
- Results for . As shown in Figure 1, UFDM, DCOR, HSIC, and MEF exhibited approximately uniform permutation p-values across all distribution pairs and dimensions, with empirical false rejection rates (FRR) remaining close to the nominal level. Isolated low KS p-values below occurred in only two cases: one for MEF in the Gaussian/Gaussian pair at dimension 5 (p-value of ) and one for UFDM in the Gaussian/Student-t pair at dimension 5 (p-value of ), suggesting minor sampling variability rather than systematic deviations from uniformity. These results show that UFDM remained comparably stable to DCOR, HSIC and MEF, in terms of type-I error control under .
- Results for . The empirical power and its -Wilson confidence intervals (CIs) are presented in Table 1 and Table 2. These results show that, in most cases, the empirical power of UFDM, DCOR, HSIC, and MEF was approximately equal to . However, Table 2 also reveals that for the sparse Circular and Interleaved Moons patterns (), MEF exhibited a noticeable decrease in empirical power. We conjecture that this reduction may stem from MEF’s comparatively higher sensitivity to kernel bandwidth selection in these specific, geometrically structured patterns. On the other hand, UFDM’s robustness in these settings may also be explained by its invariance to augmentation with independent noise (Theorem 1, Property 9), which helps to preserve the detectability of sparse geometric dependencies embedded within high-dimensional noise coordinates.
- Ablation experiment. The necessity of the SVD warm-up (Algorithm 2) is empirically demonstrated in Table A1, where the p-values obtained without SVD warm-up systematically fail to reveal dependence in many nonlinear patterns.
- Remark on the stability of the estimator. Since the UFDM objective is non-convex, different random initialisations may potentially lead to distinct local optima. To assess the impact of this issue, we investigated the numerical stability of the UFDM estimator. We computed the mean and standard deviation of the statistic across 50 independent runs for each distribution pattern and dimension (Table 1 and Table 2), as well as for the corresponding permuted patterns in which dependence is destroyed, as reported in Table 3. The obtained results align with the permutation test findings. While a slight upward shift is observed under independent (permuted) data, the proposed estimator retained consistent separation between dependent and independent settings and exhibited stable behaviour across random restarts.
4.2. Supervised Feature Extraction
- Evaluation metrics. Let us denote , if for r runs on the dataset d the average test set accuracy of baseline b is statistically significantly higher than that of with p-value threshold p. For statistical significance assessment, we used Wilcoxon’s signed-rank test [36]. We computed the win ranking (WR) and loss ranking (LR) as
- Based on these metrics, Table 4 includes full information on how many cases each baseline method statistically significantly outperformed the other method.
- Results. Using 18 datasets, we conducted 80 feature efficiency evaluations (excluding the RAW baseline) and 160 feature efficiency comparisons, of which 97 (∼60%) were statistically different. The results of the feature extraction experiments are presented in Table 4 and Table 5. They reveal that, although MEF showed best WR, UFDM also performed comparable to other measures: it statistically significantly outperformed them in cases (listed in Table 6), and was outperformed in cases (Table 4).
5. Conclusions
- Results. We proposed and analysed an IPM-based statistical dependence measure, , defined as the norm of the difference between the joint and product-marginal characteristic functions. applies to pairs of random vectors of possibly different dimensions and can be integrated into modern machine learning pipelines. In contrast to global measures (e.g., DCOR, HSIC, MEF), which aggregate information across the entire frequency domain, identifies spectrally localised dependencies by highlighting frequencies where the discrepancy is maximised, thereby offering potentially interpretable insights into the structure of dependence. We theoretically established key properties of , such as invariance under linear transformations and augmentation with independent noise, monotonicity under dimension reduction, and vanishing under independence. We also showed that UFDM’s objective aligns with the vectorial representation of CFs. In addition, we investigated the consistency of the empirical estimator and derived a finite-sample concentration bound. For practical estimation, we proposed a gradient-based estimation algorithm with SVD warm-up, and this warm-up was found to be essential for stable convergence.
- Limitations. Computing UFDM requires maximising a highly nonlinear objective, which makes the estimator sensitive to initialisation and optimisation settings. Although the proposed SVD warm-up substantially improves numerical stability, estimation may still become more challenging as dimensionality d increases or sample size n decreases. From the perspective of the effective , our empirical evaluation covers two different tasks. First, in independence testing with synthetic data and and , UFDM maintained effectiveness across diverse dependence structures. Our preliminary experiments with , and indicate a reduction in power for several dependency patterns, whereas DCOR, HSIC, and MEF remained comparatively stable. Nonetheless, UFDM preserved its performance for sparse geometrically structured dependencies (e.g., Interleaved Moons), where alternative measures often show more pronounced loss of sensitivity. Due to the high computational cost of UFDM permutation tests, we omitted systematic exploration of these regimes, leaving it to future work. On the other hand, in supervised feature extraction on real datasets, we examined substantially broader ranges, including high-dimensional settings such as USPS, Micro-Mass, and Scene. UFDM outperformed one or more baselines on several such datasets (Table 6), suggesting that it may be effective in some larger-dimensional machine learning tasks.
- Future work and potential applications. Identifying the limit distribution of the empirical could enable faster alternatives to permutation-based statistical tests, which would also facilitate the systematic analysis of previously mentioned settings. However, since the empirical is not a U- or V-statistic like HSIC or distance correlation, this would require a non-trivial analysis of the extrema of empirical processes. Possible extensions of UFDM include multivariate generalisations [23] and weighted or normalised variants to enhance empirical stability. From an application perspective, may prove useful in causality, regularisation, representation learning, and other areas of modern machine learning where statistical dependence serves as an optimisation criterion.
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
Appendix A.1. Proofs
Appendix A.2. Proof of Theorem 3
Appendix A.3. Ablation Experiment on SVD Warm-Up
| Distribution of Y | |||
|---|---|---|---|
| Linear (1.0) | 0.002 ± 0.000 | 0.002 ± 0.000 | 0.002 ± 0.000 |
| Linear (0.3) | 0.002 ± 0.000 | 0.002 ± 0.000 | 0.002 ± 0.000 |
| Logarithmic | 0.035 ± 0.097 | 0.192 ± 0.251 | 0.387 ± 0.297 |
| Quadratic | 0.023 ± 0.061 | 0.298 ± 0.291 | 0.285 ± 0.145 |
| Polynomial | 0.002 ± 0.000 | 0.062 ± 0.134 | 0.056 ± 0.078 |
| LRSO (0.05) | 0.002 ± 0.001 | 0.041 ± 0.066 | 0.026 ± 0.040 |
| Heteroscedastic | 0.004 ± 0.006 | 0.002 ± 0.001 | 0.003∗ ± 0.003 |
Appendix A.4. Dependency Patterns
| Type | Formula |
|---|---|
| Structured dependence patterns () | |
| Linear(p) | , |
| Logarithmic | |
| Quadratic | |
| Cubic | |
| LRSO(p) | |
| Heteroscedastic | , |
| Complex dependence patterns | |
| Bimodal | |
| Sparse bimodal | , |
| Sparse circular | |
| Gaussian copula | Marginals . |
| Clayton copula | |
| Interleaved Moons | |
References
- Gretton, A.; Bousquet, O.; Smola, A.; Schölkopf, B. Measuring statistical dependence with Hilbert-Schmidt norms. In Proceedings of the 16th International Conference on Algorithmic Learning Theory (ALT), Singapore, 8–11 October 2005. [Google Scholar][Green Version]
- Daniušis, P.; Vaitkus, P.; Petkevičius, L. Hilbert–Schmidt component analysis. Lith. Math. J. 2016, 57, 7–11. [Google Scholar] [CrossRef]
- Daniušis, P.; Vaitkus, P. Supervised feature extraction using Hilbert-Schmidt norms. In Proceedings of the 10th International Conference on Intelligent Data Engineering and Automated Learning (IDEAL), Burgos, Spain, 23–26 September 2009; Springer: Berlin/Heidelberg, Germany, 2009; pp. 25–33. [Google Scholar]
- Hoyer, P.; Janzing, D.; Mooij, J.M.; Peters, J.; Schölkopf, B. Nonlinear causal discovery with additive noise models. In Proceedings of the Advances in Neural Information Processing Systems 21 (NeurIPS 2008), Vancouver, BC, Canada, 8–11 December 2008. [Google Scholar]
- Li, Y.; Pogodin, R.; Sutherland, D.J.; Gretton, A. Self-Supervised Learning with Kernel Dependence Maximization. In Proceedings of the 35th Conference on Neural Information Processing Systems (NeurIPS 2021), Virtual, 6–14 December 2021. [Google Scholar]
- Ragonesi, R.; Volpi, R.; Cavazza, J.; Murino, V. Learning unbiased representations via mutual information backpropagation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) Workshops, Virtual, 19–25 June 2021; pp. 2723–2732. [Google Scholar]
- Zhen, X.; Meng, Z.; Chakraborty, R.; Singh, V. On the Versatile Uses of Partial Distance Correlation in Deep Learning. In Proceedings of the 17th European Conference on Computer Vision (ECCV), Tel Aviv, Israel, 23–27 October 2022. [Google Scholar]
- Chatterjee, S. A New Coefficient of Correlation. J. Am. Stat. Assoc. 2021, 116, 2009–2022. [Google Scholar] [CrossRef]
- Feuerverger, A. A consistent test for bivariate dependence. Int. Stat. Rev. 1993, 61, 419–433. [Google Scholar] [CrossRef]
- Póczos, B.; Ghahramani, Z.; Schneider, J.G. Copula-based kernel dependency measures. arXiv 2012, arXiv:1206.4682. [Google Scholar] [CrossRef]
- Puccetti, G. Measuring linear correlation between random vectors. Inf. Sci. 2022, 607, 1328–1347. [Google Scholar] [CrossRef]
- Shen, C.; Priebe, C.E.; Vogelstein, J.T. From Distance Correlation to Multiscale Graph Correlation. J. Am. Stat. Assoc. 2020, 115, 280–291. [Google Scholar] [CrossRef]
- Székely, G.J.; Rizzo, M.L.; Bakirov, N.K. Measuring and testing dependence by correlation of distances. Ann. Stat. 2007, 35, 2769–2794. [Google Scholar] [CrossRef]
- Tsur, D.; Goldfeld, Z.; Greenewald, K. Max-Sliced Mutual Information. In Proceedings of the Advances in Neural Information Processing Systems 36 (NeurIPS 2023), New Orleans, LA, USA, 10–16 December 2023; Curran Associates, Inc.: Red Hook, NY, USA, 2023; pp. 80338–80351. [Google Scholar]
- Sriperumbudur, B.K.; Fukumizu, K.; Gretton, A.; Schölkopf, B.; Lanckriet, G.R.G. On the empirical estimation of integral probability metrics. Electron. J. Stat. 2012, 6, 1550–1599. [Google Scholar] [CrossRef]
- Jacod, J.; Protter, P. Probability Essentials, 2nd ed.; Springer: Berlin/Heidelberg, Germany, 2003. [Google Scholar]
- Richter, W.-D. On the vector representation of characteristic functions. Stats 2023, 6, 1072–1081. [Google Scholar] [CrossRef]
- Zhang, W.; Gao, W.; Ng, H.K.T. Multivariate tests of independence based on a new class of measures of independence in Reproducing Kernel Hilbert Space. J. Multivar. Anal. 2023, 195, 105144. [Google Scholar] [CrossRef]
- Cover, T.M.; Thomas, J.A. Elements of Information Theory, 2nd ed.; Wiley-Interscience: Hoboken, NJ, USA, 2006. [Google Scholar]
- Yu, S.; Giraldo, L.G.S.; Jenssen, R.; Príncipe, J.C. Multivariate Extension of Matrix-Based Rényi’s α-Order Entropy Functional. IEEE Trans. Pattern Anal. Mach. Intell. 2020, 42, 2960–2966. [Google Scholar] [CrossRef]
- Yu, S.; Alesiani, F.; Yu, X.; Jenssen, R.; Príncipe, J.C. Measuring Dependence with Matrix-based Entropy Functional. In Proceedings of the 35th AAAI Conference on Artificial Intelligence (AAAI 2021), Virtual, 2–9 February 2021; pp. 10781–10789. [Google Scholar]
- Lopez-Paz, D.; Hennig, P.; Schölkopf, B. The Randomized Dependence Coefficient. In Proceedings of the Advances in Neural Information Processing Systems 26 (NeurIPS 2013), Lake Tahoe, NV, USA, 5–8 December 2013; Curran Associates, Inc.: Red Hook, NY, USA, 2013. [Google Scholar]
- Böttcher, B.; Keller-Ressel, M.; Schilling, R. Distance multivariance: New dependence measures for random vectors. arXiv 2018, arXiv:1711.07775. [Google Scholar] [CrossRef]
- Székely, G.J.; Rizzo, M.L. Partial distance correlation with methods for dissimilarities. Ann. Stat. 2014, 42, 2382–2412. [Google Scholar] [CrossRef]
- Schölkopf, B.; Smola, A.J.; Bach, F. Learning with Kernels: Support Vector Machines, Regularization, Optimization, and Beyond; MIT Press: Cambridge, MA, USA, 2018. [Google Scholar]
- Belghazi, M.I.; Baratin, A.; Rajeshwar, S.; Ozair, S.; Bengio, Y.; Courville, A.; Hjelm, D. Mutual Information Neural Estimation. In Proceedings of the 35th International Conference on Machine Learning (ICML 2018), Stockholm, Sweden, 10–15 July 2018; pp. 531–540. [Google Scholar]
- Sanchez Giraldo, L.G.; Rao, M.; Principe, J.C. Measures of Entropy From Data Using Infinitely Divisible Kernels. IEEE Trans. Inf. Theory 2015, 61, 535–548. [Google Scholar] [CrossRef]
- Ushakov, N.G. Selected Topics in Characteristic Functions; De Gruyter: Berlin, Germany, 2011. [Google Scholar]
- Csörgo, S.; Totik, V. On how long interval is the empirical characteristic function uniformly consistent. Acta Sci. Math. (Szeged) 1983, 45, 141–149. [Google Scholar]
- Garreau, D.; Jitkrittum, W.; Kanagawa, M. Large sample analysis of the median heuristic. arXiv 2017, arXiv:1707.07269. [Google Scholar]
- Phipson, B.; Smyth, G.K. Permutation p-values should never be zero: Calculating exact p-values when permutations are randomly drawn. Stat. Appl. Genet. Mol. Biol. 2010, 9, 39. [Google Scholar] [CrossRef]
- Vanschoren, J.; van Rijn, J.N.; Bischl, B.; Torgo, L. OpenML: Networked Science in Machine Learning. SIGKDD Explor. 2013, 15, 49–60. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhou, Z.H. Multilabel Dimensionality Reduction via Dependence Maximization. ACM Trans. Knowl. Discov. Data 2010, 4, 14:1–14:21. [Google Scholar] [CrossRef]
- McCullagh, P.; Nelder, J.A. Generalized Linear Models, 2nd ed.; Chapman and Hall/CRC Monographs on Statistics and Applied Probability Series, Chapman & Hall; Routledge: Oxfordshire, UK, 1989. [Google Scholar]
- Goldberger, J.; Hinton, G.E.; Roweis, S.; Salakhutdinov, R.R. Neighbourhood components analysis. In Proceedings of the Advances in Neural Information Processing Systems 17 (NeurIPS 2004), Vancouver, BC, Canada, 13–18 December 2004. [Google Scholar]
- Wilcoxon, F. Individual Comparisons by Ranking Methods. Biom. Bull. 1945, 1, 80–83. [Google Scholar] [CrossRef]
- Demšar, J. Statistical Comparisons of Classifiers over Multiple Data Sets. J. Mach. Learn. Res. 2006, 7, 1–30. [Google Scholar]
- Euclidean Norm of Sub-Exponential Random Vector is Sub-Exponential? MathOverflow. Version: 2025-05-06. Available online: https://mathoverflow.net/q/492045 (accessed on 10 April 2025).
- Bochner, S.; Chandrasekharan, K. Fourier Transforms (AM-19); Princeton University Press: Princeton, NJ, USA, 1949. [Google Scholar]
- Vershynin, R. High-Dimensional Probability: An Introduction with Applications in Data Science; Cambridge Series in Statistical and Probabilistic Mathematics; Cambridge University Press: Cambridge, UK, 2018. [Google Scholar] [CrossRef]

| Distribution of Y | UFDM | DCOR | HSIC | MEF | ||
|---|---|---|---|---|---|---|
| Linear (1.0) | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| Linear (0.3) | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| Logarithmic | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| Quadratic | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| Polynomial | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| LRSO (0.05) | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| Heteroscedastic | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| Linear (1.0) | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| Linear (0.3) | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| Logarithmic | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| Quadratic | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| Polynomial | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| LRSO (0.05) | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| Heteroscedastic | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| Linear (1.0) | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| Linear (0.3) | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| Logarithmic | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| Quadratic | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| Polynomial | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| LRSO (0.05) | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| Heteroscedastic | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| Linear (1.0) | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| Linear (0.3) | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| Logarithmic | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| Quadratic | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| Polynomial | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| LRSO (0.05) | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| Heteroscedastic | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| Linear (1.0) | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| Linear (0.3) | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| Logarithmic | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| Quadratic | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| Polynomial | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| LRSO (0.05) | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| Heteroscedastic | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| Linear (1.0) | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| Linear (0.3) | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| Logarithmic | 0.97 [0.92, 0.99] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| Quadratic | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| Polynomial | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| LRSO (0.05) | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| Heteroscedastic | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| Linear (1.0) | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| Linear (0.3) | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| Logarithmic | 0.98 [0.93, 0.99] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| Quadratic | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| Polynomial | 0.96 [0.90, 0.98] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| LRSO (0.05) | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| Heteroscedastic | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| Linear (1.0) | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| Linear (0.3) | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| Logarithmic | 0.98 [0.93, 0.99] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| Quadratic | 0.99 [0.95, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| Polynomial | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 0.99 [0.95, 1.00] | ||
| LRSO (0.05) | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| Heteroscedastic | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| Linear (1.0) | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| Linear (0.3) | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| Logarithmic | 0.97 [0.92, 0.99] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| Quadratic | 0.98 [0.93, 0.99] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| Polynomial | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| LRSO (0.05) | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | ||
| Heteroscedastic | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] |
| Pattern | UFDM | DCOR | HSIC | MEF | |
|---|---|---|---|---|---|
| Mixture Bimodal Marginal | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | |
| Mixture Bimodal | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | |
| Circular | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | |
| Gaussian Copula | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | |
| Clayton Copula | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | |
| Interleaved Moons | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | |
| Mixture Bimodal Marginal | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | |
| Mixture Bimodal | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | |
| Circular | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 0.87 [0.79, 0.92] | |
| Gaussian Copula | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | |
| Clayton Copula | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | |
| Interleaved Moons | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 0.49 [0.39, 0.59] | |
| Mixture Bimodal Marginal | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | |
| Mixture Bimodal | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | |
| Circular | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 0.52 [0.42, 0.62] | |
| Gaussian Copula | 0.98 [0.93, 0.99] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | |
| Clayton Copula | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | 1.00 [0.96, 1.00] | |
| Interleaved Moons | 1.00 [0.96, 1.00] | 0.97 [0.92, 0.99] | 0.96 [0.90, 0.98] | 0.27 [0.19, 0.36] |
| Dependence Pattern | ||||
|---|---|---|---|---|
| Linear (strong) | ||||
| Linear (weak) | ||||
| Logarithmic | ||||
| Quadratic | ||||
| Polynomial | ||||
| Contaminated sine | ||||
| Conditional variance | ||||
| Linear (strong) | ||||
| Linear (weak) | ||||
| Logarithmic | ||||
| Quadratic | ||||
| Polynomial | ||||
| Contaminated sine | ||||
| Conditional variance | ||||
| Linear (strong) | Student’s | |||
| Linear (weak) | ||||
| Logarithmic | ||||
| Quadratic | ||||
| Polynomial | ||||
| Contaminated sine | ||||
| Conditional variance | ||||
| Mixture bimodal marginal | Complex patterns | |||
| Mixture bimodal | ||||
| Circular | ||||
| Gaussian copula | ||||
| Clayton copula | ||||
| Interleaved moons |
| UFDM | DCOR | MEF | HSIC | NCA | |
|---|---|---|---|---|---|
| UFDM | 0 | 6 | 4 | 5 | 5 |
| DCOR | 2 | 0 | 3 | 4 | 3 |
| MEF | 4 | 8 | 0 | 9 | 7 |
| HSIC | 2 | 4 | 2 | 0 | 3 |
| NCA | 5 | 7 | 6 | 8 | 0 |
| Dataset | RAW | UFDM | DCOR | MEF | HSIC | NCA | |
|---|---|---|---|---|---|---|---|
| Australian | (690, 14, 2) | 0.710 | 0.853 | 0.846 | 0.850 | 0.824 | 0.844 |
| Collins | (500, 22, 2) | 0.840 | 0.926 | 0.906 | 0.941 | 0.927 | 0.949 |
| Heart-statlog | (270, 13, 2) | 0.621 | 0.824 | 0.823 | 0.826 | 0.816 | 0.817 |
| Mfeat-factors | (2000, 216, 10) | 0.783 | 0.968 | 0.970 | 0.968 | 0.968 | 0.969 |
| Mfeat-pixel | (2000, 240, 10) | 0.946 | 0.956 | 0.948 | 0.957 | 0.951 | 0.959 |
| Mfeat-zernike | (2000, 47, 10) | 0.741 | 0.812 | 0.810 | 0.814 | 0.811 | 0.804 |
| Micro-mass | (360, 1300, 10) | 0.874 | 0.925 | 0.919 | 0.931 | 0.923 | 0.882 |
| Optdigits | (5620, 64, 10) | 0.949 | 0.964 | 0.961 | 0.960 | 0.957 | 0.963 |
| Parkinsons | (195, 22, 2) | 0.756 | 0.827 | 0.828 | 0.850 | 0.836 | 0.837 |
| Scene | (2407, 299, 2) | 0.886 | 0.987 | 0.988 | 0.953 | 0.988 | 0.962 |
| Segment | (2310, 19, 7) | 0.760 | 0.912 | 0.911 | 0.943 | 0.936 | 0.941 |
| Sonar | (208, 60, 2) | 0.685 | 0.745 | 0.733 | 0.757 | 0.734 | 0.770 |
| Spectf | (349, 44, 2) | 0.729 | 0.737 | 0.739 | 0.738 | 0.739 | 0.750 |
| USPS | (9298, 256, 10) | 0.924 | 0.944 | 0.941 | 0.934 | 0.936 | 0.940 |
| Wdbc | (569, 30, 2) | 0.699 | 0.948 | 0.951 | 0.938 | 0.900 | 0.968 |
| Wine | (178, 13, 3) | 0.552 | 0.945 | 0.917 | 0.954 | 0.947 | 0.936 |
| 20 | 12 | 28 | 11 | 26 | |||
| 13 | 25 | 15 | 26 | 18 |
| Dataset | n | Measures Outperformed | |
|---|---|---|---|
| Australian | 690 | 14 | DCOR, HSIC, NCA |
| Collins | 500 | 22 | DCOR |
| Micro-mass | 360 | 1300 | NCA |
| Mfeat-pixel | 2000 | 240 | DCOR, HSIC |
| Mfeat-zernike | 2000 | 47 | NCA |
| Optdigits | 5620 | 64 | DCOR, MEF, HSIC |
| Scene | 2407 | 299 | MEF, NCA |
| USPS | 9298 | 256 | DCOR, MEF, HSIC, NCA |
| Wdbc | 569 | 30 | MEF, HSIC |
| Wine | 178 | 13 | DCOR |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Daniušis, P.; Juneja, S.; Kuzma, L.; Marcinkevičius, V. Measuring Statistical Dependence via Characteristic Function IPM. Entropy 2025, 27, 1254. https://doi.org/10.3390/e27121254
Daniušis P, Juneja S, Kuzma L, Marcinkevičius V. Measuring Statistical Dependence via Characteristic Function IPM. Entropy. 2025; 27(12):1254. https://doi.org/10.3390/e27121254
Chicago/Turabian StyleDaniušis, Povilas, Shubham Juneja, Lukas Kuzma, and Virginijus Marcinkevičius. 2025. "Measuring Statistical Dependence via Characteristic Function IPM" Entropy 27, no. 12: 1254. https://doi.org/10.3390/e27121254
APA StyleDaniušis, P., Juneja, S., Kuzma, L., & Marcinkevičius, V. (2025). Measuring Statistical Dependence via Characteristic Function IPM. Entropy, 27(12), 1254. https://doi.org/10.3390/e27121254






