Representative Points of the Inverse Gaussian Distribution and Their Applications
Abstract
1. Introduction
- 1.
- We establish, to the best of our knowledge, the first systematic comparative framework for three distinct types of representative points—Monte Carlo (MC-RPs), quasi-Monte Carlo (QMC-RPs), and mean square error RPs (MSE-RPs)—specifically on the Inverse Gaussian distribution. This framework provides a benchmark for evaluating discrete approximation quality in terms of moment estimation, density approximation, and resampling efficiency.
- 2.
- We introduce a novel parameter estimation methodology for the IG distribution by employing the Harrell–Davis (HD) [19] and Sfakianakis–Verginis (SV1, SV2, SV3) [20] quantile estimators. This constitutes the first application and comprehensive demonstration of these estimators for enhancing sample representativeness and significantly improving the accuracy of IG parameter estimation.
- 3.
- Through extensive simulations and real-world case studies, we provide a comprehensive performance analysis. Our results not only conclusively demonstrate the superiority of MSE-RPs in approximation tasks but also validate the practical utility and effectiveness of the proposed quantile-based estimation framework.
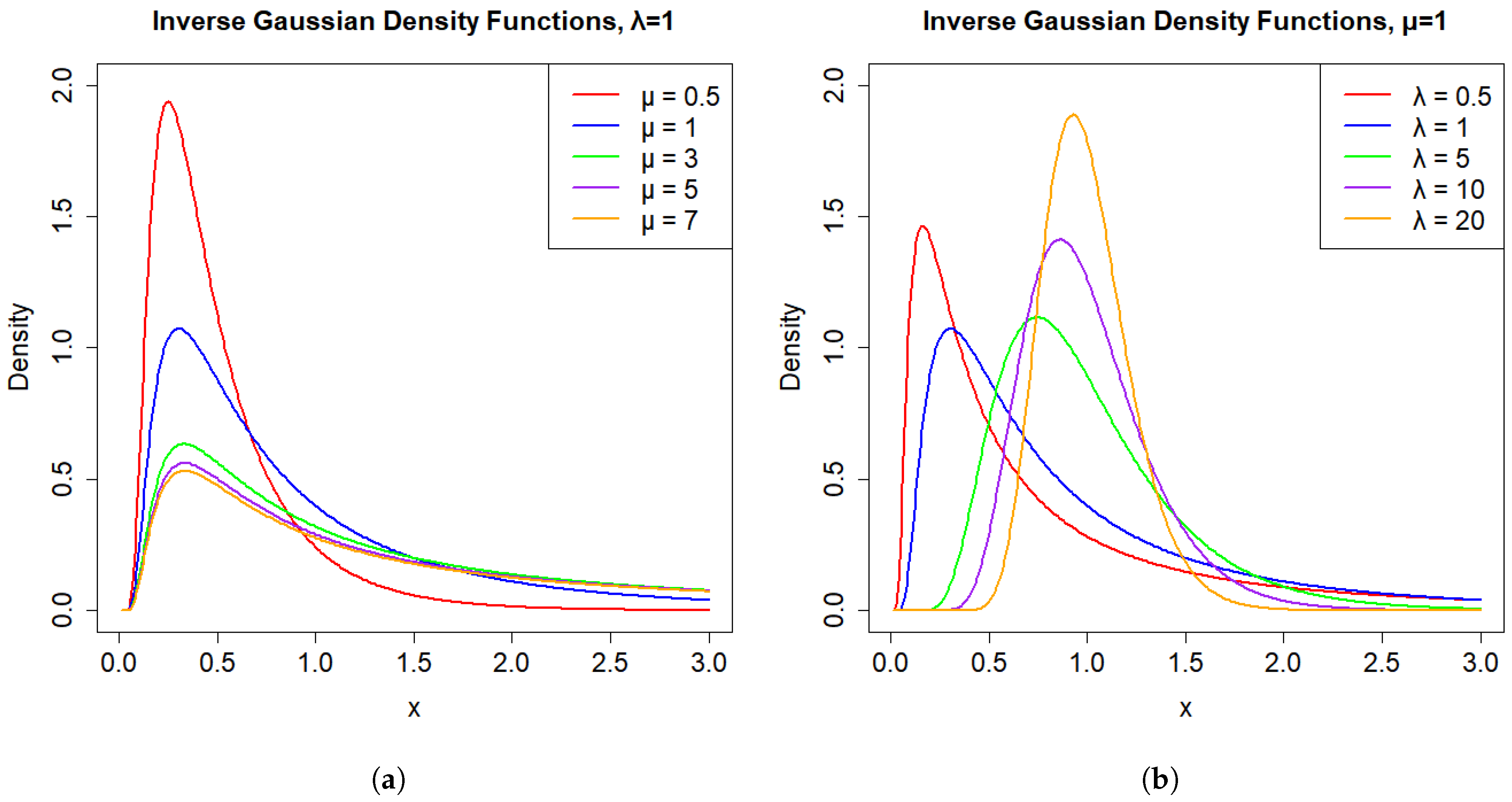
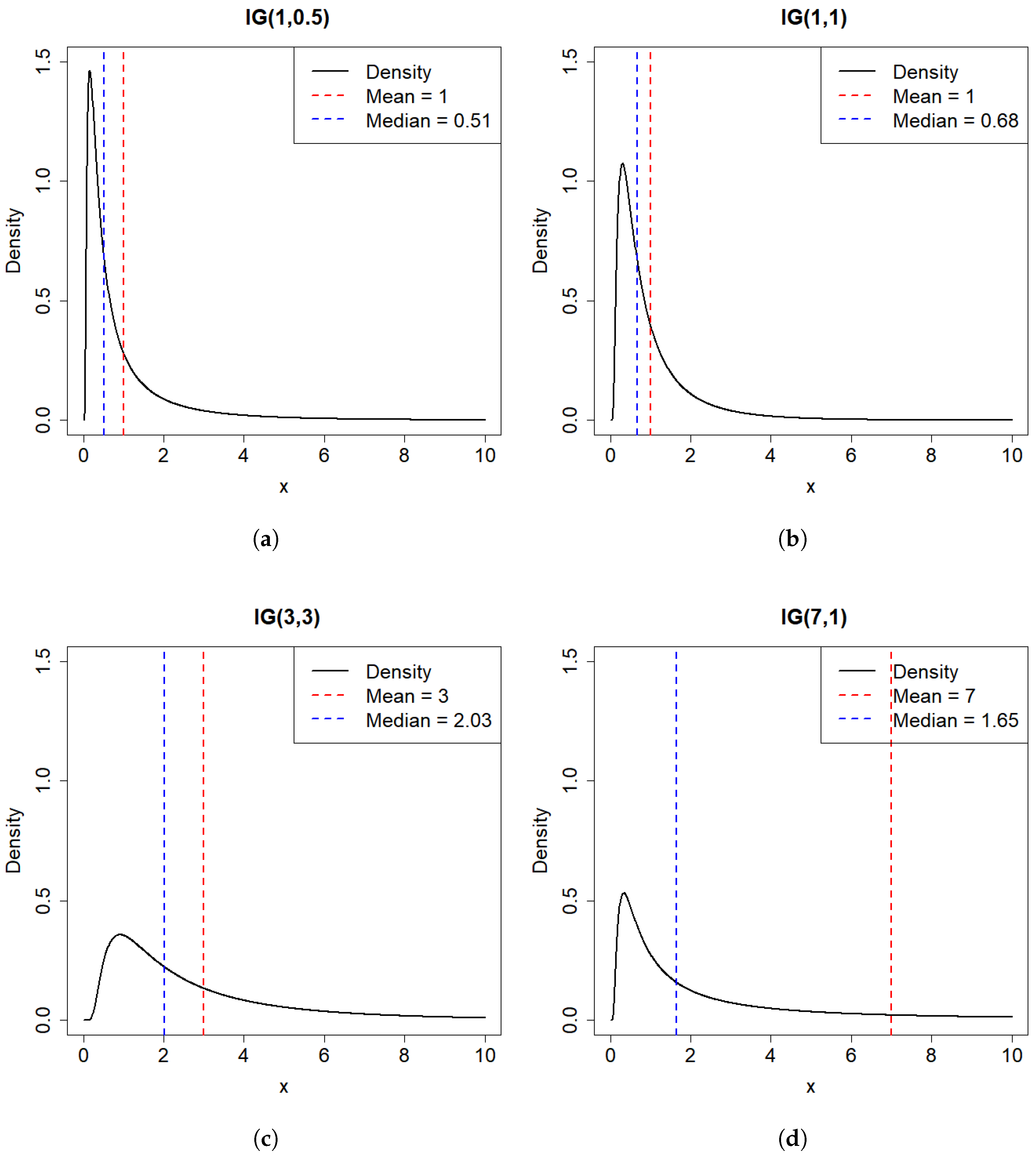
2. Basic Properties of the IG Distribution
3. Representative Points of IG Distribution
3.1. Three Types of Representative Points
3.1.1. MC-RPs
3.1.2. QMC-RPs
3.1.3. MSE-RPs
3.2. Lower Moments Estimation Based on RPs of IG Distribution
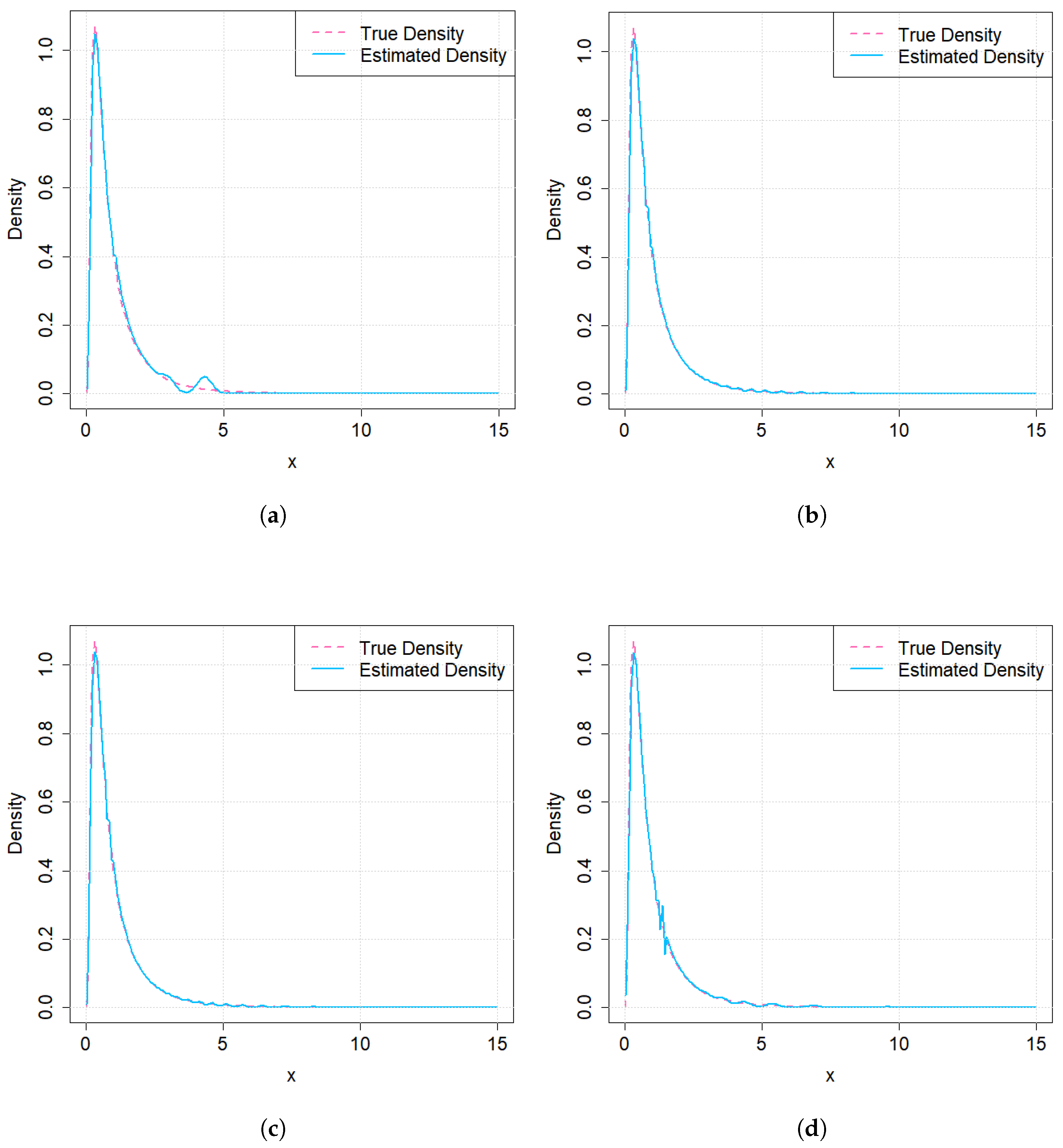
3.3. Density Estimation via RPs of IG Distribution
4. Resampling Based on RPs of IG Distribution
5. MLE via Quantile Estimators of IG Distribution
5.1. Two Nonparametric Quantile Estimators
5.1.1. HD Estimator
5.1.2. SV Estimators
5.2. Estimation Accuracy Measures
5.2.1. MLE of IG
5.2.2. MLE of IG
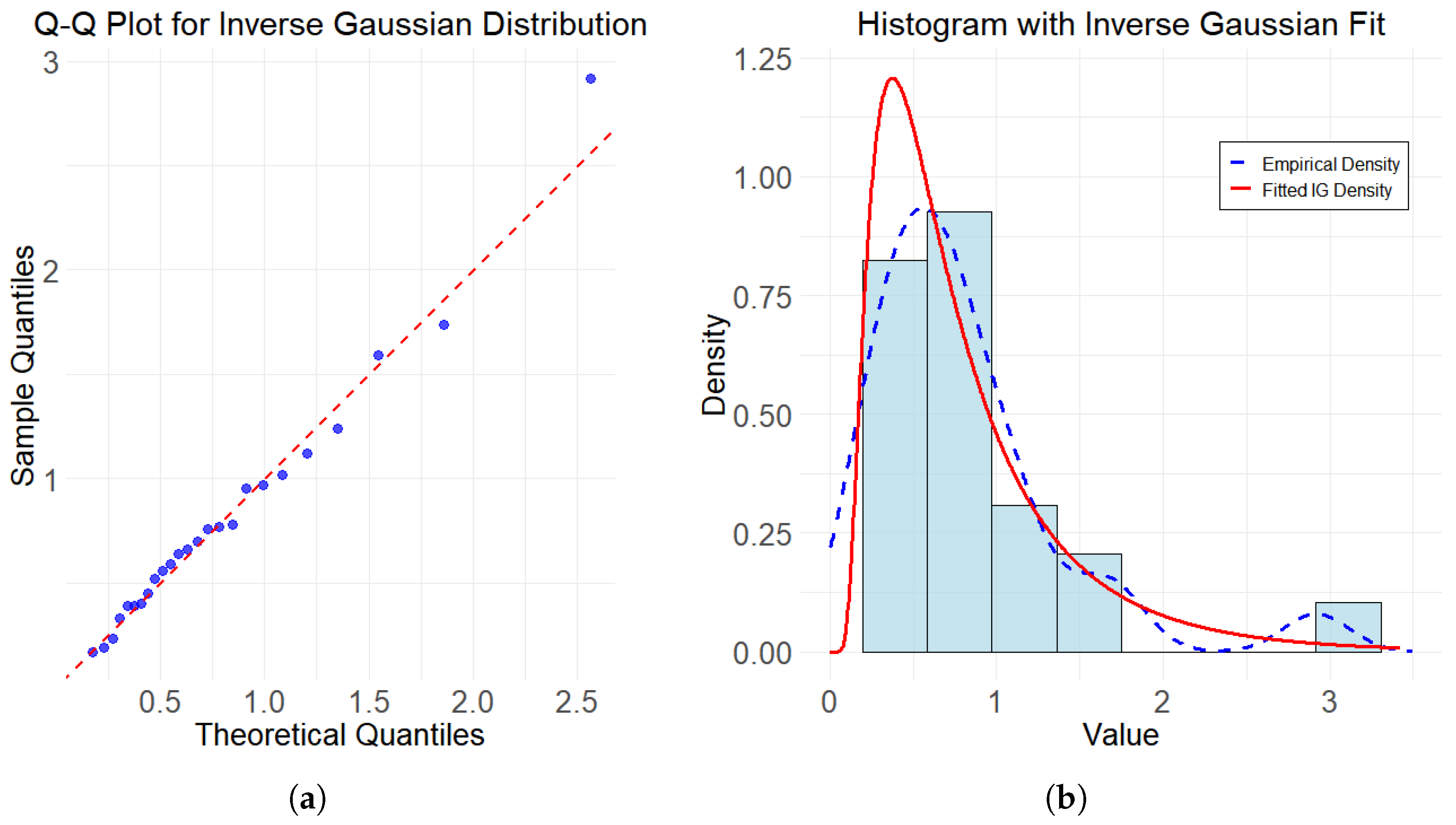
6. Case Study
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| n | Method | L2.cdf | L2.pdf | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Rank1 | Rank2 | Rank3 | Rank4 | Rank5 | Rank1 | Rank2 | Rank3 | Rank4 | Rank5 | ||
| 30 | Plain | 5 | 30 | 62 | 3 | 0 | 12 | 44 | 12 | 31 | 1 |
| 30 | HD | 9 | 39 | 12 | 40 | 0 | 4 | 24 | 67 | 5 | 0 |
| 30 | QSV1 | 13 | 15 | 17 | 22 | 33 | 16 | 12 | 9 | 35 | 28 |
| 30 | QSV2 | 39 | 5 | 1 | 7 | 48 | 26 | 11 | 9 | 24 | 30 |
| 30 | QSV3 | 34 | 11 | 8 | 28 | 19 | 42 | 9 | 3 | 5 | 41 |
| KL | ABI | ||||||||||
| 30 | Plain | 9 | 42 | 36 | 13 | 0 | 15 | 44 | 16 | 25 | 0 |
| 30 | HD | 2 | 36 | 44 | 16 | 2 | 6 | 25 | 38 | 28 | 3 |
| 30 | QSV1 | 18 | 5 | 6 | 29 | 42 | 18 | 5 | 28 | 15 | 34 |
| 30 | QSV2 | 33 | 7 | 5 | 25 | 30 | 23 | 8 | 5 | 23 | 41 |
| 30 | QSV3 | 38 | 10 | 9 | 17 | 26 | 38 | 18 | 13 | 9 | 22 |
| L2.cdf | L2.pdf | ||||||||||
| 50 | Plain | 10 | 30 | 53 | 7 | 0 | 11 | 40 | 12 | 37 | 0 |
| 50 | HD | 13 | 38 | 12 | 36 | 1 | 5 | 25 | 68 | 2 | 0 |
| 50 | QSV1 | 21 | 19 | 20 | 19 | 21 | 15 | 16 | 16 | 41 | 12 |
| 50 | QSV2 | 31 | 1 | 3 | 9 | 56 | 31 | 13 | 3 | 16 | 37 |
| 50 | QSV3 | 25 | 12 | 12 | 29 | 22 | 38 | 6 | 1 | 4 | 51 |
| KL | ABI | ||||||||||
| 50 | Plain | 10 | 44 | 32 | 13 | 1 | 8 | 51 | 17 | 23 | 1 |
| 50 | HD | 9 | 39 | 37 | 14 | 1 | 10 | 28 | 29 | 32 | 1 |
| 50 | QSV1 | 24 | 6 | 12 | 31 | 27 | 26 | 4 | 32 | 17 | 21 |
| 50 | QSV2 | 26 | 5 | 7 | 21 | 41 | 25 | 3 | 4 | 17 | 51 |
| 50 | QSV3 | 31 | 6 | 12 | 21 | 30 | 31 | 14 | 18 | 11 | 26 |
| L2.cdf | L2.pdf | ||||||||||
| 100 | Plain | 2 | 21 | 73 | 4 | 0 | 2 | 32 | 32 | 34 | 0 |
| 100 | HD | 10 | 42 | 4 | 44 | 0 | 3 | 38 | 48 | 11 | 0 |
| 100 | QSV1 | 25 | 14 | 13 | 15 | 33 | 14 | 20 | 17 | 32 | 17 |
| 100 | QSV2 | 38 | 5 | 1 | 6 | 50 | 35 | 7 | 1 | 19 | 38 |
| 100 | QSV3 | 25 | 18 | 9 | 31 | 17 | 46 | 3 | 2 | 4 | 45 |
| KL | ABI | ||||||||||
| 100 | Plain | 1 | 31 | 47 | 21 | 0 | 5 | 37 | 24 | 34 | 0 |
| 100 | HD | 7 | 43 | 26 | 24 | 0 | 12 | 34 | 19 | 35 | 0 |
| 100 | QSV1 | 24 | 13 | 7 | 25 | 31 | 18 | 15 | 33 | 11 | 23 |
| 100 | QSV2 | 36 | 5 | 4 | 11 | 44 | 29 | 7 | 7 | 5 | 52 |
| 100 | QSV3 | 32 | 8 | 16 | 19 | 25 | 36 | 7 | 17 | 15 | 25 |
| n | Method | L2.cdf | L2.pdf | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Rank1 | Rank2 | Rank3 | Rank4 | Rank5 | Rank1 | Rank2 | Rank3 | Rank4 | Rank5 | ||
| 30 | Plain | 18 | 33 | 8 | 37 | 4 | 19 | 33 | 6 | 36 | 6 |
| 30 | HD | 8 | 18 | 62 | 12 | 0 | 5 | 21 | 65 | 6 | 3 |
| 30 | QSV1 | 13 | 29 | 17 | 29 | 12 | 15 | 29 | 12 | 30 | 14 |
| 30 | QSV2 | 29 | 6 | 10 | 15 | 40 | 29 | 6 | 12 | 19 | 34 |
| 30 | QSV3 | 42 | 10 | 6 | 5 | 37 | 41 | 11 | 5 | 5 | 38 |
| KL | ABI | ||||||||||
| 30 | Plain | 18 | 34 | 6 | 36 | 6 | 18 | 32 | 9 | 37 | 4 |
| 30 | HD | 8 | 18 | 64 | 6 | 4 | 9 | 17 | 60 | 14 | 0 |
| 30 | QSV1 | 13 | 31 | 10 | 29 | 17 | 14 | 31 | 10 | 27 | 18 |
| 30 | QSV2 | 30 | 7 | 14 | 19 | 30 | 28 | 9 | 17 | 13 | 33 |
| 30 | QSV3 | 39 | 12 | 4 | 8 | 37 | 39 | 10 | 6 | 7 | 38 |
| L2.cdf | L2.pdf | ||||||||||
| 50 | Plain | 13 | 36 | 5 | 44 | 2 | 13 | 37 | 3 | 45 | 2 |
| 50 | HD | 5 | 18 | 66 | 11 | 0 | 3 | 17 | 75 | 5 | 0 |
| 50 | QSV1 | 13 | 27 | 21 | 22 | 17 | 12 | 32 | 9 | 30 | 17 |
| 50 | QSV2 | 37 | 10 | 5 | 20 | 28 | 38 | 8 | 6 | 19 | 29 |
| 50 | QSV3 | 32 | 10 | 4 | 3 | 51 | 34 | 9 | 5 | 1 | 51 |
| KL | ABI | ||||||||||
| 50 | Plain | 14 | 34 | 4 | 45 | 3 | 13 | 35 | 9 | 40 | 3 |
| 50 | HD | 4 | 19 | 74 | 3 | 0 | 7 | 20 | 63 | 9 | 1 |
| 50 | QSV1 | 13 | 29 | 10 | 29 | 19 | 16 | 22 | 16 | 27 | 19 |
| 50 | QSV2 | 36 | 12 | 5 | 20 | 27 | 32 | 14 | 8 | 16 | 30 |
| 50 | QSV3 | 33 | 8 | 6 | 3 | 50 | 32 | 9 | 7 | 6 | 46 |
| L2.cdf | L2.pdf | ||||||||||
| 100 | Plain | 2 | 37 | 19 | 42 | 0 | 4 | 42 | 11 | 43 | 0 |
| 100 | HD | 9 | 17 | 59 | 14 | 1 | 4 | 12 | 76 | 8 | 0 |
| 100 | QSV1 | 13 | 30 | 10 | 29 | 18 | 22 | 28 | 4 | 31 | 15 |
| 100 | QSV2 | 35 | 9 | 9 | 15 | 32 | 30 | 15 | 6 | 14 | 35 |
| 100 | QSV3 | 41 | 8 | 2 | 1 | 48 | 40 | 4 | 2 | 5 | 49 |
| KL | ABI | ||||||||||
| 100 | Plain | 3 | 42 | 18 | 37 | 0 | 3 | 35 | 26 | 36 | 0 |
| 100 | HD | 3 | 20 | 68 | 9 | 0 | 7 | 25 | 52 | 12 | 4 |
| 100 | QSV1 | 21 | 29 | 2 | 27 | 21 | 23 | 22 | 7 | 25 | 23 |
| 100 | QSV2 | 34 | 7 | 7 | 24 | 28 | 31 | 12 | 7 | 23 | 27 |
| 100 | QSV3 | 39 | 3 | 4 | 4 | 50 | 36 | 7 | 7 | 5 | 45 |
| n | Method | L2.cdf | L2.pdf | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Rank1 | Rank2 | Rank3 | Rank4 | Rank5 | Rank1 | Rank2 | Rank3 | Rank4 | Rank5 | ||
| 30 | Plain | 16 | 19 | 54 | 11 | 0 | 14 | 25 | 38 | 22 | 1 |
| 30 | HD | 13 | 27 | 21 | 35 | 4 | 12 | 29 | 31 | 22 | 6 |
| 30 | QSV1 | 30 | 20 | 10 | 15 | 25 | 32 | 19 | 10 | 18 | 21 |
| 30 | QSV2 | 30 | 7 | 4 | 12 | 47 | 22 | 19 | 12 | 12 | 35 |
| 30 | QSV3 | 15 | 35 | 15 | 26 | 9 | 20 | 22 | 11 | 23 | 24 |
| KL | ABI | ||||||||||
| 30 | Plain | 13 | 25 | 47 | 14 | 1 | 13 | 27 | 41 | 18 | 1 |
| 30 | HD | 14 | 29 | 21 | 30 | 6 | 13 | 26 | 29 | 24 | 8 |
| 30 | QSV1 | 31 | 16 | 15 | 10 | 28 | 34 | 16 | 10 | 13 | 27 |
| 30 | QSV2 | 26 | 9 | 8 | 19 | 38 | 22 | 11 | 9 | 19 | 39 |
| 30 | QSV3 | 20 | 30 | 10 | 29 | 11 | 21 | 33 | 11 | 23 | 12 |
| L2.cdf | L2.pdf | ||||||||||
| 50 | Plain | 6 | 13 | 72 | 9 | 0 | 11 | 19 | 43 | 27 | 0 |
| 50 | HD | 18 | 34 | 10 | 36 | 2 | 10 | 35 | 34 | 19 | 2 |
| 50 | QSV1 | 32 | 16 | 9 | 13 | 30 | 27 | 18 | 14 | 23 | 18 |
| 50 | QSV2 | 35 | 9 | 4 | 5 | 47 | 36 | 15 | 6 | 11 | 32 |
| 50 | QSV3 | 13 | 27 | 9 | 35 | 16 | 20 | 12 | 7 | 18 | 43 |
| KL | ABI | ||||||||||
| 50 | Plain | 7 | 28 | 53 | 12 | 0 | 9 | 25 | 40 | 25 | 1 |
| 50 | HD | 11 | 40 | 17 | 29 | 3 | 13 | 25 | 29 | 31 | 2 |
| 50 | QSV1 | 29 | 12 | 10 | 18 | 31 | 31 | 16 | 11 | 13 | 29 |
| 50 | QSV2 | 37 | 7 | 8 | 11 | 37 | 30 | 13 | 11 | 10 | 36 |
| 50 | QSV3 | 20 | 12 | 14 | 30 | 24 | 22 | 19 | 12 | 19 | 28 |
| L2.cdf | L2.pdf | ||||||||||
| 100 | Plain | 3 | 18 | 75 | 3 | 1 | 3 | 31 | 49 | 16 | 1 |
| 100 | HD | 9 | 40 | 7 | 43 | 1 | 5 | 37 | 28 | 29 | 1 |
| 100 | QSV1 | 33 | 11 | 6 | 15 | 35 | 23 | 21 | 8 | 25 | 23 |
| 100 | QSV2 | 40 | 2 | 3 | 4 | 51 | 36 | 4 | 5 | 10 | 45 |
| 100 | QSV3 | 15 | 31 | 8 | 34 | 12 | 33 | 9 | 9 | 19 | 30 |
| KL | ABI | ||||||||||
| 100 | Plain | 2 | 29 | 52 | 17 | 0 | 8 | 36 | 28 | 28 | 0 |
| 100 | HD | 7 | 39 | 20 | 34 | 0 | 7 | 23 | 40 | 30 | 0 |
| 100 | QSV1 | 34 | 10 | 2 | 13 | 41 | 31 | 21 | 7 | 9 | 32 |
| 100 | QSV2 | 33 | 6 | 7 | 13 | 41 | 30 | 7 | 10 | 13 | 40 |
| 100 | QSV3 | 24 | 18 | 18 | 22 | 18 | 24 | 16 | 14 | 19 | 27 |
| n | Method | L2.pdf | L2.cdf | KL | ABI | ||
|---|---|---|---|---|---|---|---|
| 30 | Plain | 0.07335 | 0.02055 | 0.00462 | 0.07528 | 0.99117 | 0.57086 |
| 30 | HD | 0.09692 | 0.03336 | 0.00737 | 0.10874 | 1.04214 | 0.58768 |
| 30 | QSV1 | 0.12600 | 0.03741 | 0.01631 | 0.15858 | 0.95335 | 0.63526 |
| 30 | QSV2 | 0.13636 | 0.06959 | 0.01630 | 0.18828 | 1.15247 | 0.61204 |
| 30 | QSV3 | 0.02076 | 0.01404 | 0.00086 | 0.04045 | 0.96127 | 0.52109 |
| 30 | Analytic formulas | 0.06303 | 0.01826 | 0.00368 | 0.07274 | 0.97843 | 0.56195 |
| 50 | Plain | 0.03534 | 0.00983 | 0.00105 | 0.03612 | 0.99346 | 0.53285 |
| 50 | HD | 0.04659 | 0.01997 | 0.00164 | 0.05583 | 1.03875 | 0.53646 |
| 50 | QSV1 | 0.06421 | 0.02064 | 0.00429 | 0.08323 | 0.96340 | 0.56493 |
| 50 | QSV2 | 0.07791 | 0.05360 | 0.00632 | 0.11825 | 1.13863 | 0.54893 |
| 50 | QSV3 | 0.01822 | 0.01106 | 0.00030 | 0.02424 | 0.97320 | 0.48916 |
| 50 | Analytic formulas | 0.05150 | 0.01489 | 0.00242 | 0.05874 | 0.98228 | 0.54989 |
| 100 | Plain | 0.01899 | 0.00528 | 0.00030 | 0.01933 | 0.99604 | 0.51735 |
| 100 | HD | 0.02247 | 0.01172 | 0.00041 | 0.02889 | 1.02662 | 0.51558 |
| 100 | QSV1 | 0.03241 | 0.01169 | 0.00116 | 0.04467 | 0.97479 | 0.53207 |
| 100 | QSV2 | 0.04274 | 0.03316 | 0.00220 | 0.06649 | 1.08829 | 0.52234 |
| 100 | QSV3 | 0.00897 | 0.00572 | 0.00008 | 0.01226 | 0.98588 | 0.49481 |
| 100 | Analytic formulas | 0.01556 | 0.00627 | 0.00029 | 0.02278 | 0.98500 | 0.51527 |
| n | Method | L2.pdf | L2.cdf | KL | ABI | ||
|---|---|---|---|---|---|---|---|
| 30 | Plain | 0.02825 | 0.03129 | 0.00166 | 0.04631 | 6.93597 | 1.08347 |
| 30 | HD | 0.04269 | 0.04957 | 0.00380 | 0.06611 | 7.02712 | 1.12835 |
| 30 | QSV1 | 0.05288 | 0.06005 | 0.00610 | 0.08786 | 6.92041 | 1.16435 |
| 30 | QSV2 | 0.05545 | 0.06837 | 0.00651 | 0.09580 | 7.15371 | 1.16965 |
| 30 | QSV3 | 0.00079 | 0.00834 | 0.00002 | 0.00855 | 6.89497 | 1.00209 |
| 30 | Analytic formulas | 0.04011 | 0.04516 | 0.00359 | 0.07640 | 6.79364 | 1.12331 |
| 50 | Plain | 0.01586 | 0.01745 | 0.00052 | 0.02738 | 6.93861 | 1.04598 |
| 50 | HD | 0.02339 | 0.02638 | 0.00110 | 0.03413 | 6.99633 | 1.06773 |
| 50 | QSV1 | 0.03187 | 0.03539 | 0.00213 | 0.05279 | 6.92572 | 1.09496 |
| 50 | QSV2 | 0.03388 | 0.04260 | 0.00230 | 0.05917 | 7.14069 | 1.09824 |
| 50 | QSV3 | 0.01103 | 0.01484 | 0.00022 | 0.02016 | 6.91901 | 0.97126 |
| 50 | Analytic formulas | 0.03340 | 0.03711 | 0.00235 | 0.05578 | 6.91821 | 1.09988 |
| 100 | Plain | 0.00949 | 0.01163 | 0.00020 | 0.02062 | 6.90626 | 1.02784 |
| 100 | HD | 0.01157 | 0.01287 | 0.00026 | 0.01685 | 6.99344 | 1.03277 |
| 100 | QSV1 | 0.01740 | 0.01949 | 0.00065 | 0.03298 | 6.89592 | 1.05109 |
| 100 | QSV2 | 0.01859 | 0.02741 | 0.00067 | 0.03783 | 7.17362 | 1.05086 |
| 100 | QSV3 | 0.00579 | 0.01180 | 0.00007 | 0.01496 | 6.89049 | 0.98572 |
| 100 | Analytic formulas | 0.01116 | 0.01402 | 0.00235 | 0.01919 | 7.05329 | 1.03077 |
| category | RP1 | RP2 | RP3 | RP4 | RP5 | RP6 | RP7 | RP8 | |
| QMC-RPs | 0.237625 | 0.429742 | 0.675841 | 1.085120 | 2.143034 | ||||
| FH-RPs | 0.412524 | 1.077368 | 2.061354 | 3.616510 | 6.523829 | ||||
| PKM-RPs | 0.412523 | 1.077366 | 2.061348 | 3.616501 | 6.523817 | ||||
| NTLBG-RPs | 0.410067 | 1.066664 | 2.035673 | 3.564004 | 6.421223 | ||||
| QMC-RPs | 0.184113 | 0.285325 | 0.379723 | 0.483171 | 0.604832 | 0.756110 | 0.956109 | 1.244060 | |
| FH-RPs | 0.282780 | 0.583336 | 0.940531 | 1.377229 | 1.918490 | 2.601214 | 3.486682 | 4.689563 | |
| PKM-RPs | 0.282778 | 0.583331 | 0.940523 | 1.377216 | 1.918472 | 2.601190 | 3.486653 | 4.689529 | |
| NTLBG-RPs | 0.261260 | 0.518738 | 0.820360 | 1.189870 | 1.653057 | 2.248230 | 3.034748 | 4.122999 | |
| QMC-RPs | 0.162420 | 0.237625 | 0.300876 | 0.363621 | 0.429742 | 0.501927 | 0.582866 | 0.675841 | |
| FH-RPs | 0.230289 | 0.428358 | 0.643997 | 0.888024 | 1.167374 | 1.489236 | 1.862458 | 2.298770 | |
| PKM-RPs | 0.230287 | 0.428352 | 0.643986 | 0.888008 | 1.167351 | 1.489207 | 1.862421 | 2.298726 | |
| NTLBG-RPs | 0.177701 | 0.297268 | 0.420456 | 0.558135 | 0.718321 | 0.909491 | 1.142247 | 1.429964 | |
| QMC-RPs | 0.149804 | 0.212175 | 0.261786 | 0.308643 | 0.355654 | 0.404373 | 0.455963 | 0.511506 | |
| FH-RPs | 0.200694 | 0.350971 | 0.506274 | 0.674670 | 0.859939 | 1.065068 | 1.293080 | 1.547407 | |
| PKM-RPs | 0.200685 | 0.350957 | 0.506254 | 0.674643 | 0.859904 | 1.065025 | 1.293027 | 1.547344 | |
| NTLBG-RPs | 0.137958 | 0.209947 | 0.277620 | 0.347437 | 0.422782 | 0.506585 | 0.601923 | 0.712768 | |
| QMC-RPs | 0.141253 | 0.195791 | 0.237625 | 0.275954 | 0.313307 | 0.350897 | 0.389506 | 0.429742 | |
| FH-RPs | 0.181246 | 0.303859 | 0.426099 | 0.554903 | 0.693050 | 0.842349 | 1.004352 | 1.180611 | |
| PKM-RPs | 0.181242 | 0.303849 | 0.426083 | 0.554879 | 0.693018 | 0.842308 | 1.004300 | 1.180549 | |
| NTLBG-RPs | 0.120763 | 0.175902 | 0.224958 | 0.272780 | 0.321183 | 0.371487 | 0.424970 | 0.482953 | |
| QMC-RPs | 0.137262 | 0.188371 | 0.226930 | 0.261786 | 0.295327 | 0.328667 | 0.362480 | 0.397263 | |
| FH-RPs | 0.172376 | 0.283337 | 0.392099 | 0.505183 | 0.625066 | 0.753237 | 0.890873 | 1.039072 | |
| PKM-RPs | 0.172371 | 0.283326 | 0.392081 | 0.505156 | 0.625031 | 0.753192 | 0.890818 | 1.039005 | |
| NTLBG-RPs | 0.115454 | 0.165381 | 0.208911 | 0.250482 | 0.291565 | 0.333181 | 0.376107 | 0.421139 | |
| QMC-RPs | 0.134937 | 0.184113 | 0.220862 | 0.253827 | 0.285325 | 0.316417 | 0.347735 | 0.379723 | |
| FH-RPs | 0.167278 | 0.271810 | 0.373259 | 0.477921 | 0.588133 | 0.705240 | 0.830258 | 0.964093 | |
| PKM-RPs | 0.167272 | 0.271798 | 0.373239 | 0.477893 | 0.588096 | 0.705193 | 0.830200 | 0.964023 | |
| NTLBG-RPs | 0.112544 | 0.160032 | 0.200761 | 0.239180 | 0.277045 | 0.314999 | 0.353415 | 0.393126 | |
| category | RP9 | RP10 | RP11 | RP12 | RP13 | RP14 | RP15 | RP16 | |
| QMC-RPs | 1.725360 | 2.922076 | |||||||
| FH-RPs | 6.466201 | 9.623196 | |||||||
| PKM-RPs | 6.466161 | 9.623159 | |||||||
| NTLBG-RPs | 5.750920 | 8.653409 | |||||||
| QMC-RPs | 0.785355 | 0.918136 | 1.085120 | 1.305995 | 1.621860 | 2.143034 | 3.411364 | ||
| FH-RPs | 2.814540 | 3.433731 | 4.193535 | 5.156277 | 6.438620 | 8.301048 | 11.562225 | ||
| PKM-RPs | 2.814488 | 3.433673 | 4.193470 | 5.156207 | 6.438548 | 8.300979 | 11.562190 | ||
| NTLBG-RPs | 1.790194 | 2.248018 | 2.841775 | 3.632491 | 4.711291 | 6.288362 | 9.129651 | ||
| QMC-RPs | 0.572167 | 0.639315 | 0.714672 | 0.800508 | 0.899950 | 1.017518 | 1.160114 | 1.339036 | |
| FH-RPs | 1.832177 | 2.152538 | 2.515086 | 2.928504 | 3.404570 | 3.959822 | 4.618516 | 5.418287 | |
| PKM-RPs | 1.832103 | 2.152453 | 2.514989 | 2.928395 | 3.404449 | 3.959688 | 4.618370 | 5.418128 | |
| NTLBG-RPs | 0.844104 | 1.001872 | 1.193483 | 1.429018 | 1.721482 | 2.089041 | 2.555589 | 3.153091 | |
| QMC-RPs | 0.472161 | 0.517322 | 0.565834 | 0.618395 | 0.675841 | 0.739201 | 0.809779 | 0.889279 | |
| FH-RPs | 1.372816 | 1.582888 | 1.813084 | 2.066105 | 2.345241 | 2.654576 | 2.999263 | 3.385945 | |
| PKM-RPs | 1.372742 | 1.582803 | 1.812987 | 2.065994 | 2.345118 | 2.654439 | 2.999113 | 3.385782 | |
| NTLBG-RPs | 0.547016 | 0.618788 | 0.700777 | 0.795894 | 0.907606 | 1.040801 | 1.201331 | 1.396686 | |
| QMC-RPs | 0.433433 | 0.471381 | 0.511506 | 0.554238 | 0.600059 | 0.649532 | 0.703326 | 0.762262 | |
| FH-RPs | 1.198965 | 1.371784 | 1.558920 | 1.761974 | 1.982824 | 2.223707 | 2.487319 | 2.776958 | |
| PKM-RPs | 1.198887 | 1.371694 | 1.558816 | 1.761856 | 1.982693 | 2.223561 | 2.487158 | 2.776784 | |
| NTLBG-RPs | 0.469147 | 0.521023 | 0.577773 | 0.640895 | 0.712346 | 0.794621 | 0.890395 | 1.003197 | |
| QMC-RPs | 0.412743 | 0.447120 | 0.483171 | 0.521227 | 0.561647 | 0.604832 | 0.651251 | 0.701455 | |
| FH-RPs | 1.107649 | 1.261883 | 1.427848 | 1.606732 | 1.799900 | 2.008942 | 2.235732 | 2.482506 | |
| PKM-RPs | 1.107567 | 1.261788 | 1.427739 | 1.606609 | 1.799763 | 2.008790 | 2.235565 | 2.482323 | |
| NTLBG-RPs | 0.434701 | 0.478539 | 0.525587 | 0.576829 | 0.633202 | 0.696344 | 0.768272 | 0.850992 | |
| category | RP17 | RP18 | RP19 | RP20 | RP21 | RP22 | RP23 | RP24 | |
| QMC-RPs | 1.574661 | 1.909444 | 2.456955 | 3.771838 | |||||
| FH-RPs | 6.422164 | 7.748047 | 9.658992 | 12.981382 | |||||
| PKM-RPs | 6.421995 | 7.747864 | 9.658797 | 12.981110 | |||||
| NTLBG-RPs | 3.934887 | 4.993269 | 6.539831 | 9.328942 | |||||
| QMC-RPs | 0.979995 | 1.085120 | 1.209288 | 1.359587 | 1.547621 | 1.794286 | 2.143034 | 2.709786 | |
| FH-RPs | 3.823382 | 4.323461 | 4.902871 | 5.586080 | 6.411034 | 7.441252 | 8.795492 | 10.738640 | |
| PKM-RPs | 3.823206 | 4.323273 | 4.902671 | 5.585868 | 6.410811 | 7.441019 | 8.795250 | 10.738390 | |
| NTLBG-RPs | 1.636442 | 1.932897 | 2.303347 | 2.772290 | 3.376042 | 4.167427 | 5.223196 | 6.759912 | |
| QMC-RPs | 0.827366 | 0.899950 | 0.981735 | 1.075041 | 1.183105 | 1.310626 | 1.464787 | 1.657343 | |
| FH-RPs | 3.096724 | 3.451793 | 3.848838 | 4.296661 | 4.807203 | 5.397224 | 6.091291 | 6.927496 | |
| PKM-RPs | 3.096535 | 3.451589 | 3.848621 | 4.296431 | 4.806960 | 5.396969 | 6.091024 | 6.927220 | |
| NTLBG-RPs | 1.138059 | 1.300997 | 1.498690 | 1.740975 | 2.041688 | 2.416649 | 2.887119 | 3.486758 | |
| QMC-RPs | 0.756110 | 0.816036 | 0.882262 | 0.956109 | 1.039311 | 1.134207 | 1.244060 | 1.373608 | |
| FH-RPs | 2.751966 | 3.047418 | 3.372972 | 3.733821 | 4.136655 | 4.590302 | 5.106733 | 5.702749 | |
| PKM-RPs | 2.751767 | 3.047204 | 3.372743 | 3.733576 | 4.136396 | 4.590028 | 5.106445 | 5.702449 | |
| NTLBG-RPs | 0.947489 | 1.061775 | 1.198136 | 1.362230 | 1.561387 | 1.805856 | 2.107466 | 2.482508 | |
| category | RP25 | RP26 | RP27 | RP28 | RP29 | RP30 | RP31 | RP32 | |
| QMC-RPs | 4.058467 | ||||||||
| FH-RPs | 14.102428 | ||||||||
| PKM-RPs | 14.102179 | ||||||||
| NTLBG-RPs | 9.558364 | ||||||||
| QMC-RPs | 1.909444 | 2.265027 | 2.841156 | 4.206263 | |||||
| FH-RPs | 7.969590 | 9.336769 | 11.294769 | 14.677905 | |||||
| PKM-RPs | 7.969308 | 9.336459 | 11.294434 | 14.677578 | |||||
| NTLBG-RPs | 4.267639 | 5.324128 | 6.847529 | 9.558364 | |||||
| QMC-RPs | 1.530093 | 1.725360 | 1.980711 | 2.340368 | 2.922076 | 4.296947 | |||
| FH-RPs | 6.402978 | 7.245596 | 8.294494 | 9.669086 | 11.635645 | 15.030035 | |||
| PKM-RPs | 6.402665 | 7.245272 | 8.294160 | 9.668744 | 11.635300 | 15.029713 | |||
| NTLBG-RPs | 2.954570 | 3.555009 | 4.328302 | 5.355569 | 6.847529 | 9.558364 |
| category | P1 | P2 | P3 | P4 | P5 | P6 | P7 | P8 | |
| QMC-RPs | 0.200000 | 0.200000 | 0.200000 | 0.200000 | 0.200000 | ||||
| FH-RPs | 0.543429 | 0.280605 | 0.122293 | 0.044281 | 0.009393 | ||||
| PKM-RPs | 0.543427 | 0.280605 | 0.122294 | 0.044281 | 0.009393 | ||||
| NTLBG-RPs | 0.539400 | 0.281200 | 0.123800 | 0.045600 | 0.010000 | ||||
| QMC-RPs | 0.100000 | 0.100000 | 0.100000 | 0.100000 | 0.100000 | 0.100000 | 0.100000 | 0.100000 | |
| FH-RPs | 0.303210 | 0.250172 | 0.171229 | 0.113110 | 0.072568 | 0.044597 | 0.025567 | 0.013023 | |
| PKM-RPs | 0.303207 | 0.250171 | 0.171229 | 0.113111 | 0.072568 | 0.044598 | 0.025567 | 0.013024 | |
| NTLBG-RPs | 0.259600 | 0.235000 | 0.174400 | 0.124400 | 0.085800 | 0.056800 | 0.034600 | 0.019000 | |
| QMC-RPs | 0.066667 | 0.066667 | 0.066667 | 0.066667 | 0.066667 | 0.066667 | 0.066667 | 0.066667 | |
| FH-RPs | 0.197129 | 0.198779 | 0.159822 | 0.123196 | 0.093408 | 0.069955 | 0.051655 | 0.037432 | |
| PKM-RPs | 0.197125 | 0.198777 | 0.159821 | 0.123197 | 0.093409 | 0.069956 | 0.051656 | 0.037433 | |
| NTLBG-RPs | 0.099000 | 0.127800 | 0.126600 | 0.118600 | 0.108000 | 0.096200 | 0.083600 | 0.070200 | |
| QMC-RPs | 0.050000 | 0.050000 | 0.050000 | 0.050000 | 0.050000 | 0.050000 | 0.050000 | 0.050000 | |
| FH-RPs | 0.139870 | 0.159043 | 0.140258 | 0.117302 | 0.096240 | 0.078195 | 0.063079 | 0.050521 | |
| PKM-RPs | 0.139858 | 0.159039 | 0.140256 | 0.117302 | 0.096241 | 0.078196 | 0.063081 | 0.050523 | |
| NTLBG-RPs | 0.041600 | 0.064000 | 0.072600 | 0.076000 | 0.077400 | 0.077400 | 0.076600 | 0.075200 | |
| QMC-RPs | 0.040000 | 0.040000 | 0.040000 | 0.040000 | 0.040000 | 0.040000 | 0.040000 | 0.040000 | |
| FH-RPs | 0.105020 | 0.129728 | 0.121821 | 0.107483 | 0.092647 | 0.078981 | 0.066896 | 0.056387 | |
| PKM-RPs | 0.105013 | 0.129722 | 0.121816 | 0.107481 | 0.092646 | 0.078981 | 0.066897 | 0.056389 | |
| NTLBG-RPs | 0.024000 | 0.040000 | 0.047200 | 0.051000 | 0.052400 | 0.053600 | 0.054000 | 0.054800 | |
| QMC-RPs | 0.035714 | 0.035714 | 0.035714 | 0.035714 | 0.035714 | 0.035714 | 0.035714 | 0.035714 | |
| FH-RPs | 0.090203 | 0.115859 | 0.112045 | 0.101330 | 0.089310 | 0.077762 | 0.067244 | 0.057882 | |
| PKM-RPs | 0.090195 | 0.115852 | 0.112040 | 0.101326 | 0.089309 | 0.077761 | 0.067245 | 0.057884 | |
| NTLBG-RPs | 0.019600 | 0.032800 | 0.039600 | 0.042600 | 0.044200 | 0.044800 | 0.045200 | 0.045400 | |
| QMC-RPs | 0.033333 | 0.033333 | 0.033333 | 0.033333 | 0.033333 | 0.033333 | 0.033333 | 0.033333 | |
| FH-RPs | 0.082054 | 0.107826 | 0.106082 | 0.097321 | 0.086883 | 0.076561 | 0.066981 | 0.058328 | |
| PKM-RPs | 0.082046 | 0.107817 | 0.106076 | 0.097317 | 0.086881 | 0.076561 | 0.066981 | 0.058329 | |
| NTLBG-RPs | 0.017400 | 0.029600 | 0.035400 | 0.038600 | 0.040400 | 0.040800 | 0.040800 | 0.041200 | |
| category | P9 | P10 | P11 | P12 | P13 | P14 | P15 | P16 | |
| QMC-RPs | 0.100000 | 0.100000 | |||||||
| FH-RPs | 0.005301 | 0.001224 | |||||||
| PKM-RPs | 0.005301 | 0.001224 | |||||||
| NTLBG-RPs | 0.008200 | 0.002200 | |||||||
| QMC-RPs | 0.066667 | 0.066667 | 0.066667 | 0.066667 | 0.066667 | 0.066667 | 0.066667 | ||
| FH-RPs | 0.026432 | 0.018001 | 0.011639 | 0.006965 | 0.003678 | 0.001544 | 0.000366 | ||
| PKM-RPs | 0.026433 | 0.018002 | 0.011640 | 0.006965 | 0.003679 | 0.001544 | 0.000366 | ||
| NTLBG-RPs | 0.056600 | 0.043400 | 0.031200 | 0.020400 | 0.011400 | 0.005400 | 0.001600 | ||
| QMC-RPs | 0.050000 | 0.050000 | 0.050000 | 0.050000 | 0.050000 | 0.050000 | 0.050000 | 0.050000 | |
| FH-RPs | 0.040125 | 0.031535 | 0.024455 | 0.018643 | 0.013901 | 0.010068 | 0.007012 | 0.004625 | |
| PKM-RPs | 0.040127 | 0.031537 | 0.024457 | 0.018645 | 0.013902 | 0.010069 | 0.007013 | 0.004626 | |
| NTLBG-RPs | 0.072800 | 0.069000 | 0.063800 | 0.057400 | 0.049600 | 0.041200 | 0.032000 | 0.023200 | |
| QMC-RPs | 0.040000 | 0.040000 | 0.040000 | 0.040000 | 0.040000 | 0.040000 | 0.040000 | 0.040000 | |
| FH-RPs | 0.047317 | 0.039514 | 0.032814 | 0.027069 | 0.022149 | 0.017945 | 0.014364 | 0.011327 | |
| PKM-RPs | 0.047319 | 0.039516 | 0.032817 | 0.027071 | 0.022151 | 0.017947 | 0.014365 | 0.011328 | |
| NTLBG-RPs | 0.055200 | 0.055600 | 0.056200 | 0.056200 | 0.055800 | 0.054800 | 0.052600 | 0.049400 | |
| QMC-RPs | 0.035714 | 0.035714 | 0.035714 | 0.035714 | 0.035714 | 0.035714 | 0.035714 | 0.035714 | |
| FH-RPs | 0.049638 | 0.042416 | 0.036105 | 0.030598 | 0.025797 | 0.021618 | 0.017984 | 0.014831 | |
| PKM-RPs | 0.049640 | 0.042418 | 0.036107 | 0.030600 | 0.025800 | 0.021620 | 0.017986 | 0.014833 | |
| NTLBG-RPs | 0.045800 | 0.046000 | 0.046400 | 0.047000 | 0.047800 | 0.048600 | 0.048800 | 0.048800 | |
| QMC-RPs | 0.033333 | 0.033333 | 0.033333 | 0.033333 | 0.033333 | 0.033333 | 0.033333 | 0.033333 | |
| FH-RPs | 0.050614 | 0.043782 | 0.037752 | 0.032439 | 0.027763 | 0.023651 | 0.020039 | 0.016869 | |
| PKM-RPs | 0.050616 | 0.043785 | 0.037755 | 0.032442 | 0.027765 | 0.023653 | 0.020041 | 0.016872 | |
| NTLBG-RPs | 0.041000 | 0.041000 | 0.041400 | 0.041800 | 0.042200 | 0.043200 | 0.044000 | 0.044600 | |
| category | P17 | P18 | P19 | P20 | P21 | P22 | P23 | P24 | |
| QMC-RPs | 0.050000 | 0.050000 | 0.050000 | 0.050000 | |||||
| FH-RPs | 0.002817 | 0.001512 | 0.000644 | 0.000154 | |||||
| PKM-RPs | 0.002818 | 0.001512 | 0.000644 | 0.000154 | |||||
| NTLBG-RPs | 0.015400 | 0.009000 | 0.004400 | 0.001400 | |||||
| QMC-RPs | 0.040000 | 0.040000 | 0.040000 | 0.040000 | 0.040000 | 0.040000 | 0.040000 | 0.040000 | |
| FH-RPs | 0.008767 | 0.006629 | 0.004862 | 0.003426 | 0.002284 | 0.001405 | 0.000761 | 0.000327 | |
| PKM-RPs | 0.008769 | 0.006629 | 0.004863 | 0.003426 | 0.002284 | 0.001405 | 0.000761 | 0.000327 | |
| NTLBG-RPs | 0.044800 | 0.039200 | 0.032800 | 0.026000 | 0.019200 | 0.012800 | 0.007400 | 0.003800 | |
| QMC-RPs | 0.035714 | 0.035714 | 0.035714 | 0.035714 | 0.035714 | 0.035714 | 0.035714 | 0.035714 | |
| FH-RPs | 0.012104 | 0.009754 | 0.007739 | 0.006025 | 0.004579 | 0.003375 | 0.002389 | 0.001600 | |
| PKM-RPs | 0.012105 | 0.009755 | 0.007740 | 0.006026 | 0.004580 | 0.003376 | 0.002390 | 0.001600 | |
| NTLBG-RPs | 0.048200 | 0.046600 | 0.043600 | 0.040000 | 0.035200 | 0.029400 | 0.023200 | 0.017200 | |
| QMC-RPs | 0.033333 | 0.033333 | 0.033333 | 0.033333 | 0.033333 | 0.033333 | 0.033333 | 0.033333 | |
| FH-RPs | 0.014094 | 0.011670 | 0.009559 | 0.007730 | 0.006153 | 0.004805 | 0.003662 | 0.002706 | |
| PKM-RPs | 0.014096 | 0.011672 | 0.009561 | 0.007731 | 0.006154 | 0.004806 | 0.003663 | 0.002707 | |
| NTLBG-RPs | 0.045200 | 0.045400 | 0.044600 | 0.043200 | 0.040600 | 0.037400 | 0.032600 | 0.027600 | |
| category | P25 | P26 | P27 | P28 | P29 | P30 | P31 | P32 | |
| QMC-RPs | 0.040000 | ||||||||
| FH-RPs | 0.000079 | ||||||||
| PKM-RPs | 0.000079 | ||||||||
| NTLBG-RPs | 0.001200 | ||||||||
| QMC-RPs | 0.035714 | 0.035714 | 0.035714 | 0.035714 | |||||
| FH-RPs | 0.000988 | 0.000537 | 0.000231 | 0.000056 | |||||
| PKM-RPs | 0.000988 | 0.000537 | 0.000231 | 0.000056 | |||||
| NTLBG-RPs | 0.011600 | 0.007000 | 0.003400 | 0.001200 | |||||
| QMC-RPs | 0.033333 | 0.033333 | 0.033333 | 0.033333 | 0.033333 | 0.033333 | |||
| FH-RPs | 0.001920 | 0.001289 | 0.000798 | 0.000434 | 0.000188 | 0.000046 | |||
| PKM-RPs | 0.001921 | 0.001289 | 0.000798 | 0.000435 | 0.000188 | 0.000046 | |||
| NTLBG-RPs | 0.021800 | 0.016200 | 0.010800 | 0.006600 | 0.003400 | 0.001200 |
References
- Schrödinger, E. Zur Theorie der Fall- und Steigversuche an Teilchen mit Brownscher Bewegung. Z. Phys. Chem. 1915, 16, 289–295. [Google Scholar]
- Tweedie, K.C.M. Inverse Statistical Variates. Nature 1945, 155, 453. [Google Scholar] [CrossRef]
- Wald, A. Sequential Analysis; Courier Corporation: Chelmsford, MA, USA, 2004. [Google Scholar]
- Cox, D.R.; Miller, H.D. The Theory of Stochastic Processes; Methuen: London, UK, 1965. [Google Scholar]
- Bartlett, M.S. An Introduction to Stochastic Processes; Cambridge University Press: London, UK, 1966. [Google Scholar]
- Moran, P.A.P. An Introduction to Probability Theory; Clarendon Press: Oxford, UK, 1968. [Google Scholar]
- Wasan, M.T.; Roy, L.K. Tables of Inverse Gaussian Percentage Points. Technometrics 1969, 11, 591–604. [Google Scholar] [CrossRef]
- Folks, J.L.; Chhikara, R.S. The Inverse Gaussian Distribution and Its Statistical Application—A Review. J. R. Stat. Soc. Ser. B (Stat. Methodol.) 1978, 40, 263–275. [Google Scholar] [CrossRef]
- Chhikara, R.S.; Folks, J.L. Estimation of Inverse Gaussian Distribution Function. J. Am. Stat. Assoc. 1974, 69, 250–254. [Google Scholar] [CrossRef]
- Chhikara, R.S.; Folks, J.L. The Inverse Gaussian Distribution as a Lifetime Model. Technometrics 1977, 19, 461–468. [Google Scholar] [CrossRef]
- Iyengar, S.; Patwardhan, G. Recent Developments in the Inverse Gaussian Distribution. In Handbook of Statistics; Elsevier: Amsterdam, The Netherlands, 1988; Volume 7, pp. 479–490. [Google Scholar]
- Kourogiorgas, C.; Panagopoulos, A.D.; Livieratos, S.N.; Chatzarakis, G.E. Rain Attenuation Time Series Synthesizer Based on Inverse Gaussian Distribution. Electron. Lett. 2015, 51, 2162–2164. [Google Scholar] [CrossRef]
- Punzo, A. A New Look at the Inverse Gaussian Distribution with Applications to Insurance and Economic Data. J. Appl. Stat. 2019, 46, 1260–1287. [Google Scholar] [CrossRef]
- Krbálek, M.; Hobza, T.; Patočka, M.; Krbálková, M.; Apeltauer, J.; Groverová, N. Statistical Aspects of Gap-Acceptance Theory for Unsignalized Intersection Capacity. Phys. A 2022, 594, 127043. [Google Scholar] [CrossRef]
- Fang, K.T.; Pan, J. A Review of Representative Points of Statistical Distributions and Their Applications. Mathematics 2023, 11, 2930. [Google Scholar] [CrossRef]
- Li, X.; He, P.; Huang, M.; Peng, X. A new class of moment-constrained mean square error representative samples for continuous distributions. J. Stat. Comput. Simul. 2025, 95, 2175–2203. [Google Scholar] [CrossRef]
- Peng, X.; Huang, M.; Li, X.; Zhou, T.; Lin, G.; Wang, X. Patient regional index: A new way to rank clinical specialties based on outpatient clinics big data. BMC Med. Res. Methodol. 2024, 24, 192. [Google Scholar] [CrossRef]
- Fang, K.T.; Ye, H.; Zhou, Y. Representative Points of Statistical Distributions and Their Applications in Statistical Inference; CRC Press: New York, NY, USA, 2025. [Google Scholar]
- Harrell, F.E.; Davis, C.E. A new distribution-free quantile estimator. Biometrika 1982, 69, 635–640. [Google Scholar] [CrossRef]
- Sfakianakis, M.E.; Verginis, D.G. A new family of nonparametric quantile estimators. Commun. Stat. Simul. Comput. 2008, 37, 337–345. [Google Scholar] [CrossRef]
- Wald, A. Sequential Analysis; John Wiley & Sons: New York, NY, USA, 1947. [Google Scholar]
- Tweedie, M.C.K. Statistical Properties of Inverse Gaussian Distributions. II. Ann. Math. Stat. 1957, 28, 696–705. [Google Scholar] [CrossRef]
- Fang, K.T.; He, S.D. The problem of selecting a given number of representative points in a normal population and a generalized Mills’ ratio. Acta Math. Appl. Sin. 1984, 7, 293–306. [Google Scholar]
- Efron, B. Bootstrap methods: Another look at the jackknife. Ann. Statist. 1979, 7, 1–26. [Google Scholar] [CrossRef]
- Hua, L.K.; Wang, Y. Applications of Number Theory to Numerical Analysis; Springer: Berlin, Germany; Science Press: Beijing, China, 1981. [Google Scholar]
- Niederreiter, H. Random Number Generation and Quasi-Monte Carlo Methods; SIAM: Philadelphia, PA, USA, 1992. [Google Scholar]
- Fang, K.T.; Wang, Y. Number-Theoretic Methods in Statistics; Chapman and Hall: London, UK, 1994. [Google Scholar]
- Barbiero, A.; Hitaj, A. Discrete approximations of continuous probability distributions obtained by minimizing Cramér–von Mises-type distances. Stat. Pap. 2022, 64, 1–29. [Google Scholar] [CrossRef]
- Flury, B.A. Principal points. Biometrika 1990, 77, 33–41. [Google Scholar] [CrossRef]
- Cox, D.R. Note on grouping. J. Am. Stat. Assoc. 1957, 52, 543–547. [Google Scholar] [CrossRef]
- Linde, Y.; Buzo, A.; Gray, R. An algorithm for vector quantizer design. IEEE Trans. Commun. 1980, 28, 84–95. [Google Scholar] [CrossRef]
- Lloyd, S.P. Least squares quantization in PCM. IEEE Trans. Inf. Theory 1982, 28, 129–137. [Google Scholar] [CrossRef]
- Tarpey, T. A parametric k-means algorithm. Comput. Stat. 2007, 22, 71–89. [Google Scholar] [CrossRef]
- Rosenblatt, M. Remarks on some nonparametric estimates of a density function. Ann. Math. Stat. 1956, 27, 832–837. [Google Scholar] [CrossRef]
- Parzen, E. On estimation of a probability density function and mode. Ann. Math. Stat. 1962, 33, 1065–1076. [Google Scholar] [CrossRef]
| Statistic | Category | 5 | 10 | 15 | 20 | 25 | 28 | 30 |
|---|---|---|---|---|---|---|---|---|
| Variance | MC-RPs (10) | −0.5105 | −0.2431 | −0.2357 | −0.3721 | −0.0064 | −0.1115 | 0.2320 |
| QMC-RPs | −0.5423 | −0.3695 | −0.2883 | −0.2396 | −0.2067 | −0.1914 | −0.1826 | |
| FH-RPs | −0.0833 | −0.0236 | −0.0109 | −0.0063 | −0.0041 | −0.0033 | −0.0029 | |
| PKM-RPs | −0.0833 | −0.0236 | −0.0109 | −0.0063 | −0.0041 | −0.0033 | −0.0029 | |
| NTLBG-RPs | −0.0846 | −0.0262 | −0.0163 | −0.0132 | −0.0115 | −0.0108 | −0.0104 | |
| Kurtosis | MC-RPs (10) | −15.5451 | −13.8874 | −13.0316 | −13.1384 | −11.6139 | −12.1894 | −11.7499 |
| QMC-RPs | −15.5670 | −13.9531 | −12.8774 | −12.0745 | −11.4366 | −11.1093 | −10.9094 | |
| FH-RPs | −4.8634 | −1.9262 | −1.0333 | −0.6452 | −0.4416 | −0.3628 | −0.3214 | |
| PKM-RPs | −4.8634 | −1.9262 | −1.0333 | −0.6452 | −0.4416 | −0.3628 | −0.3215 | |
| NTLBG-RPs | −5.0921 | −2.8455 | −2.5595 | −2.4208 | −2.2783 | −2.2534 | −2.2494 | |
| Mean | MC-RPs (10) | −0.0208 | −0.0246 | 0.0187 | −0.0755 | 0.0115 | 0.0384 | 0.0555 |
| QMC-RPs | −0.0857 | −0.0459 | −0.0316 | −0.0242 | −0.0196 | −0.0177 | −0.0165 | |
| FH-RPs | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | |
| PKM-RPs | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | |
| NTLBG-RPs | −0.0001 | −0.0001 | −0.0001 | −0.0001 | −0.0001 | −0.0001 | −0.0001 | |
| Skewness | MC-RPs (10) | −2.2837 | −1.6462 | −1.5081 | −1.5186 | −1.0662 | −1.3430 | −1.1255 |
| QMC-RPs | −2.0808 | −1.6007 | −1.3632 | −1.2107 | −1.1010 | −1.0480 | −1.0166 | |
| FH-RPs | −0.2515 | −0.0844 | −0.0420 | −0.0251 | −0.0166 | −0.0135 | −0.0119 | |
| PKM-RPs | −0.2515 | −0.0844 | −0.0420 | −0.0251 | −0.0166 | −0.0135 | −0.0119 | |
| NTLBG-RPs | −0.2723 | −0.1493 | −0.1405 | −0.1326 | −0.1240 | −0.1218 | −0.1210 |
| QMC-RPs | MSE-RPs | PKM-RPs | NTLBG-RPs | ||
|---|---|---|---|---|---|
| Zone | zone 1 | 0–1 | 0–1 | 0–1 | 0–1 |
| zone 2 | 1–15 | 1–15 | 1–15 | 1–15 | |
| h | 0.0500 | 0.0543 | 0.0543 | 0.0591 | |
| 0.2713 | 0.1417 | 0.1417 | 0.2418 | ||
| 0.03091647 | 0.03381734 | 0.03380385 | 0.05222610 | ||
| 0.03368552 | 0.00687223 | 0.00687258 | 0.00966432 |
| Statistic | Method | |||
|---|---|---|---|---|
| Variance | QMC-RPs | −0.5573 | −0.5519 | −0.5471 |
| FH-RPs | −0.7920 | −0.7917 | −0.7928 | |
| PKM-RPs | −0.7937 | −0.7922 | −0.7932 | |
| NTLBG-RPs | −0.0550 | −0.0697 | −0.0796 | |
| Kurtosis | QMC-RPs | −15.2659 | −15.3839 | −15.4725 |
| FH-RPs | 1.3644 | 1.3715 | 1.6490 | |
| PKM-RPs | 1.2462 | 1.3232 | 1.5490 | |
| NTLBG-RPs | −4.6785 | −4.8899 | −4.9691 | |
| Mean | QMC-RPs | −0.0873 | −0.0867 | −0.0870 |
| FH-RPs | −0.3691 | −0.3714 | −0.3724 | |
| PKM-RPs | −0.3690 | −0.3692 | −0.3712 | |
| NTLBG-RPs | 0.0336 | 0.0174 | 0.0077 | |
| Skewness | QMC-RPs | −2.0433 | −2.0550 | −2.0660 |
| FH-RPs | 0.1559 | 0.1836 | 0.2152 | |
| PKM-RPs | 0.1443 | 0.1639 | 0.1993 | |
| NTLBG-RPs | −0.2644 | −0.2695 | −0.2708 |
| Statistic | Method | |||
|---|---|---|---|---|
| Variance | QMC-RPs | −0.3188 | −0.3134 | −0.2980 |
| FH-RPs | −0.7672 | −0.7664 | −0.7672 | |
| PKM-RPs | −0.7693 | −0.7671 | −0.7679 | |
| NTLBG-RPs | −0.0085 | −0.0116 | −0.0125 | |
| Kurtosis | QMC-RPs | −12.9503 | −12.6811 | −12.7568 |
| FH-RPs | −1.5896 | −1.5647 | −1.1952 | |
| PKM-RPs | −1.6255 | −1.4699 | −1.2064 | |
| NTLBG-RPs | −1.7125 | −2.0773 | −2.3439 | |
| Mean | QMC-RPs | −0.0391 | −0.0414 | −0.0352 |
| FH-RPs | −0.3784 | −0.3794 | −0.3800 | |
| PKM-RPs | −0.3773 | −0.3770 | −0.3784 | |
| NTLBG-RPs | 0.0207 | 0.0128 | 0.0060 | |
| Skewness | QMC-RPs | −1.4538 | −1.3787 | −1.3668 |
| FH-RPs | −0.2725 | −0.2548 | −0.2210 | |
| PKM-RPs | −0.2858 | −0.2661 | −0.2320 | |
| NTLBG-RPs | −0.1076 | −0.1224 | −0.1335 |
| Statistic | Method | |||
|---|---|---|---|---|
| Variance | QMC-RPs | −0.2065 | −0.1950 | −0.1872 |
| FH-RPs | −0.7659 | −0.7654 | −0.7657 | |
| PKM-RPs | −0.7672 | −0.7655 | −0.7658 | |
| NTLBG-RPs | −0.0070 | −0.0051 | −0.0060 | |
| Kurtosis | QMC-RPs | −12.1054 | −11.3387 | −11.0403 |
| FH-RPs | −1.5183 | −1.5125 | −1.2570 | |
| PKM-RPs | −1.6669 | −1.5214 | −1.3705 | |
| NTLBG-RPs | −1.2137 | −1.8096 | −1.9828 | |
| Mean | QMC-RPs | −0.0204 | −0.0210 | −0.0180 |
| FH-RPs | −0.3739 | −0.3745 | −0.3760 | |
| PKM-RPs | −0.3728 | −0.3723 | −0.3749 | |
| NTLBG-RPs | 0.0052 | 0.0038 | 0.0006 | |
| Skewness | QMC-RPs | −1.2871 | −1.1344 | −1.0679 |
| FH-RPs | −0.2768 | −0.2631 | −0.2361 | |
| PKM-RPs | −0.2979 | −0.2808 | −0.2552 | |
| NTLBG-RPs | −0.1006 | −0.1127 | −0.1084 |
| SV Estimators | Construction Formula | Assumptions |
|---|---|---|
| , | ||
| Type | Method | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L2.pdf | L2.cdf | KL | ABI | L2.pdf | L2.cdf | KL | ABI | L2.pdf | L2.cdf | KL | ABI | ||
| IG(1,1) | Plain | 0.1335 | 0.0805 | 0.0423 | 0.1927 | 0.1001 | 0.0611 | 0.0208 | 0.1402 | 0.0683 | 0.0411 | 0.0104 | 0.0950 |
| IG(1,1) | HD | 0.1333 | 0.0841 | 0.0421 | 0.1997 | 0.0984 | 0.0614 | 0.0201 | 0.1408 | 0.0680 | 0.0419 | 0.0101 | 0.0966 |
| IG(1,1) | QSV1 | 0.1422 | 0.0850 | 0.0555 | 0.2088 | 0.1059 | 0.0636 | 0.0268 | 0.1514 | 0.0715 | 0.0456 | 0.0126 | 0.1001 |
| IG(1,1) | QSV2 | 0.1429 | 0.1037 | 0.0450 | 0.2295 | 0.1014 | 0.0702 | 0.0221 | 0.1553 | 0.0716 | 0.0507 | 0.0112 | 0.1064 |
| IG(1,1) | QSV3 | 0.1522 | 0.0846 | 0.0478 | 0.1962 | 0.1176 | 0.0647 | 0.0241 | 0.1497 | 0.0697 | 0.0443 | 0.0105 | 0.0946 |
| IG(1,0.5) | Plain | 0.1607 | 0.0887 | 0.0483 | 0.2396 | 0.1119 | 0.0624 | 0.0232 | 0.1596 | 0.0761 | 0.0463 | 0.0099 | 0.1101 |
| IG(1,0.5) | HD | 0.1604 | 0.0919 | 0.0485 | 0.2540 | 0.1104 | 0.0642 | 0.0217 | 0.1647 | 0.0751 | 0.0466 | 0.0095 | 0.1114 |
| IG(1,0.5) | QSV1 | 0.1757 | 0.0954 | 0.0692 | 0.2756 | 0.1174 | 0.0651 | 0.0283 | 0.1712 | 0.0781 | 0.0473 | 0.0115 | 0.1132 |
| IG(1,0.5) | QSV2 | 0.1664 | 0.1031 | 0.0512 | 0.2849 | 0.1150 | 0.0788 | 0.0230 | 0.1943 | 0.0777 | 0.0537 | 0.0106 | 0.1259 |
| IG(1,0.5) | QSV3 | 0.1865 | 0.0936 | 0.0529 | 0.2400 | 0.1427 | 0.0674 | 0.0284 | 0.1697 | 0.0783 | 0.0466 | 0.0098 | 0.1090 |
| IG(7,1) | Plain | 0.0799 | 0.1010 | 0.0182 | 0.1525 | 0.0577 | 0.0801 | 0.0103 | 0.1176 | 0.0379 | 0.0600 | 0.0049 | 0.0877 |
| IG(7,1) | HD | 0.0788 | 0.1023 | 0.0190 | 0.1579 | 0.0572 | 0.0795 | 0.0104 | 0.1184 | 0.0375 | 0.0588 | 0.0049 | 0.0859 |
| IG(7,1) | QSV1 | 0.0789 | 0.1032 | 0.0201 | 0.1619 | 0.0585 | 0.0811 | 0.0112 | 0.1231 | 0.0378 | 0.0601 | 0.0053 | 0.0885 |
| IG(7,1) | QSV2 | 0.0789 | 0.1046 | 0.0204 | 0.1632 | 0.0567 | 0.0797 | 0.0111 | 0.1220 | 0.0380 | 0.0597 | 0.0053 | 0.0886 |
| IG(7,1) | QSV3 | 0.0964 | 0.1128 | 0.0232 | 0.1587 | 0.0760 | 0.0951 | 0.0148 | 0.1305 | 0.0400 | 0.0608 | 0.0049 | 0.0872 |
| IG(3,3) | Plain | 0.0463 | 0.0946 | 0.0132 | 0.1205 | 0.0446 | 0.0902 | 0.0116 | 0.1099 | 0.0349 | 0.0710 | 0.0078 | 0.0838 |
| IG(3,3) | HD | 0.0465 | 0.0977 | 0.0133 | 0.1240 | 0.0438 | 0.0887 | 0.0110 | 0.1098 | 0.0348 | 0.0722 | 0.0075 | 0.0858 |
| IG(3,3) | QSV1 | 0.0458 | 0.0973 | 0.0141 | 0.1203 | 0.0440 | 0.0906 | 0.0123 | 0.1093 | 0.0354 | 0.0724 | 0.0087 | 0.0851 |
| IG(3,3) | QSV2 | 0.0489 | 0.1080 | 0.0137 | 0.1317 | 0.0428 | 0.0920 | 0.0105 | 0.1133 | 0.0359 | 0.0781 | 0.0078 | 0.0919 |
| IG(3,3) | QSV3 | 0.0491 | 0.0961 | 0.0136 | 0.1218 | 0.0491 | 0.0925 | 0.0123 | 0.1146 | 0.0362 | 0.0717 | 0.0081 | 0.0859 |
| n | Method | L2.cdf | L2.pdf | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Rank1 | Rank2 | Rank3 | Rank4 | Rank5 | Rank1 | Rank2 | Rank3 | Rank4 | Rank5 | ||
| 30 | Plain | 8 | 26 | 65 | 1 | 0 | 8 | 34 | 40 | 18 | 0 |
| 30 | HD | 13 | 35 | 13 | 38 | 1 | 5 | 36 | 44 | 15 | 0 |
| 30 | QSV1 | 31 | 13 | 4 | 17 | 35 | 27 | 10 | 8 | 29 | 26 |
| 30 | QSV2 | 29 | 3 | 3 | 7 | 58 | 25 | 11 | 5 | 17 | 42 |
| 30 | QSV3 | 19 | 23 | 15 | 37 | 6 | 35 | 9 | 3 | 21 | 32 |
| KL | ABI | ||||||||||
| 30 | Plain | 6 | 35 | 50 | 9 | 0 | 9 | 40 | 37 | 13 | 1 |
| 30 | HD | 7 | 40 | 31 | 22 | 0 | 9 | 24 | 45 | 22 | 0 |
| 30 | QSV1 | 31 | 2 | 9 | 11 | 47 | 32 | 8 | 5 | 15 | 40 |
| 30 | QSV2 | 27 | 6 | 5 | 27 | 35 | 21 | 9 | 5 | 24 | 41 |
| 30 | QSV3 | 29 | 17 | 5 | 31 | 18 | 29 | 19 | 8 | 26 | 18 |
| L2.cdf | L2.pdf | ||||||||||
| 50 | Plain | 5 | 20 | 72 | 3 | 0 | 8 | 36 | 29 | 27 | 0 |
| 50 | HD | 12 | 40 | 12 | 36 | 0 | 5 | 35 | 50 | 10 | 0 |
| 50 | QSV1 | 26 | 21 | 7 | 10 | 36 | 21 | 12 | 12 | 34 | 21 |
| 50 | QSV2 | 36 | 5 | 2 | 6 | 51 | 30 | 13 | 7 | 19 | 31 |
| 50 | QSV3 | 21 | 14 | 7 | 45 | 13 | 36 | 4 | 2 | 10 | 48 |
| KL | ABI | ||||||||||
| 50 | Plain | 5 | 41 | 43 | 11 | 0 | 5 | 50 | 17 | 28 | 0 |
| 50 | HD | 8 | 44 | 29 | 19 | 0 | 10 | 25 | 44 | 21 | 0 |
| 50 | QSV1 | 26 | 6 | 13 | 19 | 36 | 24 | 15 | 17 | 17 | 27 |
| 50 | QSV2 | 32 | 7 | 4 | 18 | 39 | 26 | 5 | 12 | 19 | 38 |
| 50 | QSV3 | 29 | 2 | 11 | 33 | 25 | 35 | 5 | 10 | 15 | 35 |
| L2.cdf | L2.pdf | ||||||||||
| 100 | Plain | 3 | 14 | 78 | 5 | 0 | 2 | 34 | 44 | 20 | 0 |
| 100 | HD | 5 | 44 | 11 | 40 | 0 | 2 | 44 | 33 | 21 | 0 |
| 100 | QSV1 | 30 | 14 | 4 | 16 | 36 | 20 | 16 | 10 | 27 | 27 |
| 100 | QSV2 | 40 | 1 | 2 | 5 | 52 | 33 | 5 | 3 | 20 | 39 |
| 100 | QSV3 | 22 | 27 | 5 | 34 | 12 | 43 | 1 | 10 | 12 | 34 |
| KL | ABI | ||||||||||
| 100 | Plain | 1 | 31 | 49 | 19 | 0 | 6 | 43 | 15 | 36 | 0 |
| 100 | HD | 4 | 45 | 21 | 30 | 0 | 5 | 24 | 47 | 24 | 0 |
| 100 | QSV1 | 32 | 8 | 4 | 14 | 42 | 27 | 20 | 10 | 12 | 31 |
| 100 | QSV2 | 31 | 6 | 6 | 19 | 38 | 29 | 4 | 9 | 19 | 39 |
| 100 | QSV3 | 32 | 10 | 20 | 18 | 20 | 33 | 9 | 19 | 9 | 30 |
| n | Method | L2.pdf | L2.cdf | KL | ABI | ||
|---|---|---|---|---|---|---|---|
| 30 | Plain | 0.04980 | 0.01671 | 0.00286 | 0.05779 | 1.00509 | 1.11049 |
| 30 | HD | 0.06393 | 0.02833 | 0.00440 | 0.08200 | 1.03902 | 1.12497 |
| 30 | QSV1 | 0.08129 | 0.03122 | 0.01076 | 0.12560 | 0.95727 | 1.20847 |
| 30 | QSV2 | 0.10074 | 0.06753 | 0.01303 | 0.14175 | 1.13144 | 1.15207 |
| 30 | QSV3 | 0.01142 | 0.01127 | 0.00043 | 0.02244 | 0.97528 | 1.02015 |
| 30 | Analytic formulas | 0.05243 | 0.01728 | 0.00358 | 0.06668 | 0.99020 | 1.12355 |
| 50 | Plain | 0.02534 | 0.00914 | 0.00092 | 0.03520 | 0.98907 | 1.05946 |
| 50 | HD | 0.03076 | 0.01272 | 0.00098 | 0.03766 | 1.01584 | 1.05948 |
| 50 | QSV1 | 0.05169 | 0.02467 | 0.00505 | 0.08667 | 0.95599 | 1.12933 |
| 50 | QSV2 | 0.05874 | 0.04148 | 0.00454 | 0.07973 | 1.08267 | 1.07678 |
| 50 | QSV3 | 0.01877 | 0.01447 | 0.00052 | 0.02331 | 0.97068 | 0.98270 |
| 50 | Analytic formulas | 0.04398 | 0.01474 | 0.00221 | 0.05065 | 1.00447 | 1.09683 |
| 100 | Plain | 0.01481 | 0.00584 | 0.00034 | 0.02172 | 0.99134 | 1.03478 |
| 100 | HD | 0.01450 | 0.00672 | 0.00021 | 0.01794 | 1.01011 | 1.02578 |
| 100 | QSV1 | 0.02855 | 0.01487 | 0.00160 | 0.04883 | 0.97173 | 1.06940 |
| 100 | QSV2 | 0.03223 | 0.02508 | 0.00152 | 0.04259 | 1.05160 | 1.03357 |
| 100 | QSV3 | 0.01188 | 0.00900 | 0.00020 | 0.01492 | 0.98183 | 0.98833 |
| 100 | Analytic formulas | 0.00624 | 0.01302 | 0.00030 | 0.02123 | 0.98855 | 1.03101 |
| n | Method | L2.pdf | L2.cdf | KL | ABI | ||
|---|---|---|---|---|---|---|---|
| 30 | Plain | 0.00711 | 0.00734 | 0.00016 | 0.01392 | 3.00712 | 3.07637 |
| 30 | HD | 0.01284 | 0.02280 | 0.00056 | 0.02875 | 3.07231 | 3.10020 |
| 30 | QSV1 | 0.01683 | 0.02596 | 0.00166 | 0.04962 | 2.91507 | 3.21281 |
| 30 | QSV2 | 0.02414 | 0.05617 | 0.00254 | 0.05589 | 3.20040 | 3.13492 |
| 30 | QSV3 | 0.00643 | 0.01504 | 0.00019 | 0.01394 | 2.94697 | 2.96938 |
| 30 | Analytic formulas | 0.03027 | 0.02994 | 0.00358 | 0.06668 | 2.97059 | 3.37065 |
| 50 | Plain | 0.00519 | 0.00796 | 0.00015 | 0.01506 | 2.97368 | 3.06402 |
| 50 | HD | 0.00777 | 0.01213 | 0.00019 | 0.01698 | 3.03533 | 3.06654 |
| 50 | QSV1 | 0.01484 | 0.02948 | 0.00159 | 0.04801 | 2.89459 | 3.18262 |
| 50 | QSV2 | 0.01959 | 0.04771 | 0.00178 | 0.04483 | 3.17242 | 3.09655 |
| 50 | QSV3 | 0.01123 | 0.02132 | 0.00044 | 0.02437 | 2.93001 | 2.92375 |
| 50 | Analytic formulas | 0.02539 | 0.02553 | 0.00221 | 0.05065 | 3.01341 | 3.29050 |
| 100 | Plain | 0.00492 | 0.00875 | 0.00015 | 0.01519 | 2.96921 | 3.06037 |
| 100 | HD | 0.00469 | 0.00720 | 0.00007 | 0.01018 | 3.02073 | 3.04032 |
| 100 | QSV1 | 0.01117 | 0.02341 | 0.00094 | 0.03670 | 2.91492 | 3.13509 |
| 100 | QSV2 | 0.01357 | 0.03421 | 0.00090 | 0.03048 | 3.12448 | 3.05841 |
| 100 | QSV3 | 0.00817 | 0.01695 | 0.00026 | 0.01784 | 2.94240 | 2.95058 |
| 100 | Analytic formulas | 0.00752 | 0.01082 | 0.00030 | 0.02123 | 2.96566 | 3.09304 |
| n | Method | L2.pdf | L2.cdf | KL | ABI | |||
|---|---|---|---|---|---|---|---|---|
| 30 | Plain | 0.06271 | 0.01494 | 0.00928 | 0.02120 | 0.98465 | 0.52369 | 1.00088 |
| 30 | HD | 0.07647 | 0.02574 | 0.01829 | 0.03954 | 1.01264 | 0.53394 | 0.96188 |
| 30 | QSV1 | 0.05848 | 0.06164 | 0.00823 | 0.05831 | 1.15003 | 0.49167 | 1.00825 |
| 30 | QSV2 | 0.12524 | 0.04358 | 0.04314 | 0.05009 | 1.02293 | 0.53946 | 1.04841 |
| 30 | QSV3 | 0.01719 | 0.01117 | 0.00073 | 0.03444 | 0.98345 | 0.49372 | 1.07421 |
| 50 | Plain | 0.04974 | 0.01349 | 0.00482 | 0.01919 | 0.99446 | 0.51553 | 1.02097 |
| 50 | HD | 0.05489 | 0.02092 | 0.00781 | 0.02797 | 1.01616 | 0.52314 | 0.97853 |
| 50 | QSV1 | 0.03694 | 0.04825 | 0.00538 | 0.05698 | 1.11840 | 0.48912 | 1.03078 |
| 50 | QSV2 | 0.08767 | 0.03141 | 0.01780 | 0.03404 | 1.02009 | 0.52820 | 1.02574 |
| 50 | QSV3 | 0.02644 | 0.00981 | 0.00084 | 0.03741 | 0.99845 | 0.49272 | 1.09612 |
| 100 | Plain | 0.04974 | 0.00804 | 0.00482 | 0.01876 | 0.98980 | 0.50272 | 1.04063 |
| 100 | HD | 0.00727 | 0.00777 | 0.00095 | 0.00687 | 1.00415 | 0.50791 | 1.00065 |
| 100 | QSV1 | 0.03694 | 0.02574 | 0.00240 | 0.05140 | 1.06915 | 0.48445 | 1.05395 |
| 100 | QSV2 | 0.04340 | 0.01254 | 0.00350 | 0.01594 | 0.99890 | 0.51301 | 1.02071 |
| 100 | QSV3 | 0.01859 | 0.00846 | 0.00091 | 0.04493 | 0.99796 | 0.48500 | 1.10276 |
| 0.17 | 0.19 | 0.23 | 0.33 | 0.39 |
| 0.39 | 0.40 | 0.45 | 0.52 | 0.56 |
| 0.59 | 0.64 | 0.66 | 0.70 | 0.76 |
| 0.77 | 0.78 | 0.95 | 0.97 | 1.02 |
| 1.12 | 1.24 | 1.59 | 1.74 | 2.92 |
| Method | K-S | p-Value | Bias | SSE | R2 | |||
|---|---|---|---|---|---|---|---|---|
| Plain | 0.8032 | 1.4397 | −14.3916 | 0.0621 | 0.9976 | 0.0134 | 0.0261 | 0.9875 |
| HD | 0.8305 | 1.4418 | −14.4158 | 0.078 | 0.9981 | 0.0247 | 0.036 | 0.9828 |
| QSV1 | 0.7691 | 1.6801 | −14.5999 | 0.0733 | 0.9993 | 0.0097 | 0.0214 | 0.9897 |
| QSV2 | 0.928 | 1.4146 | −14.7918 | 0.126 | 0.8225 | 0.0582 | 0.1134 | 0.9476 |
| QSV3 | 0.7762 | 1.3708 | −14.4322 | 0.0584 | 0.9688 | −0.002 | 0.0272 | 0.9869 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, W.-W.; Fang, K.-T.; Peng, X.-L. Representative Points of the Inverse Gaussian Distribution and Their Applications. Entropy 2025, 27, 1190. https://doi.org/10.3390/e27121190
Hu W-W, Fang K-T, Peng X-L. Representative Points of the Inverse Gaussian Distribution and Their Applications. Entropy. 2025; 27(12):1190. https://doi.org/10.3390/e27121190
Chicago/Turabian StyleHu, Wen-Wen, Kai-Tai Fang, and Xiao-Ling Peng. 2025. "Representative Points of the Inverse Gaussian Distribution and Their Applications" Entropy 27, no. 12: 1190. https://doi.org/10.3390/e27121190
APA StyleHu, W.-W., Fang, K.-T., & Peng, X.-L. (2025). Representative Points of the Inverse Gaussian Distribution and Their Applications. Entropy, 27(12), 1190. https://doi.org/10.3390/e27121190






