Levy Sooty Tern Optimization Algorithm Builds DNA Storage Coding Sets for Random Access
Abstract
1. Introduction
2. Coding Constraints
2.1. GC Content Constraint
2.2. Edit Distance Constraint
2.3. No-Runlength Constraint
3. Algorithm Description
3.1. Sooty Tern Optimization Algorithm
3.2. Levy Sooty Tern Optimization
3.3. Benchmark Function
4. Result and Analysis
4.1. Algorithm Performance Comparison
4.2. DNA Storage Code Set
5. Conclusions
Funding
Data Availability Statement
Conflicts of Interest
References
- Cao, B.; Wang, B.; Zhang, Q. GCNSA: DNA storage encoding with a graph convolutional network and self-attention. iScience 2023, 26, 106231. [Google Scholar] [CrossRef] [PubMed]
- Mu, Z.; Cao, B.; Wang, P.; Wang, B.; Zhang, Q. RBS: A Rotational Coding Based on Blocking Strategy for DNA Storage. IEEE Trans. NanoBioscience 2023, 22, 912–922. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Cao, B.; Ma, T.; Zhao, Y.; Zheng, Y.; Wang, B.; Zhou, S.; Zhang, Q. Storing Images in DNA via base128 Encoding. J. Chem. Inf. Model. 2024, 64, 1719–1729. [Google Scholar] [CrossRef] [PubMed]
- Cao, B.; Zheng, Y.; Shao, Q.; Liu, Z.; Xie, L.; Zhao, Y.; Wang, B.; Zhang, Q.; Wei, X. Efficient data reconstruction: The bottleneck of large-scale application of DNA storage. Cell Rep. 2024, 43. [Google Scholar] [CrossRef]
- Church, G.M.; Gao, Y.; Kosuri, S. Next-generation digital information storage in DNA. Science 2012, 337, 1628. [Google Scholar] [CrossRef]
- Li, X.; Zhou, S.; Zou, L. Design of DNA Storage Coding with Enhanced Constraints. Entropy 2022, 24, 1151. [Google Scholar] [CrossRef]
- Khuat, T.-H.; Kim, S. A Quaternary Code Correcting a Burst of at Most Two Deletion or Insertion Errors in DNA Storage. Entropy 2021, 23, 1592. [Google Scholar] [CrossRef]
- Goldman, N.M.; Bertone, P.; Chen, S.; Dessimoz, C.; Leproust, E.M.; Sipos, B.; Birney, E. Towards practical, high-capacity, low-maintenance information storage in synthesized DNA. Nature 2013, 494, 77–80. [Google Scholar] [CrossRef]
- Cao, B.; Zhang, X.; Cui, S.; Zhang, Q. Adaptive coding for DNA storage with high storage density and low coverage. NPJ Syst. Biol. Appl. 2022, 8, 23. [Google Scholar] [CrossRef]
- Tabatabaei, S.K.; Pham, B.; Pan, C.; Liu, J.Q.; Chandak, S.; Shorkey, S.A.; Hernandez, A.G.; Aksimentiev, A.; Chen, M.; Schroeder, C.M.; et al. Expanding the Molecular Alphabet of DNA-Based Data Storage Systems with Neural Network Nanopore Readout. Nano Lett. 2022, 22, 1905–1914. [Google Scholar] [CrossRef]
- Pan, C.; Tabatabaei, S.K.; Tabatabaei Yazdi, S.; Hernandez, A.G.; Schroeder, C.M.; Milenkovic, O. Rewritable two-dimensional DNA-based data storage with machine learning reconstruction. Nat. Commun. 2022, 13, 2984. [Google Scholar] [CrossRef]
- Cao, B.; Shi, P.; Zheng, Y.; Zhang, Q. FMG: An observable DNA storage coding method based on frequency matrix game graphs. Comput. Biol. Med. 2022, 151, 106269. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Cao, B.; Wu, J.; Wang, B.; Zhang, Q. High Net Information Density DNA Data Storage by the MOPE Encoding Algorithm. IEEE/ACM Trans. Comput. Biol. Bioinform. 2023, 20, 2992–3000. [Google Scholar] [CrossRef] [PubMed]
- Grass, R.N.; Heckel, R.; Puddu, M.; Paunescu, D.; Stark, W.J. Robust Chemical Preservation of Digital Information on DNA in Silica with Error-Correcting Codes. Angew. Chem. 2015, 54, 2552–2555. [Google Scholar] [CrossRef] [PubMed]
- Blawat, M.; Gaedke, K.; Hutter, I.; Chen, X.; Turczyk, B.M.; Inverso, S.A.; Pruitt, B.W.; Church, G.M. Forward Error Correction for DNA Data Storage. Procedia Comput. Sci. 2016, 80, 1011–1022. [Google Scholar] [CrossRef]
- Deng, M.; Yu, C.; Liang, Q.; He, R.L.; Yau, S.S.-T. A novel method of characterizing genetic sequences: Genome space with biological distance and applications. PLoS ONE 2011, 6, e17293. [Google Scholar] [CrossRef]
- Erlich, Y.; Zielinski, D. DNA Fountain enables a robust and efficient storage architecture. Science 2017, 355, 950–953. [Google Scholar] [CrossRef]
- Press, W.H.; Hawkins, J.A.; Jones, S.K.; Schaub, J.M.; Finkelstein, I.J. HEDGES error-correcting code for DNA storage corrects indels and allows sequence constraints. Proc. Natl. Acad. Sci. USA 2020, 117, 18489–18496. [Google Scholar] [CrossRef]
- Cai, K.; Chee, Y.M.; Gabrys, R.; Kiah, H.M.; Nguyen, T.T. Correcting a Single Indel/Edit for DNA-Based Data Storage: Linear-Time Encoders and Order-Optimality. IEEE Trans. Inf. Theory 2021, 67, 3438–3451. [Google Scholar] [CrossRef]
- Yang, S.; Bögels, B.W.A.; Wang, F.; Xu, C.; Dou, H.; Mann, S.; Fan, C.; de Greef, T.F.A. DNA as a universal chemical substrate for computing and data storage. Nat. Rev. Chem. 2024, 8, 179–194. [Google Scholar] [CrossRef]
- Organick, L.; Ang, S.D.; Chen, Y.; Lopez, R.; Yekhanin, S.; Makarychev, K.; Racz, M.Z.; Kamath, G.M.; Gopalan, P.; Nguyen, B.H. Random access in large-scale DNA data storage. Nat. Biotechnol. 2018, 36, 242–248. [Google Scholar] [CrossRef]
- Banal, J.L.; Shepherd, T.R.; Berleant, J.; Huang, H.; Reyes, M.; Ackerman, C.M.; Blainey, P.C.; Bathe, M. Random access DNA memory using Boolean search in an archival file storage system. Nat. Mater. 2021, 20, 1272–1280. [Google Scholar] [CrossRef] [PubMed]
- Anavy, L.; Vaknin, I.; Atar, O.; Amit, R.; Yakhini, Z. Data storage in DNA with fewer synthesis cycles using composite DNA letters. Nat. Biotechnol. 2019, 37, 1229–1236. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Lim, D.; Kim, J.; Song, Y. Processing DNA Storage through Programmable Assembly in a Droplet-Based Fluidics System. Adv. Sci. 2023, 10, 2303197. [Google Scholar] [CrossRef] [PubMed]
- Cao, B.; Li, X.; Zhang, X.; Wang, B.; Zhang, Q.; Wei, X. Designing Uncorrelated Address Constrain for DNA Storage by DMVO Algorithm. IEEE/ACM Trans. Comput. Biol. Bioinform. 2022, 19, 866–877. [Google Scholar] [CrossRef]
- Cao, B.; Zhang, X.; Wu, J.; Wang, B.; Zhang, Q. Minimum free energy coding for DNA storage. IEEE Trans. Nanobioscience 2021, 2, 212–222. [Google Scholar] [CrossRef]
- Yin, Q.; Cao, B.; Li, X.; Wang, B.; Zhang, Q.; Wei, X. An Intelligent Optimization Algorithm for Constructing a DNA Storage Code: NOL-HHO. Int. J. Mol. Sci. 2020, 21, 2191. [Google Scholar] [CrossRef]
- Rasool, A.; Hong, J.; Jiang, Q.; Chen, H.; Qu, Q. BO-DNA: Biologically optimized encoding model for a highly-reliable DNA data storage. Comput. Biol. Med. 2023, 165, 107404. [Google Scholar] [CrossRef]
- Cao, B.; Zhao, S.; Li, X.; Wang, B. K-Means Multi-Verse Optimizer (KMVO) Algorithm to Construct DNA Storage Codes. IEEE Access 2020, 8, 29547–29556. [Google Scholar] [CrossRef]
- Dhiman, G.; Kaur, A. STOA: A bio-inspired based optimization algorithm for industrial engineering problems. Eng. Appl. Artif. Intell. 2019, 82, 148–174. [Google Scholar] [CrossRef]
- Viswanathan, G.M.; Buldyrev, S.V.; Havlin, S.; Da Luz, M.; Raposo, E.; Stanley, H.E. Optimizing the success of random searches. Nature 1999, 401, 911–914. [Google Scholar] [CrossRef] [PubMed]
- Faramarzi, A.; Heidarinejad, M.; Stephens, B.; Mirjalili, S. Equilibrium optimizer: A novel optimization algorithm. Knowl. Based Syst. 2020, 191, 105190. [Google Scholar] [CrossRef]
- Zheng, Y.; Cao, B.; Zhang, X.; Cui, S.; Wang, B.; Zhang, Q. DNA-QLC: An efficient and reliable image encoding scheme for DNA storage. BMC Genom. 2024, 25, 266. [Google Scholar] [CrossRef]
- Limbachiya, D.; Gupta, M.K.; Aggarwal, V. Family of Constrained Codes for Archival DNA Data Storage. IEEE Commun. Lett. 2018, 22, 1972–1975. [Google Scholar] [CrossRef]
- Wang, P.; Cao, B.; Ma, T.; Wang, B.; Zhang, Q.; Zheng, P. DUHI: Dynamically updated hash index clustering method for DNA storage. Comput. Biol. Med. 2023, 164, 107244. [Google Scholar] [CrossRef]
| Function | Dim | Range | Fmin |
|---|---|---|---|
| 50 | [−100,100] | 0 | |
| 50 | [−10,10] | 0 | |
| 50 | [−100,100] | 0 | |
| 50 | [−100,100] | 0 | |
| 50 | [−30,30] | 0 | |
| 50 | [−100,100] | 0 | |
| 50 | [−1.28,1.28] | 0 |
| Function | Dim | Range | Fmin |
|---|---|---|---|
| 50 | [−500,500] | ||
| 50 | [−5.12,5.12] | 0 | |
| 50 | [−32,32] | 0 | |
| 50 | [−600,600] | 0 | |
| 50 | [−50,50] | 0 | |
| 50 | [−50,50] | 0 |
| F | LSTOA | STOA [30] | PSO [32] | GWO [32] | GA [32] | GSA [32] | SSA [32] |
|---|---|---|---|---|---|---|---|
| Ave | Ave | Ave | Ave | Ave | Ave | Ave | |
| F1 | 8.68 × 10−18 | 2.66 × 10−17 | 9.59 × 10−6 | 6.59 × 10−28 | 5.55 × 10−1 | 2.53 × 10−16 | 1.58 × 10−7 |
| F2 | 2.45 × 10−12 | 6.76 × 10−12 | 2.56 × 10−2 | 7.18 × 10−17 | 5.66 × 10−3 | 5.57 × 10−2 | 2.66 |
| F3 | 2.13 × 10−7 | 6.26 × 10−8 | 8.23 × 101 | 3.29 × 10−6 | 8.46 × 102 | 8.97 × 102 | 1.71 × 103 |
| F4 | 1.89 × 10−4 | 2.46 × 10−5 | 4.26 | 5.61 × 10−7 | 4.56 | 7.35 | 1.17 × 101 |
| F5 | 2.76 × 101 | 2.77 × 101 | 9.24 × 101 | 2.68 × 101 | 2.68 × 102 | 6.75 × 101 | 2.96 × 102 |
| F6 | 2.40 | 2.44 | 8.89 × 10−6 | 8.17 × 10−1 | 5.63 × 10−1 | 2.50 × 10−16 | 1.80 × 10−7 |
| F7 | 2.75 × 10−3 | 1.94 × 10−3 | 2.72 × 10−2 | 2.21 × 10−3 | 4.29 × 10−2 | 8.94 × 10−2 | 1.76 × 10−1 |
| F8 | −5.47 × 103 | −5.39 × 103 | −6.08 × 103 | −6.12 × 103 | −1.05 × 104 | −2.82 × 103 | −7.46 × 103 |
| F9 | 8.45 × 10−1 | 2.75 | 5.28 × 101 | 3.11 × 10−1 | 3.08 × 101 | 2.60 × 101 | 5.84 × 101 |
| F10 | 2.00 × 101 | 2.00 × 101 | 5.01 × 10−3 | 1.06 × 10−13 | 1.64 | 6.21 × 10−2 | 2.68 |
| F11 | 1.69 × 10−2 | 9.55 × 10−3 | 2.38 × 10−2 | 4.48 × 10−3 | 5.61 × 10−1 | 2.77 × 101 | 1.60 × 10−2 |
| F12 | 2.15 × 10−1 | 1.85 × 10−1 | 2.76 × 10−2 | 5.34 × 10−2 | 3.09 × 10−2 | 1.80 | 6.99 |
| F13 | 1.62 | 1.73 | 7.32 × 10−3 | 6.54 × 10−1 | 3.62 × 10−1 | 8.90 | 1.59 × 101 |
| F | LSTOA | STOA [30] | PSO [32] | GWO [32] | GA [32] | GSA [32] | SSA [32] |
|---|---|---|---|---|---|---|---|
| SD | SD | SD | SD | SD | SD | SD | |
| F1 | 2.86 × 10−34 | 9.40 × 10−33 | 3.35 × 10−5 | 1.58 × 10−28 | 1.23 | 9.67 × 10−17 | 1.71 × 10−7 |
| F2 | 1.32 × 10−23 | 5.89 × 10−23 | 4.60 × 10−2 | 7.28 × 10−17 | 1.44 × 10−2 | 1.94 × 10−1 | 1.67 |
| F3 | 1.61 × 10−13 | 1.15 × 10−14 | 9.72 × 101 | 1.61 × 10−5 | 1.61 × 102 | 3.19 × 102 | 1.12 × 104 |
| F4 | 4.82 × 10−7 | 4.57 × 10−10 | 6.77 × 10−1 | 1.04 × 10−6 | 5.92 × 10−1 | 1.74 | 4.18 |
| F5 | 4.95 × 10−1 | 5.52 × 10−1 | 7.45 × 101 | 7.93 × 10−1 | 3.38 × 102 | 6.22 × 101 | 5.09 × 102 |
| F6 | 2.28 × 10−1 | 3.49 × 10−1 | 9.91 × 10−6 | 4.82 × 10−1 | 1.72 | 1.74 × 10−16 | 3.00 × 10−7 |
| F7 | 4.69 × 10−6 | 3.93 × 10−6 | 8.04 × 10−3 | 2.00 × 10−3 | 5.94 × 10−3 | 4.34 × 10−2 | 6.29 × 10−2 |
| F8 | 1.71 × 105 | 1.71 × 105 | 7.55 × 102 | 9.10 × 102 | 3.53 × 102 | 4.93 × 102 | 7.73 × 102 |
| F9 | 5.06 | 6.70 × 101 | 1.67 × 101 | 3.52 × 10−1 | 7.57 | 7.47 | 2.00 × 101 |
| F10 | 2.34 × 10−6 | 3.97 × 10−6 | 1.26 × 10−2 | 2.24 × 10−13 | 4.62 × 10−1 | 2.36 × 10−1 | 8.28 × 10−1 |
| F11 | 1.37 × 10−3 | 3.05 × 10−4 | 2.87 × 10−2 | 6.65 × 10−3 | 2.69 × 10−1 | 5.04 | 1.12 × 10−2 |
| F12 | 2.88 × 10−2 | 5.17 × 10−3 | 5.40 × 10−2 | 2.07 × 10−2 | 4.09 × 10−2 | 9.51 × 10−1 | 4.42 |
| F13 | 2.61 × 10−2 | 7.23 × 10−2 | 1.05 × 10−2 | 4.47 × 10−3 | 3.10 × 10−1 | 7.13 | 1.61 × 101 |
| N\D | 3 | 4 | 5 | 6 | 7 | 8 | 9 |
|---|---|---|---|---|---|---|---|
| 4 | 6 b | ||||||
| 6 l | |||||||
| 5 | 12 b | 5 b | |||||
| 12 l | 5 l | ||||||
| 6 | 30 b | 11 b | 4 b | ||||
| 30 l | 11 l | 4 l | |||||
| 7 | 53 b | 19 b | 6 b | 3 b | |||
| 53 l | 19 l | 6 l | 3 l | ||||
| 8 | 101 b | 38 b | 12 b | 5 b | 3 b | ||
| 101 l | 41 l | 12 l | 5 l | 3 l | |||
| 9 | 167 b | 58 b | 19 b | 7 b | 3 b | 2 b | |
| 170 l | 63 l | 19 l | 7 l | 3 l | 2 l | ||
| 10 | 250 b | 110 b | 34 b | 11 b | 5 b | 3 b | 2 b |
| 250 l | 114 l | 37 l | 11 l | 5 l | 3 l | 2 l |
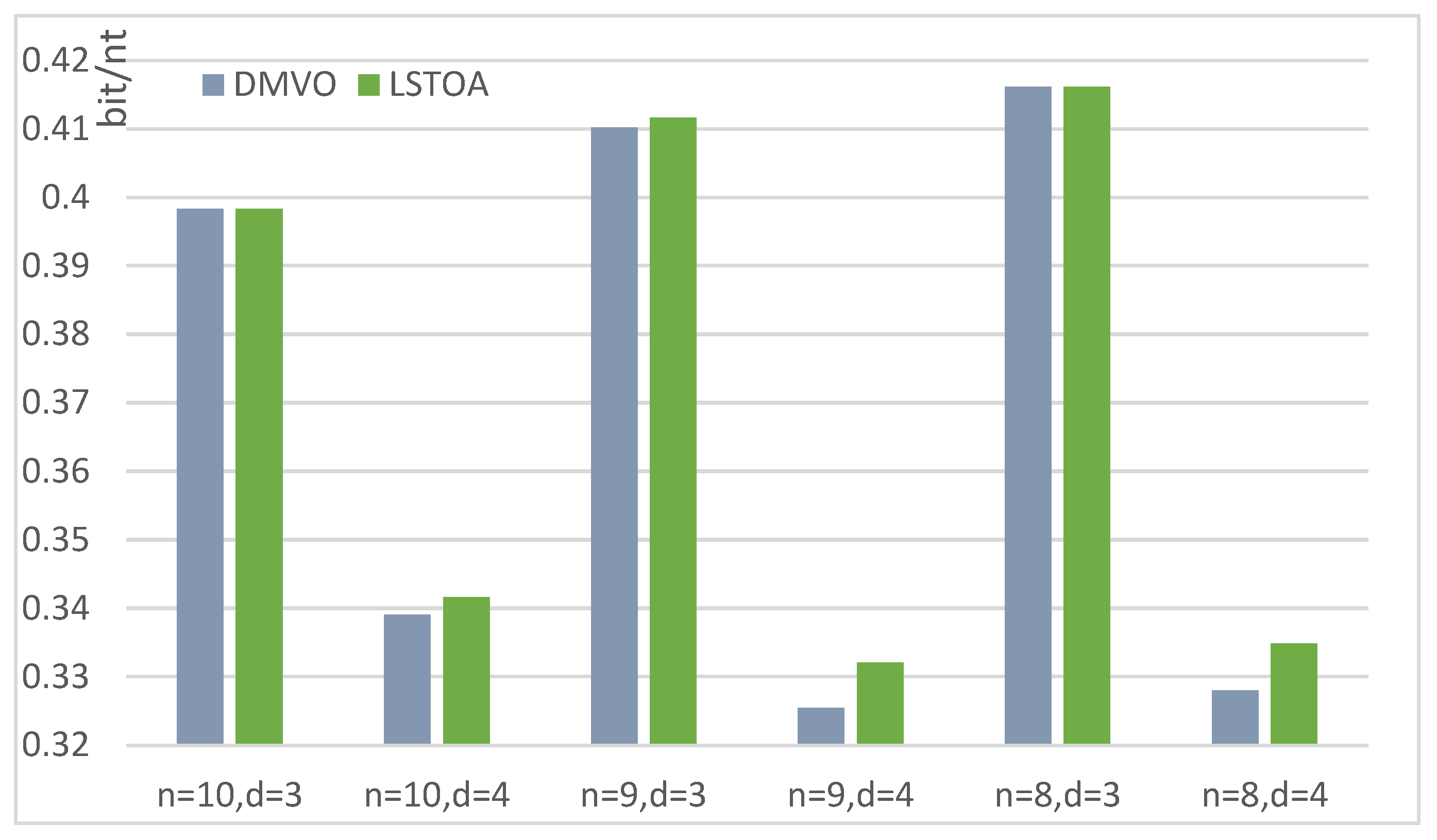
| n = 8, d = 3 | n = 8, d = 4 | n = 9, d = 3 | n = 9, d = 4 | n = 10, d = 5 | n = 10, d = 6 | ||
|---|---|---|---|---|---|---|---|
| DMVO [25] | Size | 101 | 38 | 167 | 58 | 110 | 34 |
| time (min) | 22 | 21.5 | 23.1 | 20.9 | 24 | 24 | |
| LSTOA | Size | 101 | 41 | 188 | 63 | 114 | 37 |
| time (min) | 19 | 17.6 | 20.3 | 20.5 | 19.8 | 20.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J. Levy Sooty Tern Optimization Algorithm Builds DNA Storage Coding Sets for Random Access. Entropy 2024, 26, 778. https://doi.org/10.3390/e26090778
Zhang J. Levy Sooty Tern Optimization Algorithm Builds DNA Storage Coding Sets for Random Access. Entropy. 2024; 26(9):778. https://doi.org/10.3390/e26090778
Chicago/Turabian StyleZhang, Jianxia. 2024. "Levy Sooty Tern Optimization Algorithm Builds DNA Storage Coding Sets for Random Access" Entropy 26, no. 9: 778. https://doi.org/10.3390/e26090778
APA StyleZhang, J. (2024). Levy Sooty Tern Optimization Algorithm Builds DNA Storage Coding Sets for Random Access. Entropy, 26(9), 778. https://doi.org/10.3390/e26090778





