Sleep Apnea Detection Using Wavelet Scattering Transformation and Random Forest Classifier
Abstract
1. Introduction
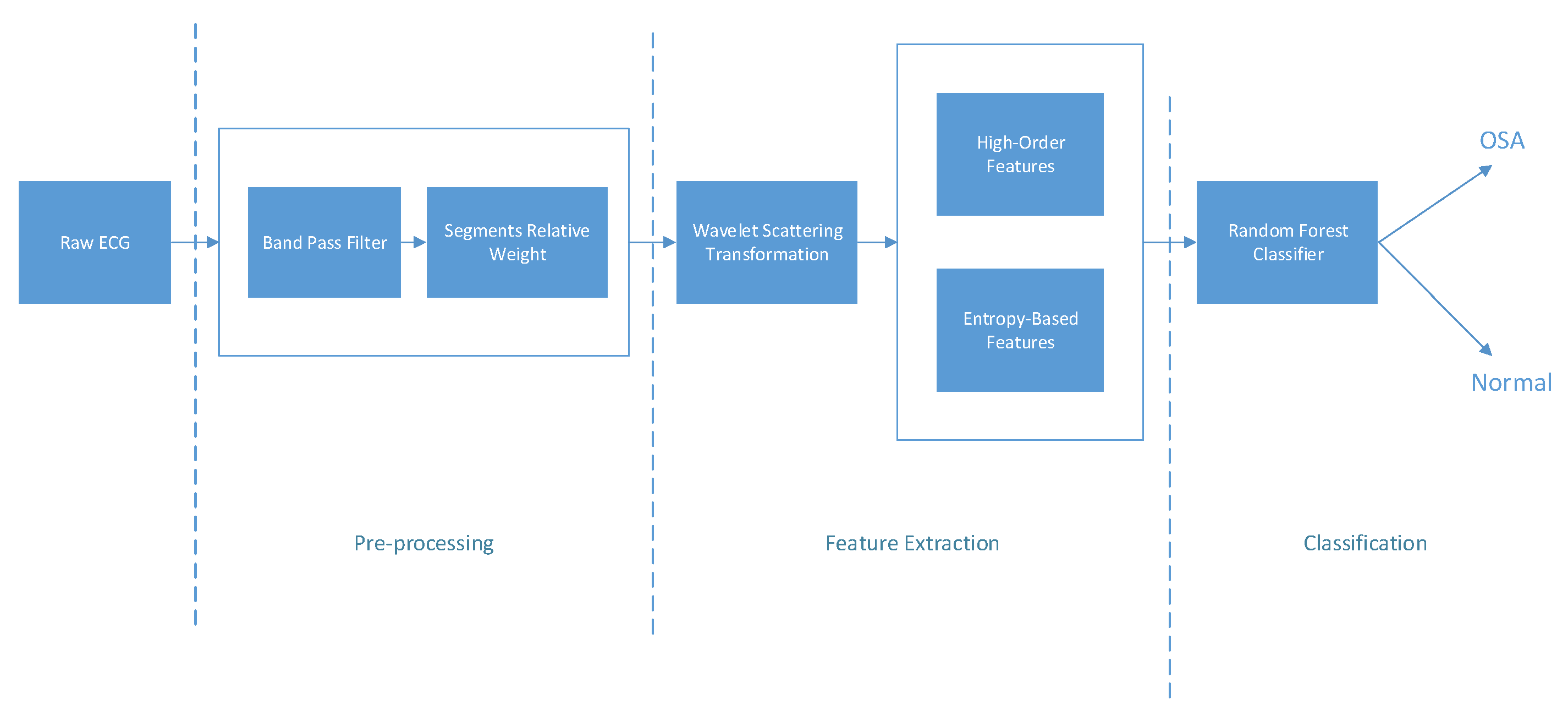
2. Materials and Methods
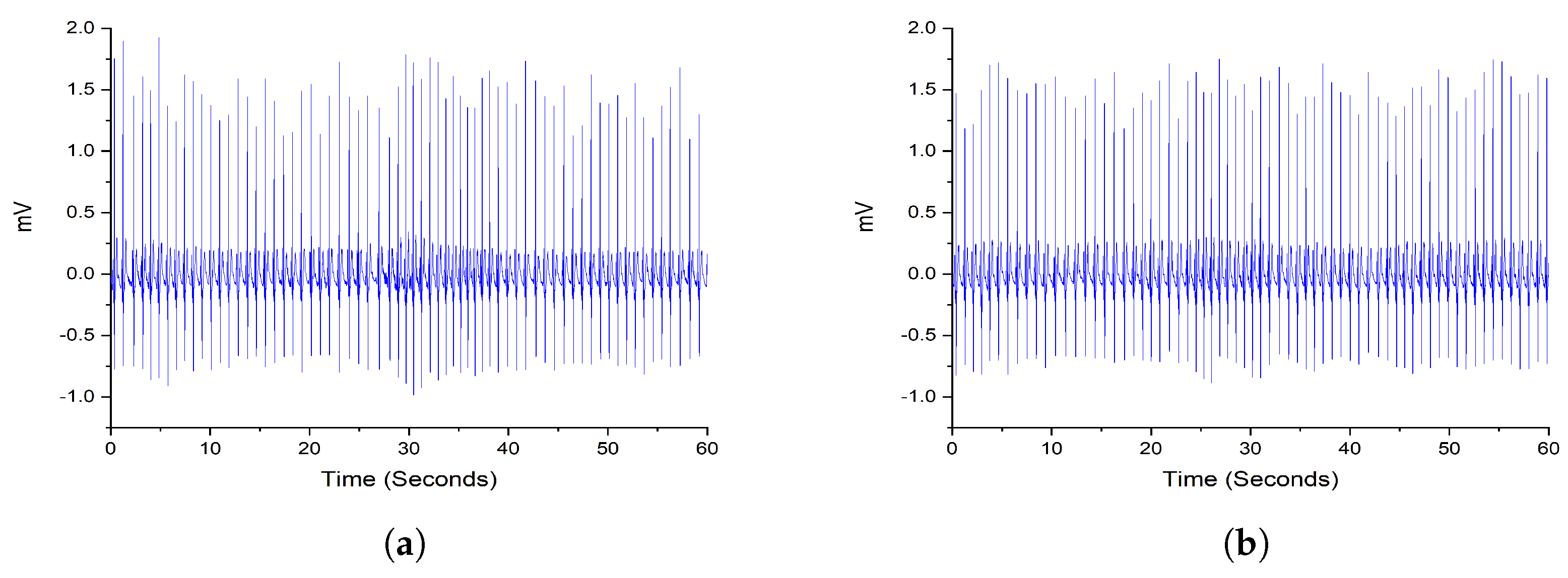
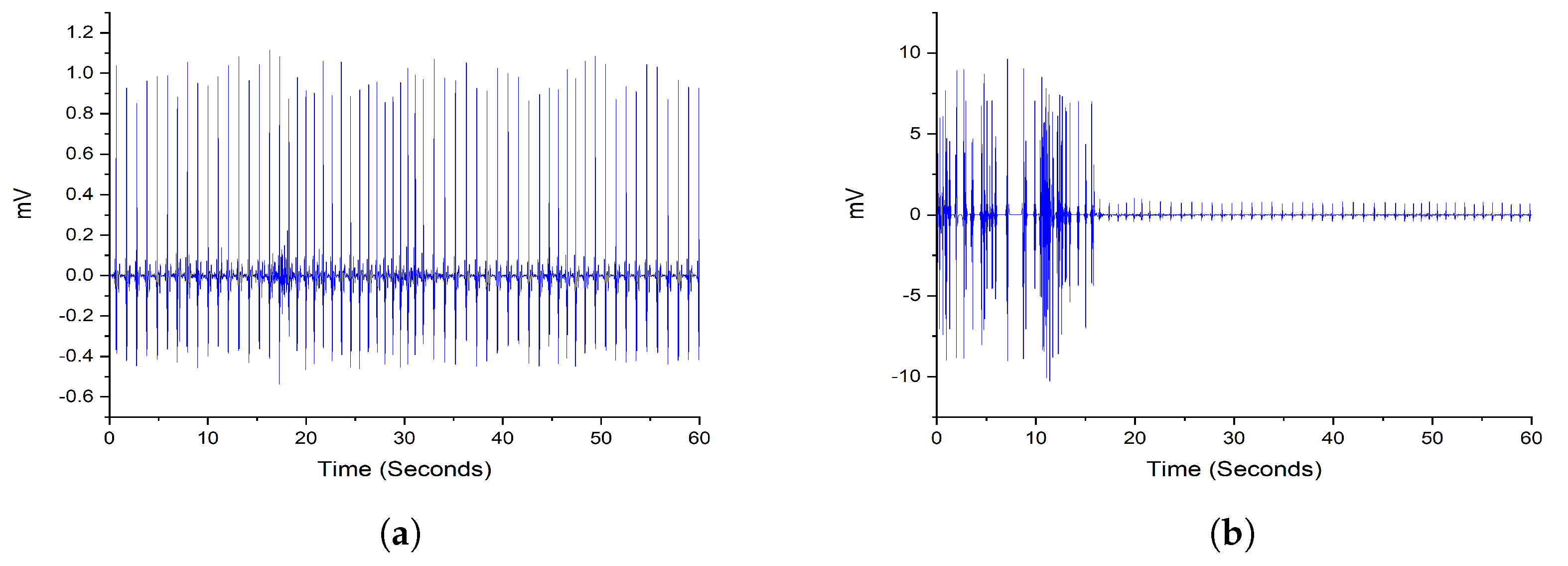
2.1. Dataset Description
- Normal: AHI .
- Mild: AHI .
- Moderate: AHI .
- Severe: AHI .
2.2. Preprocessing
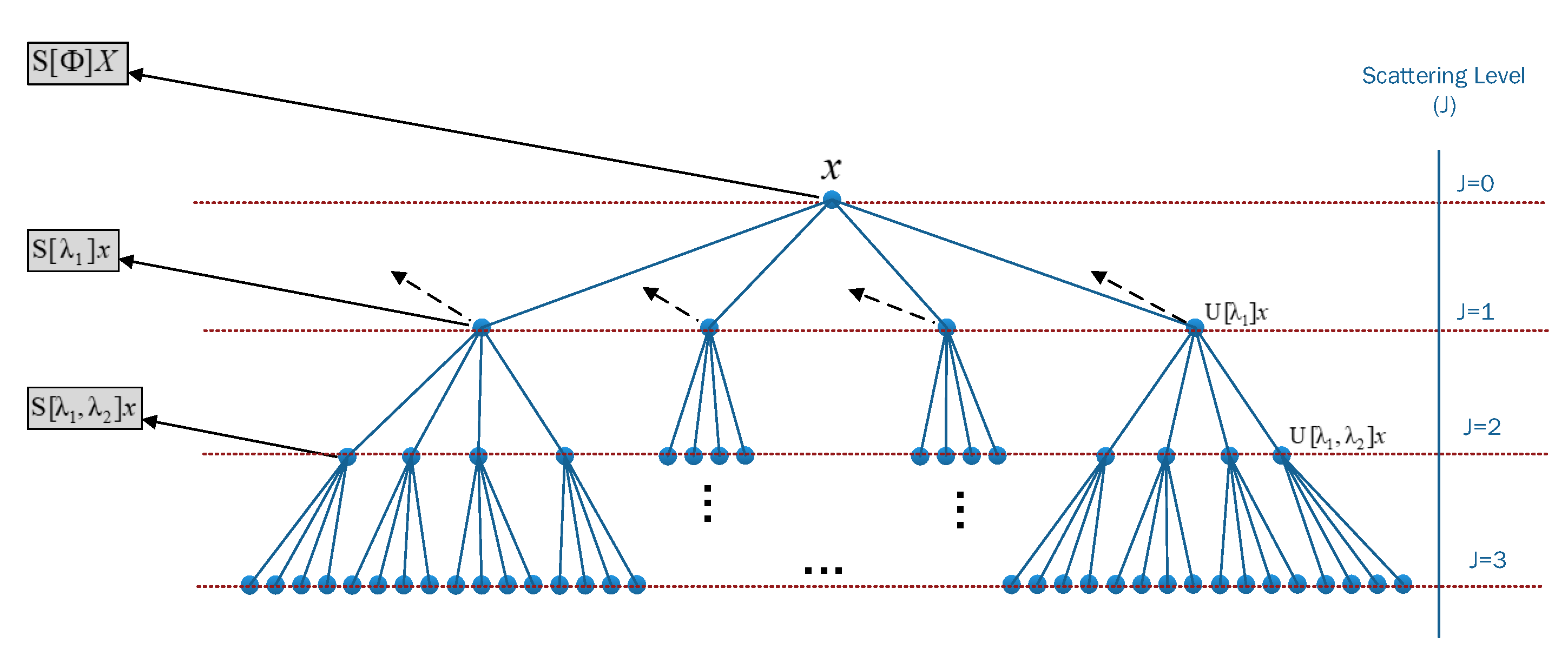
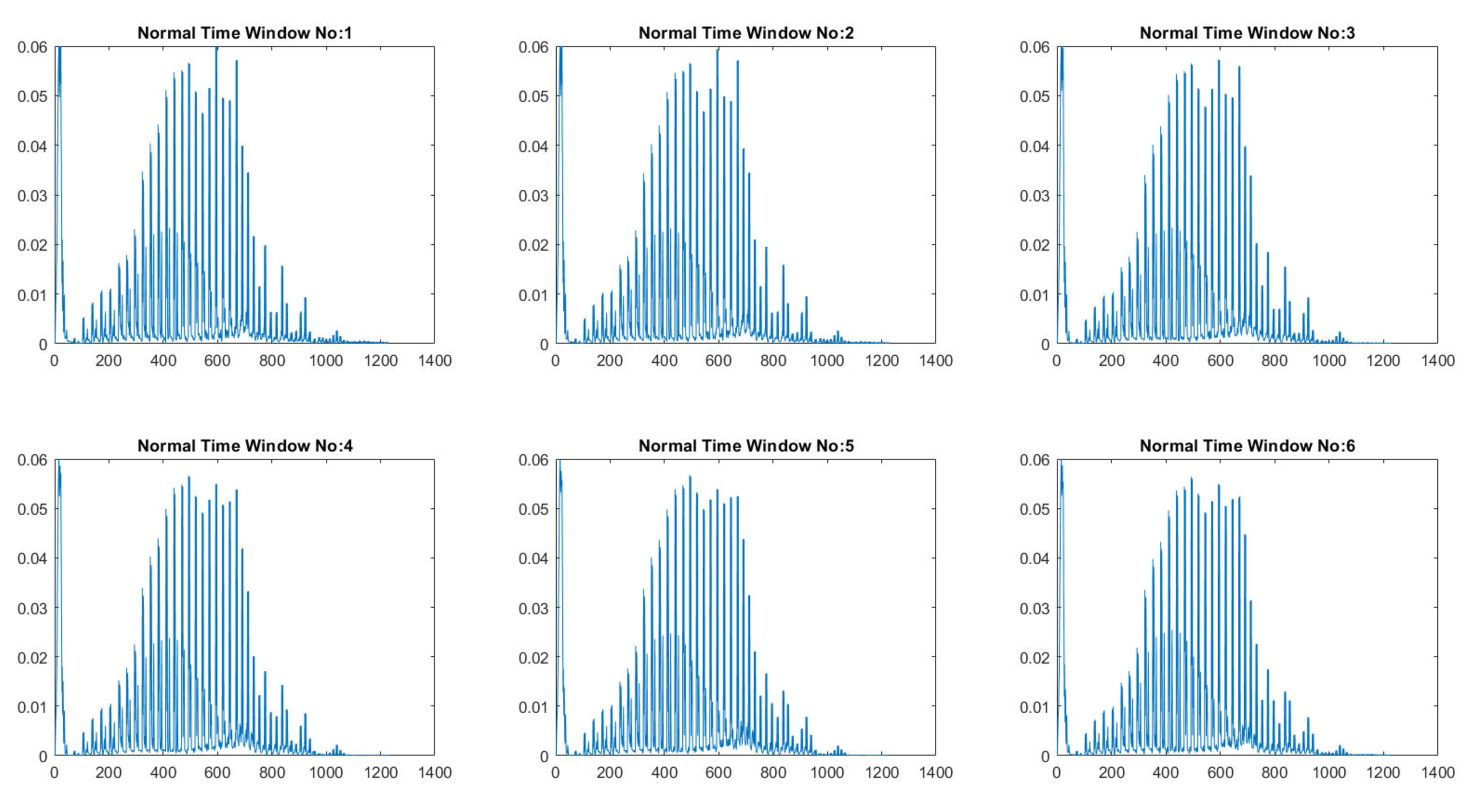
2.3. Wavelet Decomposition Using Scattering Wavelet
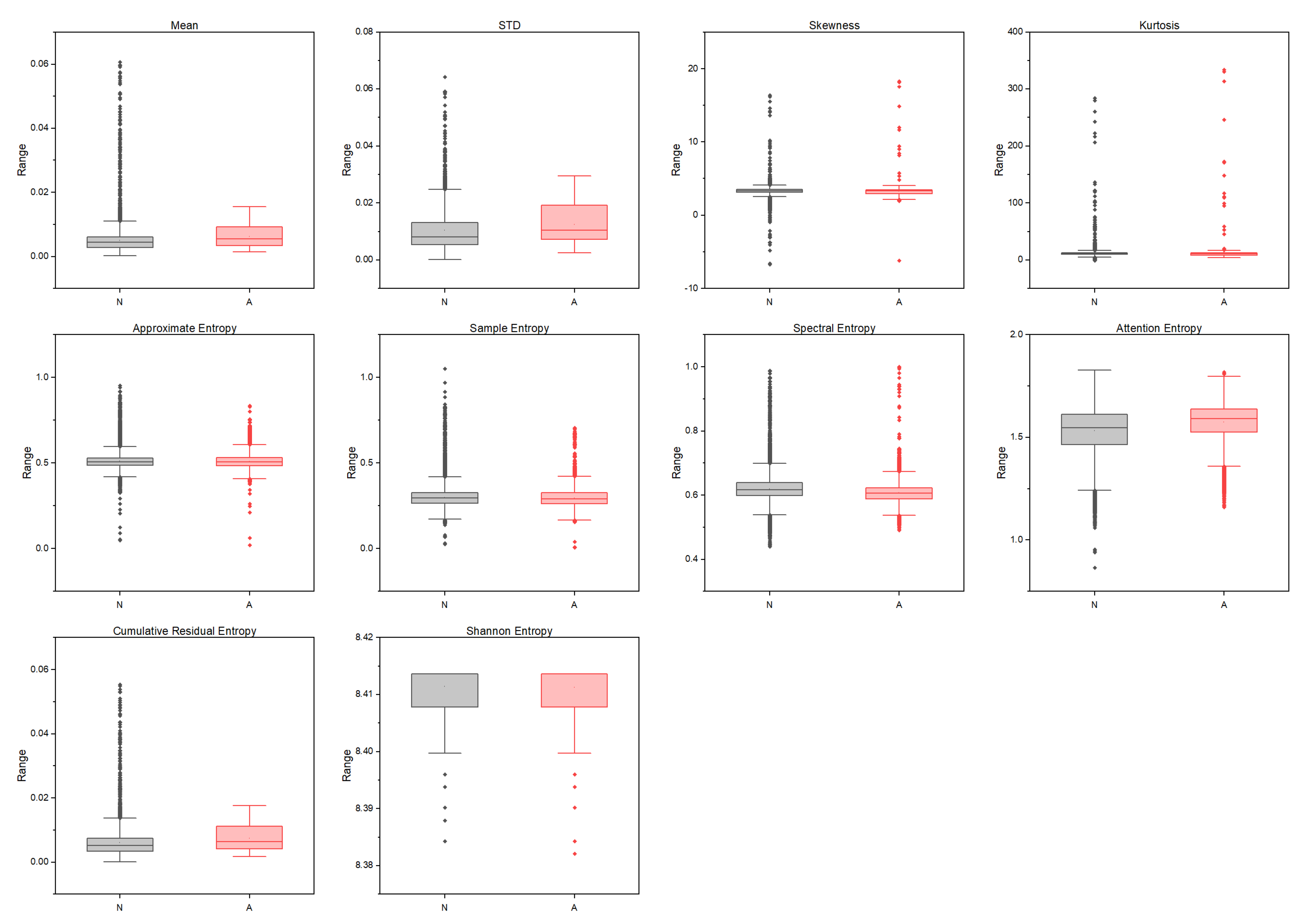
2.4. High-Order and Entropy-Based Features
2.4.1. High-Order Moments Features
2.4.2. Shannon Entropy
- The patterns in the time-series data must be complex enough to model the data. Therefore, it requires a lot of data to populate all histogram bins to obtain a dense histogram.
- The calculation of the Shannon entropy is a time-consuming process.
2.4.3. Approximate Entropy
2.4.4. Sample Entropy
- (1)
- The value of the ApEn depends on the value of r, which does not guarantee consistency.
- (2)
- The value of the ApEn depends on the length of the data segment.
2.4.5. Spectral Entropy
2.4.6. Attention Entropy
2.4.7. Cumulative Residual Entropy
2.5. OSA Classification
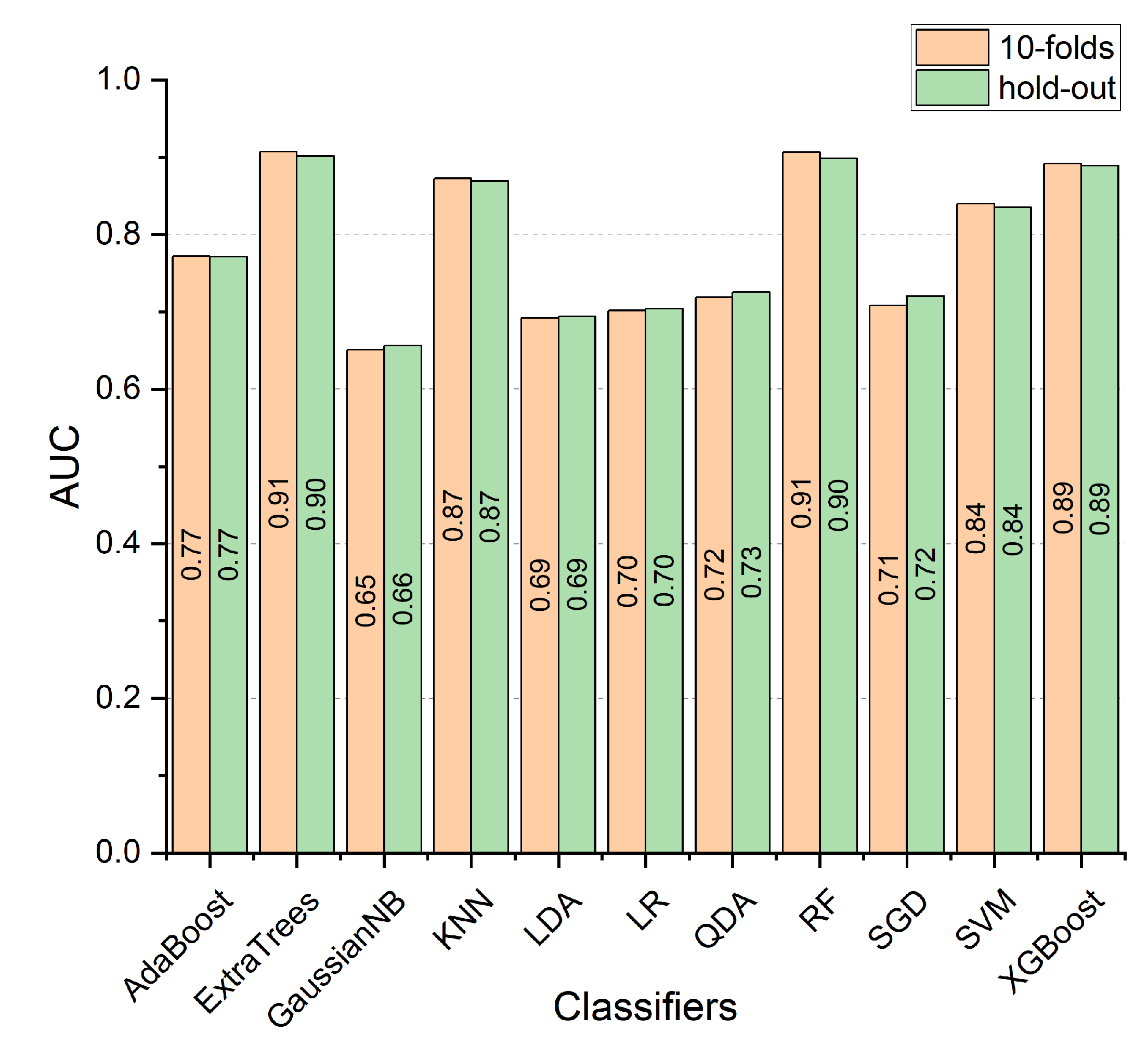
3. Results
3.1. Performance Measures
3.2. Experimental Results
4. Discussion
Comparative Analysis
5. Conclusions
Funding
Conflicts of Interest
References
- Pepin, J.L.; Bailly, S.; Tamisier, R. Big Data in sleep apnoea: Opportunities and challenges. Respirology 2020, 25, 486–494. [Google Scholar] [CrossRef]
- Mendonca, F.; Mostafa, S.S.; Ravelo-Garcia, A.G.; Morgado-Dias, F.; Penzel, T. A Review of Obstructive Sleep Apnea Detection Approaches. IEEE J. Biomed. Health Inform. 2019, 23, 825–837. [Google Scholar] [CrossRef]
- Haidar, R.; Koprinska, I.; Jeffries, B. Sleep Apnea Event Detection from Nasal Airflow Using Convolutional Neural Networks. In Proceedings of the International Conference on Neural Information Processing, Long Beach, CA, USA, 4–9 December 2017; Springer: Berlin/Heidelberg, Germany, 2017; Volume 10638, pp. 819–827. [Google Scholar] [CrossRef]
- Alvarez, D.; Cerezo-Hernandez, A.; Crespo, A.; Gutierrez-Tobal, G.C.; Vaquerizo-Villar, F.; Barroso-Garcia, V.; Moreno, F.; Arroyo, C.A.; Ruiz, T.; Hornero, R.; et al. A machine learning-based test for adult sleep apnoea screening at home using oximetry and airflow. Sci. Rep. 2020, 10, 5332. [Google Scholar] [CrossRef]
- Bertoni, D.; Sterni, L.M.; Pereira, K.D.; Das, G.; Isaiah, A. Predicting polysomnographic severity thresholds in children using machine learning. Pediatr. Res. 2020, 88, 404–411. [Google Scholar] [CrossRef]
- Bozkurt, F.; Uçar, M.K.; Bozkurt, M.R.; Bilgin, C. Detection of Abnormal Respiratory Events with Single Channel ECG and Hybrid Machine Learning Model in Patients with Obstructive Sleep Apnea. IRBM 2020, 41, 241–251. [Google Scholar] [CrossRef]
- Bozkurt, F.; Ucar, M.K.; Bilgin, C.; Zengin, A. Sleep-wake stage detection with single channel ECG and hybrid machine learning model in patients with obstructive sleep apnea. Phys. Eng. Sci. Med. 2021, 44, 63–77. [Google Scholar] [CrossRef]
- Penzel, T.; McNames, J.; de Chazal, P.; Raymond, B.; Murray, A.; Moody, G. Systematic comparison of different algorithms for apnoea detection based on electrocardiogram recordings. Med. Biol. Eng. Comput. 2002, 40, 402–407. [Google Scholar] [CrossRef]
- Chang, H.Y.; Yeh, C.Y.; Lee, C.T.; Lin, C.C. A Sleep Apnea Detection System Based on a One-Dimensional Deep Convolution Neural Network Model Using Single-Lead Electrocardiogram. Sensors 2020, 20, 4157. [Google Scholar] [CrossRef]
- Almazaydeh, L.; Elleithy, K.; Faezipour, M. Detection of obstructive sleep apnea through ECG signal features. In Proceedings of the International Conference on Electro/Information Technology, Indianapolis, IN, USA, 6–8 May 2012; pp. 1–6. [Google Scholar] [CrossRef]
- Kesper, K.; Canisius, S.; Penzel, T.; Ploch, T.; Cassel, W. ECG signal analysis for the assessment of sleep-disordered breathing and sleep pattern. Med. Biol. Eng. Comput. 2012, 50, 135–144. [Google Scholar] [CrossRef]
- Jafari, A. Sleep apnoea detection from ECG using features extracted from reconstructed phase space and frequency domain. Biomed. Signal Process. Control 2013, 8, 551–558. [Google Scholar] [CrossRef]
- Sadr, N.; de Chazal, P.; van Schaik, A.; Breen, P. Sleep apnoea episodes recognition by a committee of ELM classifiers from ECG signal. In Proceedings of the 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Milan, Italy, 25–29 August 2015; Volume 2015, pp. 7675–7678. [Google Scholar] [CrossRef]
- Hassan, A.R.; Bashar, S.K.; Bhuiyan, M.I.H. Computerized obstructive sleep apnea diagnosis from single-lead ECG signals using dual-tree complex wavelet transform. In Proceedings of the IEEE Region 10 Humanitarian Technology Conference (R10-HTC), Dhaka, Bangladesh, 21–23 December 2017; pp. 43–46. [Google Scholar] [CrossRef]
- Zarei, A.; Asl, B.M. Automatic Detection of Obstructive Sleep Apnea Using Wavelet Transform and Entropy-Based Features From Single-Lead ECG Signal. IEEE J. Biomed. Health Inform. 2019, 23, 1011–1021. [Google Scholar] [CrossRef]
- Koda, T.; Sakamoto, T.; Okumura, S.; Taki, H.; Hamada, S.; Chin, K. Radar-Based Automatic Detection of Sleep Apnea Using Support Vector Machine. In Proceedings of the 2020 International Symposium on Antennas and Propagation (ISAP), Osaka, Japan, 25–28 January 2021. [Google Scholar] [CrossRef]
- Choi, J.W.; Kim, D.H.; Koo, D.L.; Park, Y.; Nam, H.; Lee, J.H.; Kim, H.J.; Hong, S.N.; Jang, G.; Lim, S.; et al. Automated Detection of Sleep Apnea-Hypopnea Events Based on 60 GHz Frequency-Modulated Continuous-Wave Radar Using Convolutional Recurrent Neural Networks: A Preliminary Report of a Prospective Cohort Study. Sensors 2022, 22, 7177. [Google Scholar] [CrossRef]
- Mukherjee, D.; Dhar, K.; Schwenker, F.; Sarkar, R. Ensemble of Deep Learning Models for Sleep Apnea Detection: An Experimental Study. Sensors 2021, 21, 5425. [Google Scholar] [CrossRef]
- Gupta, K.; Bajaj, V.; Ansari, I.A. OSACN-Net: Automated Classification of Sleep Apnea Using Deep Learning Model and Smoothed Gabor Spectrograms of ECG Signal. IEEE Trans. Instrum. Meas. 2022, 71, 1–9. [Google Scholar] [CrossRef]
- Aljabri, M.; AlGhamdi, M. A review on the use of deep learning for medical images segmentation. Neurocomputing 2022, 506, 311–335. [Google Scholar] [CrossRef]
- Mallat, S. Understanding deep convolutional networks. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2016, 374, 20150203. [Google Scholar] [CrossRef] [PubMed]
- Souli, S.; Lachiri, Z. Audio sounds classification using scattering features and support vectors machines for medical surveillance. Appl. Acoust. 2018, 130, 270–282. [Google Scholar] [CrossRef]
- Liu, Z.; Yao, G.; Zhang, Q.; Zhang, J.; Zeng, X. Wavelet Scattering Transform for ECG Beat Classification. Comput. Math. Methods Med. 2020, 2020, 1–11. [Google Scholar] [CrossRef]
- Penzel, T.; Moody, G.B.; Mark, R.G.; Goldberger, A.L.; Peter, J.H. Apnea-ECG Database. In Proceedings of the Computers in Cardiology 2000, Cambridge, MA, USA, 24–27 September 2000. [Google Scholar] [CrossRef]
- de Chazal, P.; Penzel, T.; Heneghan, C. Automated detection of obstructive sleep apnoea at different time scales using the electrocardiogram. Physiol. Meas. 2004, 25, 967–983. [Google Scholar] [CrossRef]
- Bruna, J.; Mallat, S. Invariant scattering convolution networks. IEEE Trans. Pattern Anal. Mach. Intell. 2012, 35, 1872–1886. [Google Scholar] [CrossRef]
- Bruna, J.; Mallat, S. Classification with scattering operators. In Proceedings of the CVPR 2011, Colorado Springs, CO, USA, 20–25 June 2011. [Google Scholar] [CrossRef]
- Ahmad, M.Z.; Kamboh, A.M.; Saleem, S.; Khan, A.A. Mallat’s Scattering Transform Based Anomaly Sensing for Detection of Seizures in Scalp EEG. IEEE Access 2017, 5, 16919–16929. [Google Scholar] [CrossRef]
- Delgado-Bonal, A.; Marshak, A. Approximate Entropy and Sample Entropy: A Comprehensive Tutorial. Entropy 2019, 21, 541. [Google Scholar] [CrossRef] [PubMed]
- Blanco, S.; Garay, A.; Coulombie, D. Comparison of Frequency Bands Using Spectral Entropy for Epileptic Seizure Prediction. ISRN Neurol. 2013, 2013, 1–5. [Google Scholar] [CrossRef]
- Yang, J.; Choudhary, G.I.; Rahardja, S.; Franti, P. Classification of Interbeat Interval Time-series Using Attention Entropy. IEEE Trans. Affect. Comput. 2021. [Google Scholar] [CrossRef]
- Rao, M.; Chen, Y.; Vemuri, B.; Wang, F. Cumulative Residual Entropy: A New Measure of Information. IEEE Trans. Inf. Theory 2004, 50, 1220–1228. [Google Scholar] [CrossRef]
- Alshamrani, M. IoT and artificial intelligence implementations for remote healthcare monitoring systems: A survey. J. King Saud Univ. Comput. Inf. Sci. 2022, 34, 4687–4701. [Google Scholar] [CrossRef]
- Hamza, M.A.; Hashim, A.H.A.; Alsolai, H.; Gaddah, A.; Othman, M.; Yaseen, I.; Rizwanullah, M.; Zamani, A.S. Wearables-Assisted Smart Health Monitoring for Sleep Quality Prediction Using Optimal Deep Learning. Sustainability 2023, 15, 1084. [Google Scholar] [CrossRef]
- Alotaibi, M.; Alotaibi, S.S. Optimal Disease Diagnosis in Internet of Things (IoT) Based Healthcare System Using Energy Efficient Clustering. Appl. Sci. 2022, 12, 3804. [Google Scholar] [CrossRef]
- Duhayyim, M.A.; Mohamed, H.G.; Aljebreen, M.; Nour, M.K.; Mohamed, A.; Abdelmageed, A.A.; Yaseen, I.; Mohammed, G.P. Artificial Ecosystem-Based Optimization with an Improved Deep Learning Model for IoT-Assisted Sustainable Waste Management. Sustainability 2022, 14, 11704. [Google Scholar] [CrossRef]
- Hassan, A.R.; Haque, M.A. An expert system for automated identification of obstructive sleep apnea from single-lead ECG using random under sampling boosting. Neurocomputing 2017, 235, 122–130. [Google Scholar] [CrossRef]
- Varon, C.; Caicedo, A.; Testelmans, D.; Buyse, B.; Huffel, S.V. A Novel Algorithm for the Automatic Detection of Sleep Apnea From Single-Lead ECG. IEEE Trans. Biomed. Eng. 2015, 62, 2269–2278. [Google Scholar] [CrossRef]
- Nguyen, H.D.; Wilkins, B.A.; Cheng, Q.; Benjamin, B.A. An Online Sleep Apnea Detection Method Based on Recurrence Quantification Analysis. IEEE J. Biomed. Health Inform. 2014, 18, 1285–1293. [Google Scholar] [CrossRef]
- Hassan, A.R. Computer-aided obstructive sleep apnea detection using normal inverse Gaussian parameters and adaptive boosting. Biomed. Signal Process. Control 2016, 29, 22–30. [Google Scholar] [CrossRef]
- Hassan, A.R.; Haque, M.A. Computer-aided obstructive sleep apnea screening from single-lead electrocardiogram using statistical and spectral features and bootstrap aggregating. Biocybern. Biomed. Eng. 2016, 36, 256–266. [Google Scholar] [CrossRef]
- Hassan, A.R.; Haque, M.A. Computer-aided obstructive sleep apnea identification using statistical features in the EMD domain and extreme learning machine. Biomed. Phys. Eng. Express 2016, 2, 035003. [Google Scholar] [CrossRef]
- Sharma, M.; Raval, M.; Acharya, U.R. A new approach to identify obstructive sleep apnea using an optimal orthogonal wavelet filter bank with ECG signals. Inform. Med. Unlocked 2019, 16, 100170. [Google Scholar] [CrossRef]
- Tripathy, R. Application of intrinsic band function technique for automated detection of sleep apnea using HRV and EDR signals. Biocybern. Biomed. Eng. 2018, 38, 136–144. [Google Scholar] [CrossRef]
- Pinho, A.; Pombo, N.; Silva, B.M.C.; Bousson, K.; Garcia, N. Towards an accurate sleep apnea detection based on ECG signal: The quintessential of a wise feature selection. Appl. Soft Comput. 2019, 83, 105568. [Google Scholar] [CrossRef]
- Padovano, D.; Martinez-Rodrigo, A.; Pastor, J.M.; Rieta, J.J.; Alcaraz, R. On the Generalization of Sleep Apnea Detection Methods Based on Heart Rate Variability and Machine Learning. IEEE Access 2022, 10, 92710–92725. [Google Scholar] [CrossRef]


| Classifier | ACC | SEN | SPE | Precision | F1 | Kappa |
|---|---|---|---|---|---|---|
| AdaBoost | ||||||
| ExtraTrees | ||||||
| GaussianNB | ||||||
| KNN | ||||||
| LDA | ||||||
| LR | ||||||
| QDA | ||||||
| RF | ||||||
| SGD | ||||||
| SVM | ||||||
| XgBoost |
| Classifier | ACC | SEN | SPE | Precision | F1 | Kappa |
|---|---|---|---|---|---|---|
| AdaBoost | ||||||
| ExtraTrees | ||||||
| GaussianNB | ||||||
| KNN | ||||||
| LDA | ||||||
| LR | ||||||
| QDA | ||||||
| RF | ||||||
| SGD | ||||||
| SVM | ||||||
| XgBoost |
| Classifier | ACC | SEN | SPE | Precision | F1 | Kappa |
|---|---|---|---|---|---|---|
| AdaBoost | ||||||
| ExtraTrees | ||||||
| GaussianNB | ||||||
| KNN | ||||||
| LDA | ||||||
| LR | ||||||
| QDA | ||||||
| RF | ||||||
| SGD | ||||||
| SVM | ||||||
| XgBoost |
| Classifier | ACC | SEN | SPE | Precision | F1 | Kappa |
|---|---|---|---|---|---|---|
| AdaBoost | ||||||
| ExtraTrees | ||||||
| GaussianNB | ||||||
| KNN | ||||||
| LDA | ||||||
| LR | ||||||
| QDA | ||||||
| RF | ||||||
| SGD | ||||||
| SVM | ||||||
| XgBoost |
| Methodology | ACC | SEN | SPE |
|---|---|---|---|
| QRS + RR intervals [10] | 96.5 | 92.9 | 100 |
| QRS [11] | 80.5 | - | - |
| QRS + Reconstructed Phase Space [12] | 94.8 | 94.16 | 95.42 |
| RR + extreme learning machine [13] | 82.5 | 81.9 | 82.8 |
| dual-tree complex wavelet transform [14] | 84.4 | 90.38 | 74.84 |
| TQWT + Statistical Features [37] | 88.88 | 87.58 | 91.49 |
| DWT + Non-linear features [15] | 92.98 | 91.74 | 93.75 |
| QRS [38] | 84.74 | 84.71 | 84.69 |
| RQA + HRV [39] | 85.26 | 86.37 | 83.37 |
| TQWT + normal inverse Gaussian [40] | 87.33 | 81.99 | 90.72 |
| statistical and spectral features [41] | 85.97 | 84.14 | 86.83 |
| Empirical Mode Decomposition [42] | 83.77 | 85.2 | 82.79 |
| wavelet filter banks + non-linear [43] | 90.87 | 92.43 | 88.33 |
| HRV + Non-linear features [44] | 77.27 | 79.25 | 75.32 |
| QRS [45] | 82.12 | 88.41 | 72.29 |
| Bandpass Filtering + CNN [9] | 87.9 | 81.1 | 92 |
| RRI + CNN [18] | 85.58 | - | 88.26 |
| deep learning model [19] | 94.81 | - | - |
| HRV + Ensemble Learning [46] | 84.61 | 84.78 | 84.44 |
| WST + Entropy-based Features | 91.65 | 88.83 | 92.47 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharaf, A.I. Sleep Apnea Detection Using Wavelet Scattering Transformation and Random Forest Classifier. Entropy 2023, 25, 399. https://doi.org/10.3390/e25030399
Sharaf AI. Sleep Apnea Detection Using Wavelet Scattering Transformation and Random Forest Classifier. Entropy. 2023; 25(3):399. https://doi.org/10.3390/e25030399
Chicago/Turabian StyleSharaf, Ahmed I. 2023. "Sleep Apnea Detection Using Wavelet Scattering Transformation and Random Forest Classifier" Entropy 25, no. 3: 399. https://doi.org/10.3390/e25030399
APA StyleSharaf, A. I. (2023). Sleep Apnea Detection Using Wavelet Scattering Transformation and Random Forest Classifier. Entropy, 25(3), 399. https://doi.org/10.3390/e25030399






