Deep Learning-Based Segmentation of Head and Neck Organs-at-Risk with Clinical Partially Labeled Data
Abstract
1. Introduction
1.1. Contouring in HN Radiotherapy
1.2. Literature Review
2. Materials and Methods
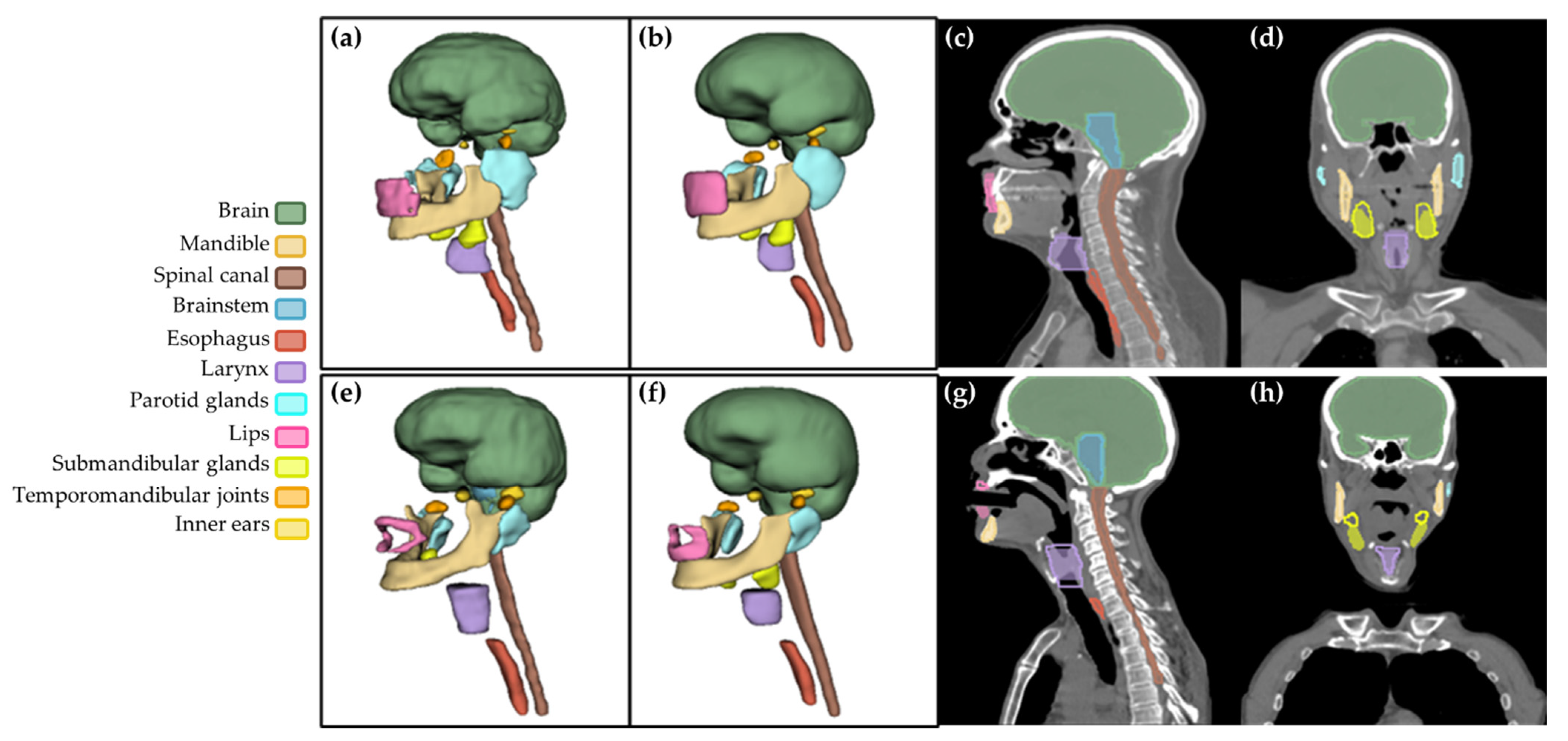
2.1. Database
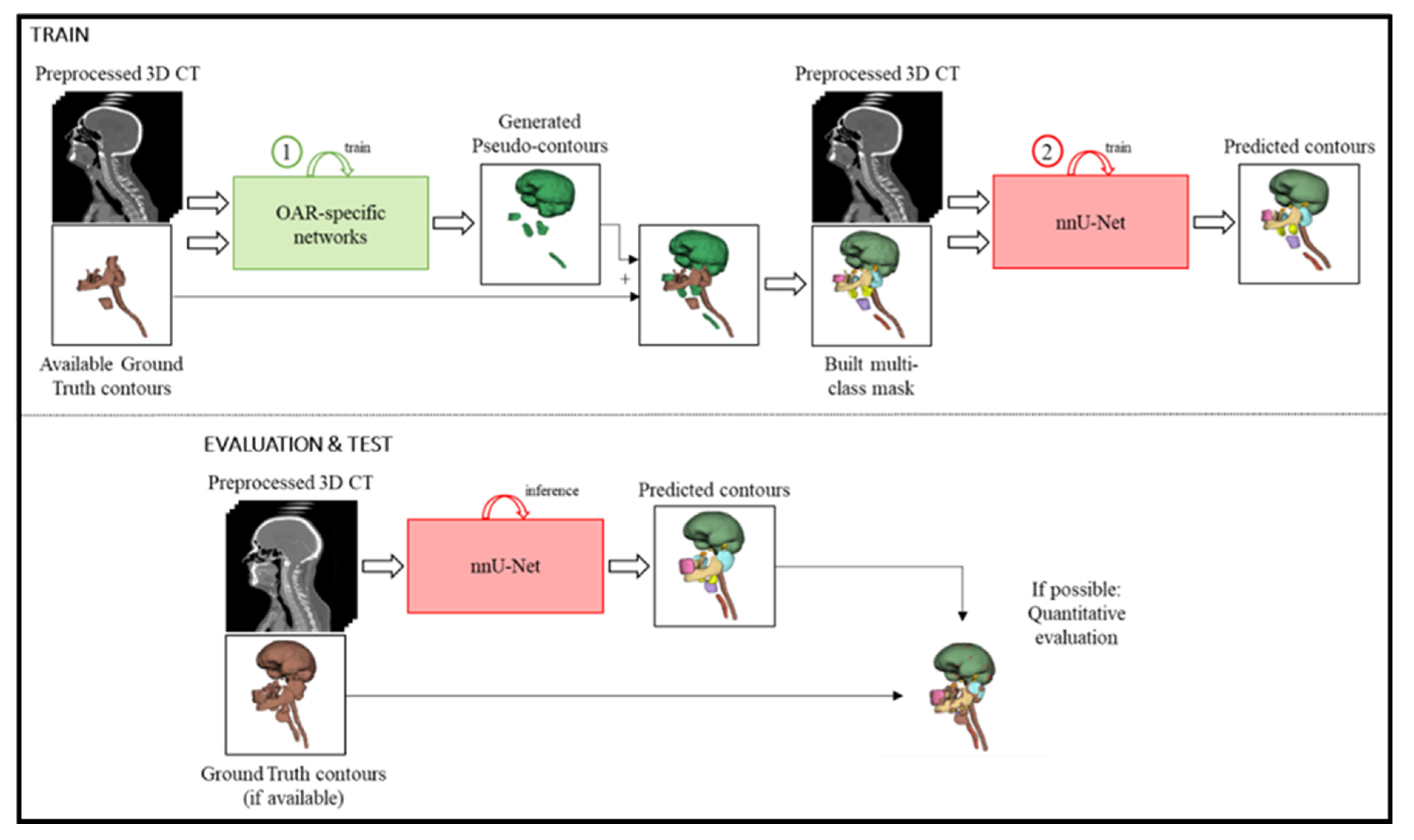
2.2. Proposed Framework
2.2.1. Preprocessing
2.2.2. OAR-Specific Models
2.2.3. Pseudo-Contour Generation
2.2.4. Multi-Class Network
3. Results
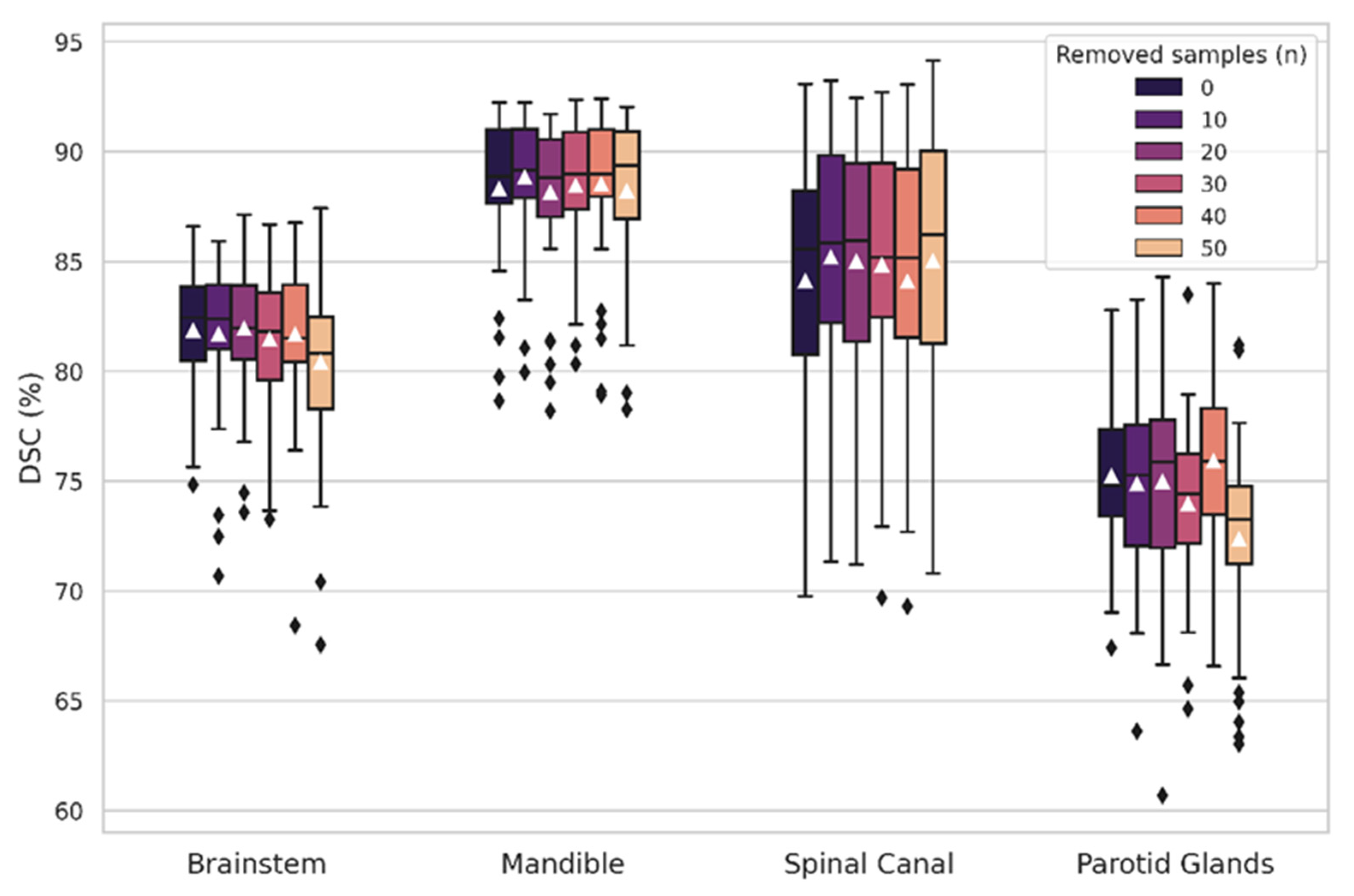
3.1. OAR-Specific Models
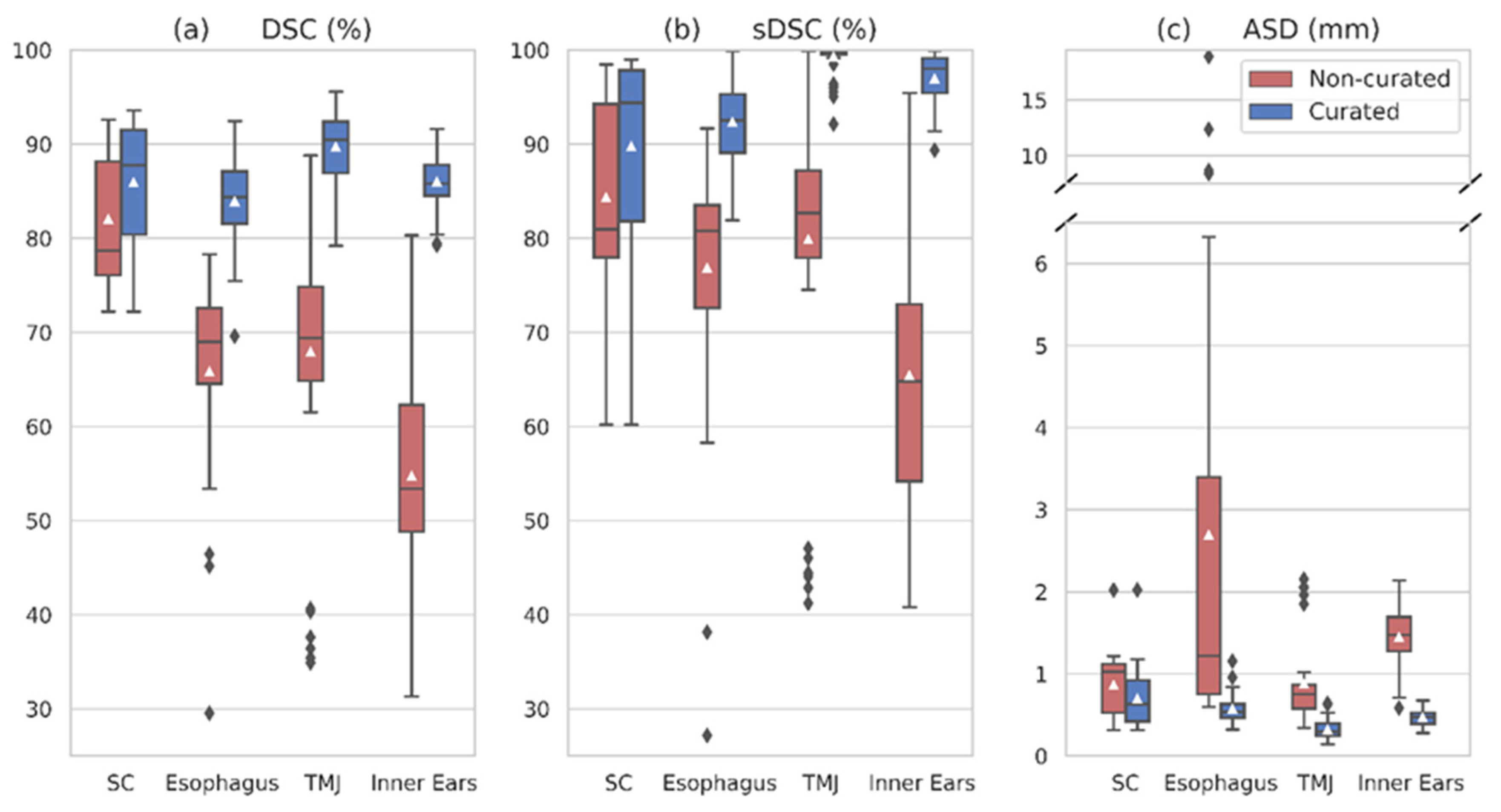
3.2. Multi-Class Network
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- International Agency for Research on Cancer; World Health Organization. Estimated Number of New Cases in 2020, World, Males, Ages 45+ (Excl. NMSC). Cancer Today. Available online: https://gco.iarc.fr/today/ (accessed on 17 September 2022).
- Kawakita, D.; Oze, I.; Iwasaki, S.; Matsuda, T.; Matsuo, K.; Ito, H. Trends in the Incidence of Head and Neck Cancer by Subsite between 1993 and 2015 in Japan. Cancer Med. 2022, 11, 1553–1560. [Google Scholar] [CrossRef] [PubMed]
- Nikolov, S.; Blackwell, S.; Zverovitch, A.; Mendes, R.; Livne, M.; De Fauw, J.; Patel, Y.; Meyer, C.; Askham, H.; Romera-Paredes, B.; et al. Deep Learning to Achieve Clinically Applicable Segmentation of Head and Neck Anatomy for Radiotherapy. arXiv 2021, arXiv:1809.04430. [Google Scholar]
- Sherer, M.V.; Lin, D.; Elguindi, S.; Duke, S.; Tan, L.-T.; Cacicedo, J.; Dahele, M.; Gillespie, E.F. Metrics to Evaluate the Performance of Auto-Segmentation for Radiation Treatment Planning: A Critical Review. Radiother. Oncol. 2021, 160, 185–191. [Google Scholar] [CrossRef]
- Willems, S.; Crijns, W.; La Greca Saint-Esteven, A.; Van Der Veen, J.; Robben, D.; Depuydt, T.; Nuyts, S.; Haustermans, K.; Maes, F. Clinical Implementation of DeepVoxNet for Auto-Delineation of Organs at Risk in Head and Neck Cancer Patients in Radiotherapy. In OR 2.0 Context-Aware Operating Theaters, Computer Assisted Robotic Endoscopy, Clinical Image-Based Procedures, and Skin Image Analysis; Stoyanov, D., Taylor, Z., Sarikaya, D., McLeod, J., González Ballester, M.A., Codella, N.C.F., Martel, A., Maier-Hein, L., Malpani, A., Zenati, M.A., et al., Eds.; Lecture Notes in Computer Science; Springer International Publishing: Cham, Switzerland, 2018; Volume 11041, pp. 223–232. [Google Scholar] [CrossRef]
- Robert, C.; Munoz, A.; Moreau, D.; Mazurier, J.; Sidorski, G.; Gasnier, A.; Beldjoudi, G.; Grégoire, V.; Deutsch, E.; Meyer, P.; et al. Clinical Implementation of Deep-Learning Based Auto-Contouring Tools—Experience of Three French Radiotherapy Centers. Cancer/Radiothér. 2021, 25, 607–616. [Google Scholar] [CrossRef] [PubMed]
- Chi, W.; Ma, L.; Wu, J.; Chen, M.; Lu, W.; Gu, X. Deep Learning-Based Medical Image Segmentation with Limited Labels. Phys. Med. Biol. 2020, 65, 235001. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Yang, Y.; Fang, Y.; Wang, J.; Hu, W. A Preliminary Experience of Implementing Deep-Learning Based Auto-Segmentation in Head and Neck Cancer: A Study on Real-World Clinical Cases. Front. Oncol. 2021, 11, 638197. [Google Scholar] [CrossRef] [PubMed]
- Harrison, K.; Pullen, H.; Welsh, C.; Oktay, O.; Alvarez-Valle, J.; Jena, R. Machine Learning for Auto-Segmentation in Radiotherapy Planning. Clin. Oncol. 2022, 34, 74–88. [Google Scholar] [CrossRef]
- Oktay, O.; Nanavati, J.; Schwaighofer, A.; Carter, D.; Bristow, M.; Tanno, R.; Jena, R.; Barnett, G.; Noble, D.; Rimmer, Y.; et al. Evaluation of Deep Learning to Augment Image-Guided Radiotherapy for Head and Neck and Prostate Cancers. JAMA Netw. Open 2020, 3, e2027426. [Google Scholar] [CrossRef]
- Liu, C.; Zhang, X.; Si, W.; Ni, X. Multiview Self-Supervised Segmentation for OARs Delineation in Radiotherapy. Evid. Based Complement. Altern. Med. 2021, 2021, 8894222. [Google Scholar] [CrossRef]
- Li, Y.; Rao, S.; Chen, W.; Azghadi, S.F.; Nguyen, K.N.B.; Moran, A.; Usera, B.M.; Dyer, B.A.; Shang, L.; Chen, Q.; et al. Evaluating Automatic Segmentation for Swallowing-Related Organs for Head and Neck Cancer. Technol. Cancer Res. Treat. 2022, 21, 153303382211057. [Google Scholar] [CrossRef]
- van Rooij, W.; Dahele, M.; Ribeiro Brandao, H.; Delaney, A.R.; Slotman, B.J.; Verbakel, W.F. Deep Learning-Based Delineation of Head and Neck Organs at Risk: Geometric and Dosimetric Evaluation. Int. J. Radiat. Oncol. 2019, 104, 677–684. [Google Scholar] [CrossRef] [PubMed]
- Brouwer, C.L.; Boukerroui, D.; Oliveira, J.; Looney, P.; Steenbakkers, R.J.H.M.; Langendijk, J.A.; Both, S.; Gooding, M.J. Assessment of Manual Adjustment Performed in Clinical Practice Following Deep Learning Contouring for Head and Neck Organs at Risk in Radiotherapy. Phys. Imaging Radiat. Oncol. 2020, 16, 54–60. [Google Scholar] [CrossRef] [PubMed]
- van Dijk, L.V.; Van den Bosch, L.; Aljabar, P.; Peressutti, D.; Both, S.; Steenbakkers, R.J.H.M.; Langendijk, J.A.; Gooding, M.J.; Brouwer, C.L. Improving Automatic Delineation for Head and Neck Organs at Risk by Deep Learning Contouring. Radiother. Oncol. 2020, 142, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.; Fong, A.; McVicar, N.; Smith, S.; Giambattista, J.; Wells, D.; Kolbeck, C.; Giambattista, J.; Gondara, L.; Alexander, A. Comparing Deep Learning-Based Auto-Segmentation of Organs at Risk and Clinical Target Volumes to Expert Inter-Observer Variability in Radiotherapy Planning. Radiother. Oncol. 2020, 144, 152–158. [Google Scholar] [CrossRef]
- Thor, M.; Iyer, A.; Jiang, J.; Apte, A.; Veeraraghavan, H.; Allgood, N.B.; Kouri, J.A.; Zhou, Y.; LoCastro, E.; Elguindi, S.; et al. Deep Learning Auto-Segmentation and Automated Treatment Planning for Trismus Risk Reduction in Head and Neck Cancer Radiotherapy. Phys. Imaging Radiat. Oncol. 2021, 19, 96–101. [Google Scholar] [CrossRef]
- Rhee, D.J.; Cardenas, C.E.; Elhalawani, H.; McCarroll, R.; Zhang, L.; Yang, J.; Garden, A.S.; Peterson, C.B.; Beadle, B.M.; Court, L.E. Automatic Detection of Contouring Errors Using Convolutional Neural Networks. Med. Phys. 2019, 46, 5086–5097. [Google Scholar] [CrossRef]
- Xu, J.; Zeng, B.; Egger, J.; Wang, C.; Smedby, Ö.; Jiang, X.; Chen, X. A Review on AI-Based Medical Image Computing in Head and Neck Surgery. Phys. Med. Biol. 2022, 67, 17TR01. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. arXiv 2015, arXiv:1505.04597. [Google Scholar]
- Koo, J.; Caudell, J.J.; Latifi, K.; Jordan, P.; Shen, S.; Adamson, P.M.; Moros, E.G.; Feygelman, V. Comparative Evaluation of a Prototype Deep Learning Algorithm for Autosegmentation of Normal Tissues in Head and Neck Radiotherapy. Radiother. Oncol. 2022, 174, 52–58. [Google Scholar] [CrossRef]
- Zhong, T.; Huang, X.; Tang, F.; Liang, S.; Deng, X.; Zhang, Y. Boosting-based Cascaded Convolutional Neural Networks for the Segmentation of CT Organs-at-risk in Nasopharyngeal Carcinoma. Med. Phys. 2019, 46, 5602–5611. [Google Scholar] [CrossRef]
- Liang, S.; Tang, F.; Huang, X.; Yang, K.; Zhong, T.; Hu, R.; Liu, S.; Yuan, X.; Zhang, Y. Deep-Learning-Based Detection and Segmentation of Organs at Risk in Nasopharyngeal Carcinoma Computed Tomographic Images for Radiotherapy Planning. Eur. Radiol. 2019, 29, 1961–1967. [Google Scholar] [CrossRef] [PubMed]
- Sultana, S.; Robinson, A.; Song, D.Y.; Lee, J. Automatic Multi-Organ Segmentation in Computed Tomography Images Using Hierarchical Convolutional Neural Network. J. Med. Imaging 2020, 7, 055001. [Google Scholar]
- Tang, H.; Chen, X.; Liu, Y.; Lu, Z.; You, J.; Yang, M.; Yao, S.; Zhao, G.; Xu, Y.; Chen, T.; et al. Clinically Applicable Deep Learning Framework for Organs at Risk Delineation in CT Images. Nat. Mach. Intell. 2019, 1, 480–491. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, H.; Tian, S.; Zhang, X.; Li, J.; Lei, R.; Gao, M.; Liu, C.; Yang, L.; Bi, X.; et al. A Slice Classification Model-Facilitated 3D Encoder–Decoder Network for Segmenting Organs at Risk in Head and Neck Cancer. J. Radiat. Res. 2021, 62, 94–103. [Google Scholar] [CrossRef] [PubMed]
- Guo, D.; Jin, D.; Zhu, Z.; Ho, T.-Y.; Harrison, A.P.; Chao, C.-H.; Xiao, J.; Lu, L. Organ at Risk Segmentation for Head and Neck Cancer Using Stratified Learning and Neural Architecture Search. In Proceedings of the 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 13–19 June 2020; pp. 4222–4231. [Google Scholar] [CrossRef]
- Fang, Y.; Wang, J.; Ou, X.; Ying, H.; Hu, C.; Zhang, Z.; Hu, W. The Impact of Training Sample Size on Deep Learning-Based Organ Auto-Segmentation for Head-and-Neck Patients. Phys. Med. Biol. 2021, 66, 185012. [Google Scholar] [CrossRef] [PubMed]
- Rao, D.; Prakashini, K.; Singh, R.; Vijayananda, J. Automated Segmentation of the Larynx on Computed Tomography Images: A Review. Biomed. Eng. Lett. 2022, 12, 175–183. [Google Scholar] [CrossRef] [PubMed]
- Tryggestad, E.; Anand, A.; Beltran, C.; Brooks, J.; Cimmiyotti, J.; Grimaldi, N.; Hodge, T.; Hunzeker, A.; Lucido, J.J.; Laack, N.N.; et al. Scalable Radiotherapy Data Curation Infrastructure for Deep-Learning Based Autosegmentation of Organs-at-Risk: A Case Study in Head and Neck Cancer. Front. Oncol. 2022, 12, 936134. [Google Scholar] [CrossRef]
- Cubero, L.; Cabezas, E.; Simon, A.; Castelli, J.; de Crevoisier, R.; Serrano, J.; Acosta, O.; Pascau, J. Deep Learning-Based Segmentation of Head and Neck Organs on CT for Radiotherapy Treatment: Lessons Learned with Clinical Data. In Proceedings of the XXXIX Congreso Anual de la Sociedad Española de Ingeniería Biomédica (CASEIB), Madrid, Spain, 25 November 2021; pp. 127–130. [Google Scholar]
- Zhu, W.; Huang, Y.; Zeng, L.; Chen, X.; Liu, Y.; Qian, Z.; Du, N.; Fan, W.; Xie, X. AnatomyNet: Deep Learning for Fast and Fully Automated Whole-volume Segmentation of Head and Neck Anatomy. Med. Phys. 2019, 46, 576–589. [Google Scholar] [CrossRef]
- Castelli, J.; Benezery, K.; Hasbini, A.; Gery, B.; Berger, A.; Liem, X.; Guihard, S.; Chapet, S.; Thureau, S.; Auberdiac, P.; et al. OC-0831 Results of ARTIX phase III study: Adaptive radiotherapy versus standard IMRT in head and neck cancer. Radiother. Oncol. 2022, 170 (Suppl. 1), S749–S750. [Google Scholar] [CrossRef]
- Isensee, F.; Petersen, J.; Klein, A.; Zimmerer, D.; Jaeger, P.F.; Kohl, S.; Wasserthal, J.; Koehler, G.; Norajitra, T.; Wirkert, S.; et al. NnU-Net: Self-Adapting Framework for U-Net-Based Medical Image Segmentation. arXiv 2018, arXiv:1809.10486. [Google Scholar]
- Brouwer, C.L.; Steenbakkers, R.J.H.M.; Bourhis, J.; Budach, W.; Grau, C.; Grégoire, V.; van Herk, M.; Lee, A.; Maingon, P.; Nutting, C.; et al. CT-Based Delineation of Organs at Risk in the Head and Neck Region: DAHANCA, EORTC, GORTEC, HKNPCSG, NCIC CTG, NCRI, NRG Oncology and TROG Consensus Guidelines. Radiother. Oncol. 2015, 117, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Concepción-Brito, M.; Moreta-Martínez, R.; Serrano, J.; García-Mato, D.; García-Sevilla, M.; Pascau, J. Segmentation of Organs at Risk in Head and Neck Radiation Therapy with 3D Convolutional Networks. Int. J. Comput. Assist. Radiol. Surg. 2019, 14 (Suppl. S1), s27–s28. [Google Scholar]
- Kerfoot, E.; Clough, J.; Oksuz, I.; Lee, J.; King, A.P.; Schnabel, J.A. Left-Ventricle Quantification Using Residual U-Net. In Statistical Atlases and Computational Models of the Heart. Atrial Segmentation and LV Quantification Challenges; Pop, M., Sermesant, M., Zhao, J., Li, S., McLeod, K., Young, A., Rhode, K., Mansi, T., Eds.; Lecture Notes in Computer Science; Springer International Publishing: Cham, Switzerland, 2019; Volume 11395, pp. 371–380. ISBN 978-3-030-12028-3. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Delving Deep into Rectifiers: Surpassing Human-Level Performance on ImageNet Classification. arXiv 2015, arXiv:1502.01852. [Google Scholar]
- MONAI. Available online: https://monai.io/index.html (accessed on 18 May 2022).
- Loshchilov, I.; Hutter, F. Decoupled Weight Decay Regularization. arXiv 2019, arXiv:1711.05101. [Google Scholar]
- Dice, L.R. Measures of the Amount of Ecologic Association Between Species. Ecology 1945, 26, 297–302. [Google Scholar] [CrossRef]
- Yeghiazaryan, V.; Voiculescu, I. An Overview of Current Evaluation Methods Used in Medical Image Segmentation; University of Oxford, Department of Computer Science: Oxford, UK, 2015. [Google Scholar]
- Kim, N.; Chun, J.; Chang, J.S.; Lee, C.G.; Keum, K.C.; Kim, J.S. Feasibility of Continual Deep Learning-Based Segmentation for Personalized Adaptive Radiation Therapy in Head and Neck Area. Cancers 2021, 13, 702. [Google Scholar] [CrossRef] [PubMed]
- Brunenberg, E.J.L.; Steinseifer, I.K.; van den Bosch, S.; Kaanders, J.H.A.M.; Brouwer, C.L.; Gooding, M.J.; van Elmpt, W.; Monshouwer, R. External Validation of Deep Learning-Based Contouring of Head and Neck Organs at Risk. Phys. Imaging Radiat. Oncol. 2020, 15, 8–15. [Google Scholar] [CrossRef]
- Dai, X.; Lei, Y.; Wang, T.; Zhou, J.; Roper, J.; McDonald, M.; Beitler, J.J.; Curran, W.J.; Liu, T.; Yang, X. Automated Delineation of Head and Neck Organs at Risk Using Synthetic MRI-aided Mask Scoring Regional Convolutional Neural Network. Med. Phys. 2021, 48, 5862–5873. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhao, T.; Gay, H.; Zhang, W.; Sun, B. Weaving Attention U-net: A Novel Hybrid CNN and Attention-based Method for Organs-at-risk Segmentation in Head and Neck CT Images. Med. Phys. 2021, 48, 7052–7062. [Google Scholar] [CrossRef]
- Tappeiner, E.; Welk, M.; Schubert, R. Tackling the Class Imbalance Problem of Deep Learning-Based Head and Neck Organ Segmentation. Int. J. Comput. Assist. Radiol. Surg. 2022, 17, 2103–2111. [Google Scholar] [CrossRef]
- Siciarz, P.; McCurdy, B. U-Net Architecture with Embedded Inception-ResNet-v2 Image Encoding Modules for Automatic Segmentation of Organs-at-Risk in Head and Neck Cancer Radiation Therapy Based on Computed Tomography Scans. Phys. Med. Biol. 2022, 67, 115007. [Google Scholar] [CrossRef] [PubMed]
- Gibbons, E.; Hoffmann, M.; Westhuyzen, J.; Hodgson, A.; Chick, B.; Last, A. Clinical Evaluation of Deep Learning and Atlas-based Auto-segmentation for Critical Organs at Risk in Radiation Therapy. J. Med. Radiat. Sci. 2022, jmrs.618. [Google Scholar] [CrossRef] [PubMed]
- Asbach, J.C.; Singh, A.K.; Matott, L.S.; Le, A.H. Deep Learning Tools for the Cancer Clinic: An Open-Source Framework with Head and Neck Contour Validation. Radiat. Oncol. 2022, 17, 28. [Google Scholar] [CrossRef] [PubMed]
| OAR | # Annotations Used to Train the Workflow | # CT Images in OAR-Specific Models | ||
|---|---|---|---|---|
| Patients (n = 40) | CTs (n = 225) | ntrain | ntest | |
| Brain | 33 | 165 | 132 | 33 |
| Mandible | 40 | 224 | 179 | 45 |
| Spinal canal | 40 | 221 | 176 | 45 |
| Brainstem | 40 | 225 | 180 | 45 |
| Esophagus | 34 | 179 | 143 | 36 |
| Larynx | 36 | 198 | 158 | 40 |
| Parotid glands | 40 | 217 | 173 | 44 |
| Lips | 33 | 181 | 144 | 37 |
| Submandibular glands | 34 | 183 | 146 | 37 |
| Temporomandibular joints | 38 | 205 | 164 | 41 |
| Inner ears | 39 | 217 | 173 | 44 |
| OAR | Test | Independent Test (nCT = 44) | ||||
|---|---|---|---|---|---|---|
| DSC (%) | ASD (mm) | DSC (%) Uncurated | DSC (%) Curated | ASD (mm) Uncurated | ASD (mm) Curated | |
| Brain | 98.04 ± 1.19 | 0.440 ± 0.070 | 98.16 ± 0.29 | 0.494 ± 0.088 | ||
| Mandible | 91.09 ± 1.75 | 0.411 ± 0.078 | 88.31 ± 3.42 | 0.553 ± 0.132 | ||
| Spinal canal | 88.87 ± 4.54 | 0.903 ± 1.302 | 80.21 ± 6.53 | 84.11 ± 5.67 | 1.007 ± 0.454 | 0.833 ± 0.401 |
| Brainstem | 89.07 ± 4.19 | 0.636 ± 0.225 | 81.86 ± 2.76 | 1.084 ± 0.214 | ||
| Esophagus | 81.39 ± 4.61 | 0.844 ± 0.492 | 63.07 ± 11.87 | 73.50 ± 10.36 | 2.818 ± 3.178 | 1.116 ± 0.743 |
| Larynx | 82.64 ± 9.11 | 0.990 ± 0.545 | 66.73 ± 9.31 | 2.026 ± 0.753 | ||
| PG | 86.05 ± 5.79 | 1.162 ± 2.701 | 75.22 ± 3.55 | 1.113 ± 0.246 | ||
| Lips | 71.88 ± 15.11 | 2.177 ± 3.801 | 45.17 ± 18.18 | 2.346 ± 1.748 | ||
| SMG | 79.39 ± 7.72 | 0.782 ± 0.331 | 55.40 ± 21.14 | 2.489 ± 2.475 | ||
| TMJ | 87.31 ± 3.57 | 0.430 ± 0.201 | 67.26 ± 12.36 | 83.34 ± 4.72 | 0.916 ± 0.467 | 0.523 ± 0.132 |
| Inner ears | 87.39 ± 3.03 | 0.514 ± 0.703 | 55.49 ± 12.98 | 83.76 ± 3.69 | 1.414 ± 0.398 | 0.531 ± 0.129 |
| OAR | (a) Semi-Supervised Model | (b) Self-Supervised Model | |||||
|---|---|---|---|---|---|---|---|
| DSC (%) | sDSC (%) | ASD (mm) | DSC (%) | sDSC (%) | ASD (mm) | ||
| Brain | 97.99 ± 0.29 | 96.68 ± 1.27 | 0.475 ± 0.067 | 98.02 ± 0.29 | 96.97 ± 1.17 | 0.470 ± 0.068 | |
| Mandible | 90.10 ± 2.76 | 96.46 ± 1.72 | 0.465 ± 0.108 | 90.60 ± 2.34 | 97.21 ± 1.38 | 0.440 ± 0.082 | |
| Spinal canal | U | 82.03 ± 6.74 | 84.33 ± 10.61 | 0.864 ± 0.364 | 83.53 ± 6.03 | 87.26 ± 8.58 | 0.822 ± 0.354 |
| C | 85.95 ± 6.25 | 89.75 ± 10.29 | 0.696 ± 0.339 | 86.92 ± 5.04 | 91.86 ± 7.29 | 0.669 ± 0.287 | |
| Brainstem | 86.14 ± 3.32 | 98.15 ± 4.27 | 0.827 ± 0.184 | 87.05 ± 3.63 | 98.33 ± 4.71 | 0.776 ± 0.194 | |
| Esophagus | U | 65.83 ± 12.19 | 76.86 ± 12.25 | 2.694 ± 3.552 | 66.04 ± 13.92 | 77.11 ± 14.15 | 2.987 ± 3.929 |
| C | 83.92 ± 4.56 | 92.35 ± 4.27 | 0.569 ± 0.156 | 86.77 ± 4.43 | 95.24 ± 3.53 | 0.507 ± 0.139 | |
| Larynx | 71.57 ± 7.00 | 73.73 ± 12.15 | 1.630 ± 0.779 | 79.47 ± 6.50 | 85.67 ± 11.16 | 1.254 ± 0.719 | |
| PG | 82.56 ± 2.81 | 96.29 ± 2.37 | 0.802 ± 0.142 | 83.70 ± 2.20 | 97.48 ± 1.84 | 0.753 ± 0.118 | |
| Lips | 51.20 ± 15.80 | 58.05 ± 18.76 | 1.449 ± 0.420 | 68.96 ± 9.09 | 78.83 ± 13.28 | 0.957 ± 0.301 | |
| SMG | 61.29 ± 25.61 | 78.44 ± 26.64 | 2.245 ± 2.690 | 65.80 ± 22.34 | 83.79 ± 22.04 | 1.912 ± 2.223 | |
| TMJ | U | 67.92 ± 14.16 | 79.89 ± 15.96 | 0.877 ± 0.489 | 67.83 ± 14.92 | 79.16 ± 16.18 | 0.882 ± 0.546 |
| C | 89.73 ± 3.81 | 99.27 ± 1.62 | 0.321 ± 0.119 | 91.38 ± 2.17 | 99.59 ± 1.02 | 0.256 ± 0.073 | |
| Inner ears | U | 54.74 ± 12.69 | 65.43 ± 14.35 | 1.446 ± 0.378 | 53.12 ± 13.42 | 61.71 ± 16.18 | 1.534 ± 0.416 |
| C | 86.03 ± 2.88 | 96.98 ± 2.77 | 0.467 ± 0.097 | 88.16 ± 2.56 | 98.18 ± 2.03 | 0.414 ± 0.094 | |
| Brain | Mandible | Spinal Canal | Brainstem | Esophagus | Larynx | PG | Lips | SMG | ATM | Inner Ears | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L | R | L | R | L | R | L | R | ||||||||
| Nikolov (2018) [3] | 99 | 96 | 95 | 88 | 85 | 85 | 85 | 85 | 65 | 75 | |||||
| Zhu (2019) [32] | 93 | 87 | 88 | 87 | 81 | 81 | |||||||||
| Liang (2019) [23] | 91 | 90 | 87 | 85 | 85 | 85 | 84 | ||||||||
| van Rooij (2019) [13] | 64 | 60 | 78 | 83 | 83 | 82 | |||||||||
| Tang * (2019) [25] | 93 | 86 | 89 | 85 | 85 | 81 | |||||||||
| Zhong (2019) [22] | 92 | ||||||||||||||
| van Dijk (2020) [15] | 95 1 | 83 1 | 84 1 | 83 1 | 77 1 | 78 1 | |||||||||
| Guo (2020) [27] | 95 | 88 | 88 | 88 | 84 | 84 | |||||||||
| Brunenberg (2020) [44] | 90 | 78 | 83 | 83 | 79 | 78 | |||||||||
| Sultana (2020) [24] | 87 | 86 | 87 | 85 | |||||||||||
| Oktay (2020) [10] | 94 | 85 | 84 | 85 | 83 | 78 | |||||||||
| Chi * (2020) [7] | 88 | 73 | 73 | 63 | 61 | ||||||||||
| Zhang (2021) [26] | 89 | 87 | 71 | 77 | 70 | 70 | |||||||||
| Liu * (2021) [11] | 97 | 92 | 89 | 86 | 90 | 84 | 87 | 90 | 90 | ||||||
| Dai (2021) [45] | 89 | 90 | 85 | 88 | 83 | 82 | 67 | 67 | |||||||
| Zhang (2021) [46] | 95 | 92 | 86 | 88 | 87 | 85 | |||||||||
| Li (2022) [12] | 84 | ||||||||||||||
| Tappeiner (2022) [47] | 94 | 88 | 88 | 87 | |||||||||||
| Siciarz (2022) [48] | 97 | 89 | 86 | 87 | 84 | 86 | 80 | 81 | 71 | 77 | 76 | 72 | 74 | ||
| Koo (2022) [21] | 87 | 88 | 82 | 82 | 83 | 83 | 81 | 83 | |||||||
| Gibbons (2022) [49] | 91 2 | 83 2 | 48 2 | 80 2 | 80 2 | 69 2 | 67 2 | ||||||||
| Asbach (2022) [50] | 93 | 85 | 43 | 81 | 81 | 72 | 70 | 45 | 47 | ||||||
| OURS | 98 | 90 | 86 | 86 | 84 | 72 | 83 | 51 | 61 | 90 | 86 | ||||
| OURS—Self-Supervised | 98 | 91 | 87 | 87 | 87 | 77 | 84 | 69 | 66 | 91 | 88 | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cubero, L.; Castelli, J.; Simon, A.; de Crevoisier, R.; Acosta, O.; Pascau, J. Deep Learning-Based Segmentation of Head and Neck Organs-at-Risk with Clinical Partially Labeled Data. Entropy 2022, 24, 1661. https://doi.org/10.3390/e24111661
Cubero L, Castelli J, Simon A, de Crevoisier R, Acosta O, Pascau J. Deep Learning-Based Segmentation of Head and Neck Organs-at-Risk with Clinical Partially Labeled Data. Entropy. 2022; 24(11):1661. https://doi.org/10.3390/e24111661
Chicago/Turabian StyleCubero, Lucía, Joël Castelli, Antoine Simon, Renaud de Crevoisier, Oscar Acosta, and Javier Pascau. 2022. "Deep Learning-Based Segmentation of Head and Neck Organs-at-Risk with Clinical Partially Labeled Data" Entropy 24, no. 11: 1661. https://doi.org/10.3390/e24111661
APA StyleCubero, L., Castelli, J., Simon, A., de Crevoisier, R., Acosta, O., & Pascau, J. (2022). Deep Learning-Based Segmentation of Head and Neck Organs-at-Risk with Clinical Partially Labeled Data. Entropy, 24(11), 1661. https://doi.org/10.3390/e24111661








