An Approach to Growth Delimitation of Straight Line Segment Classifiers Based on a Minimum Bounding Box
Abstract
:1. Introduction
2. Supervised Learning Based on Distances
2.1. Related Classifiers
2.1.1. K-Nearest Neighbor Classifier (k-NN)
2.1.2. Learning Vector Quantization (LVQ)
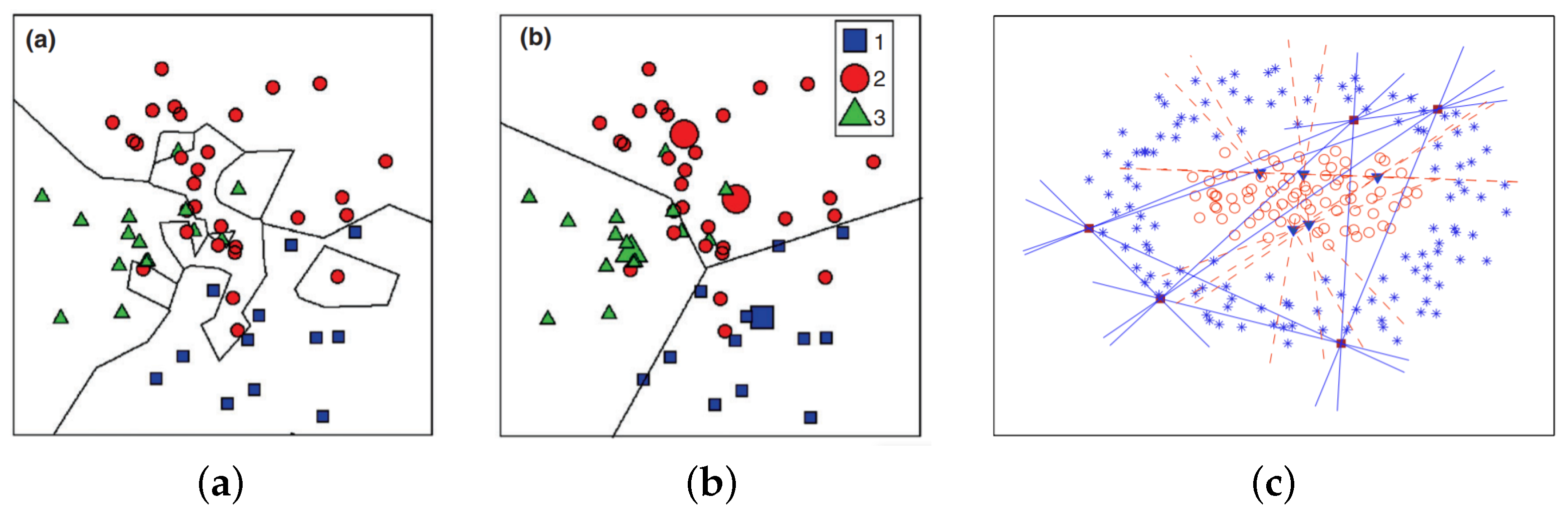
2.1.3. Nearest Feature Line (NFL)
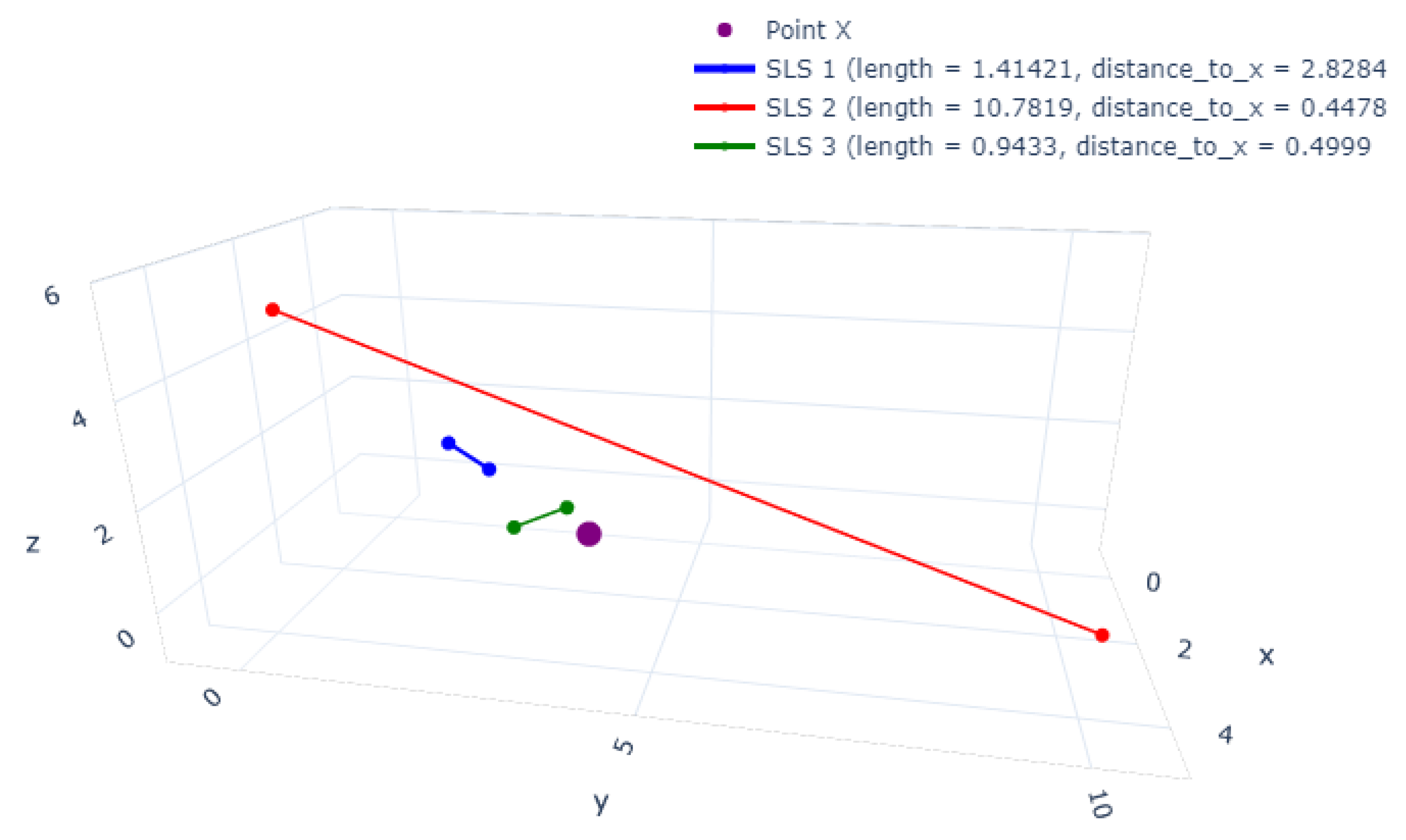
3. The Straight-Line Segment Classifier
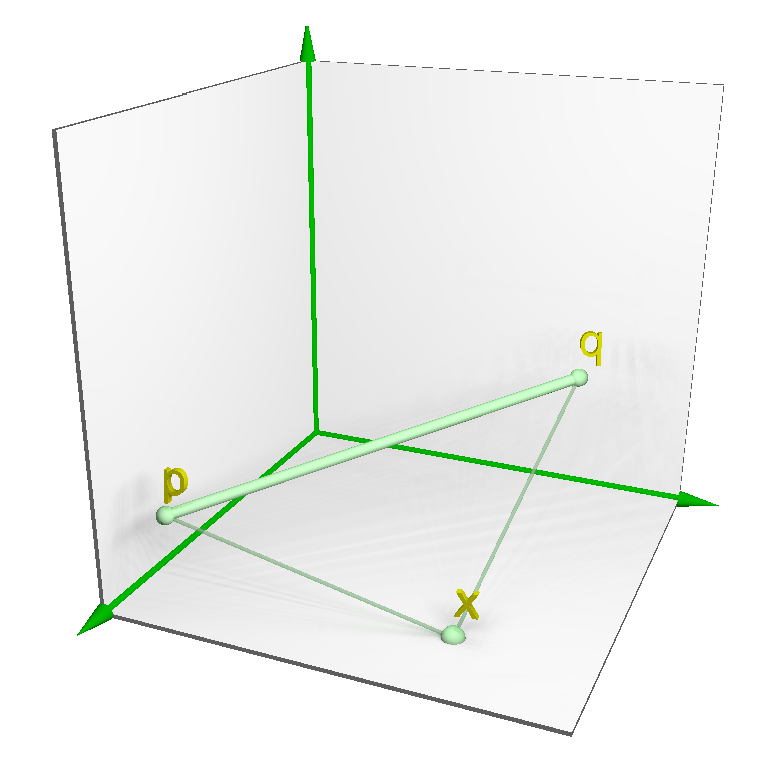
3.1. Notation and Definitions
3.2. Training Algorithm
| Algorithm 1: Training algorithm |
| Require: |
| gdParams = |
| Ensure: |
| 1: and |
| 2: and |
| 3: for class ← to 1 do |
| 4: KMeans(,m) |
| 5: for z ← 0 to m − 1 do |
| 6: KMeans(,2) |
| 7: |
| 8: |
| 9: add to |
| 10: end for |
| 11: end for |
| 12: |
| 13: , |
| 14: , GradDesc |
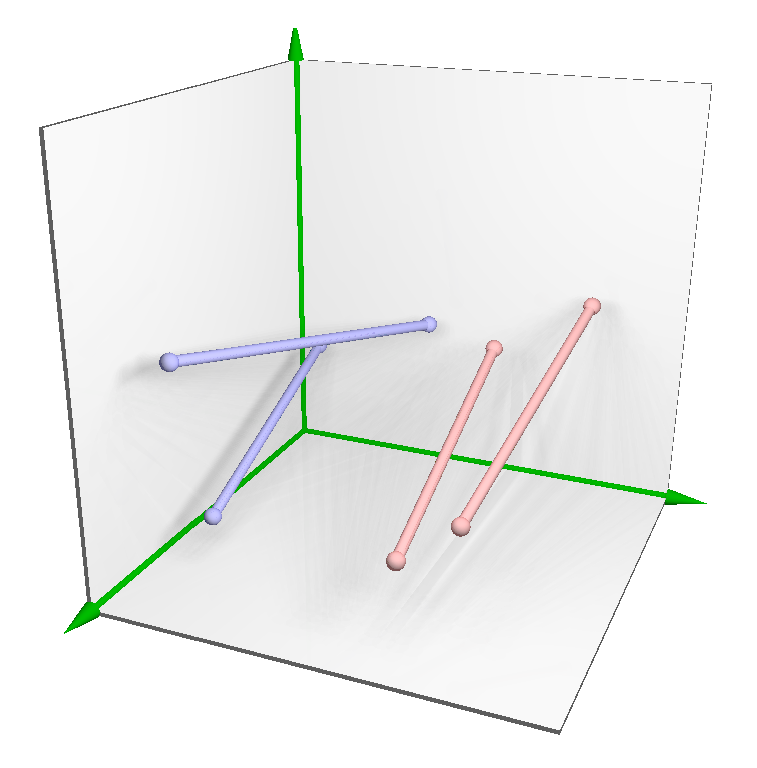
3.2.1. Placing
3.2.2. Tuning
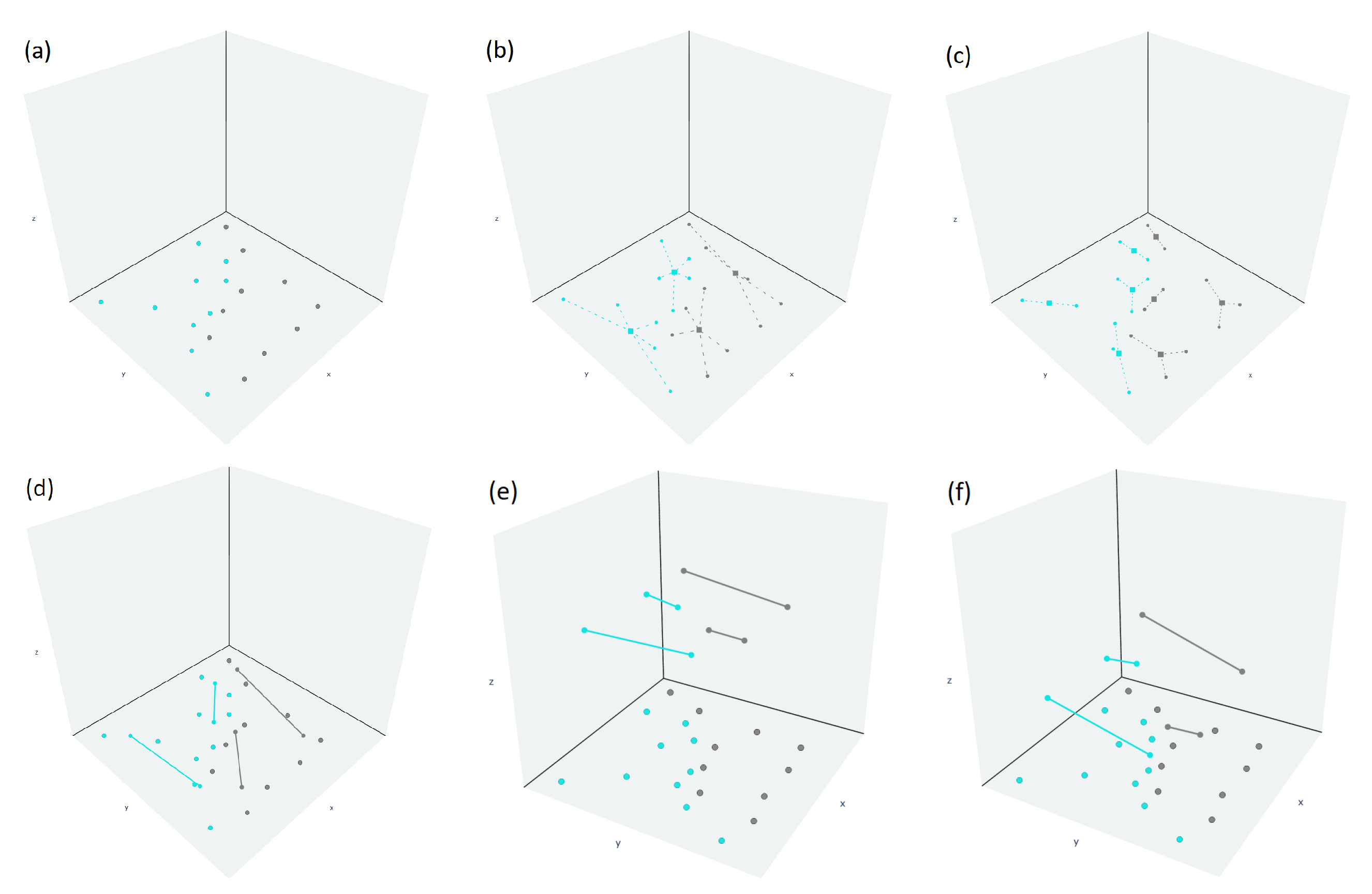
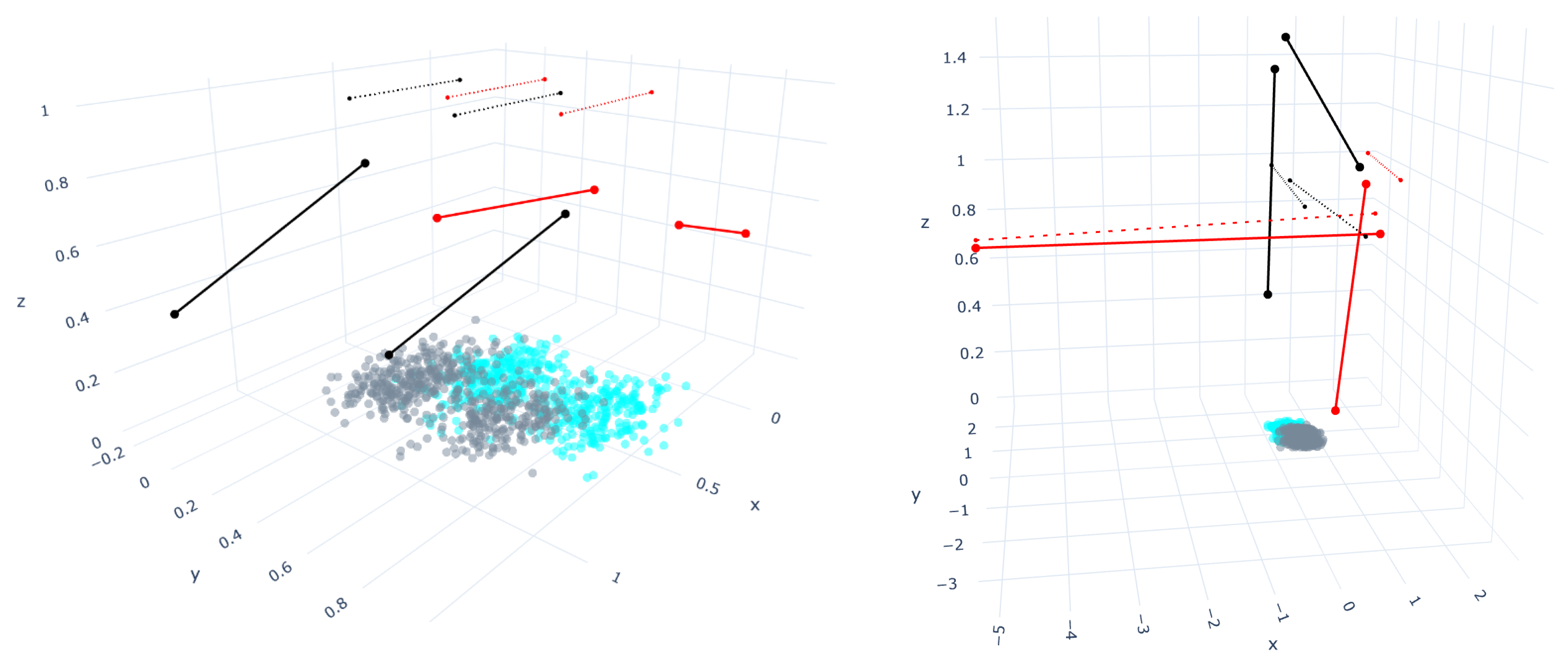
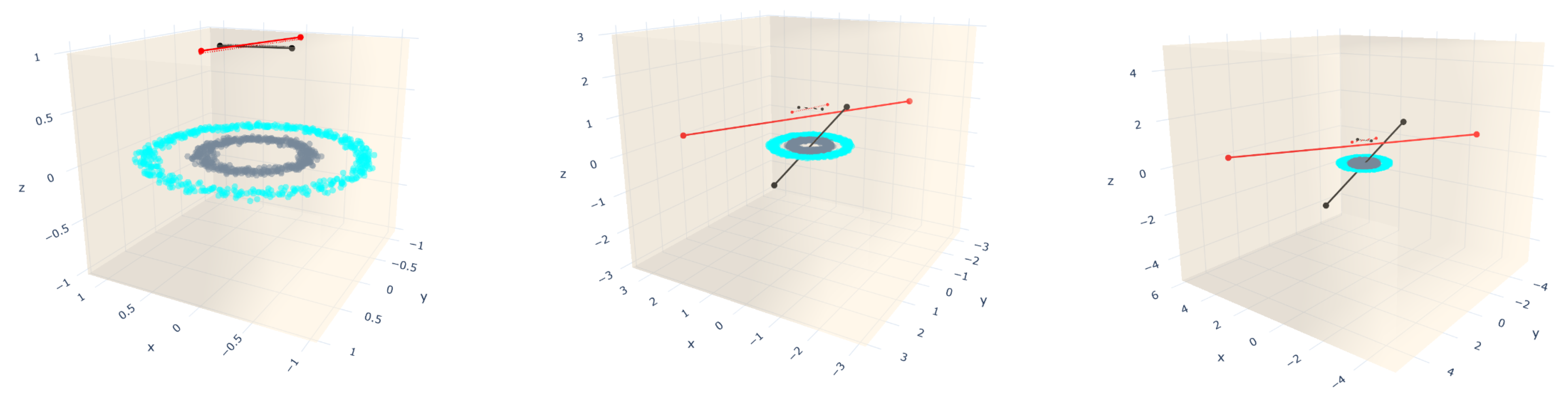
4. Bounding Box Approach for Straight-Line Segments Growth Restriction
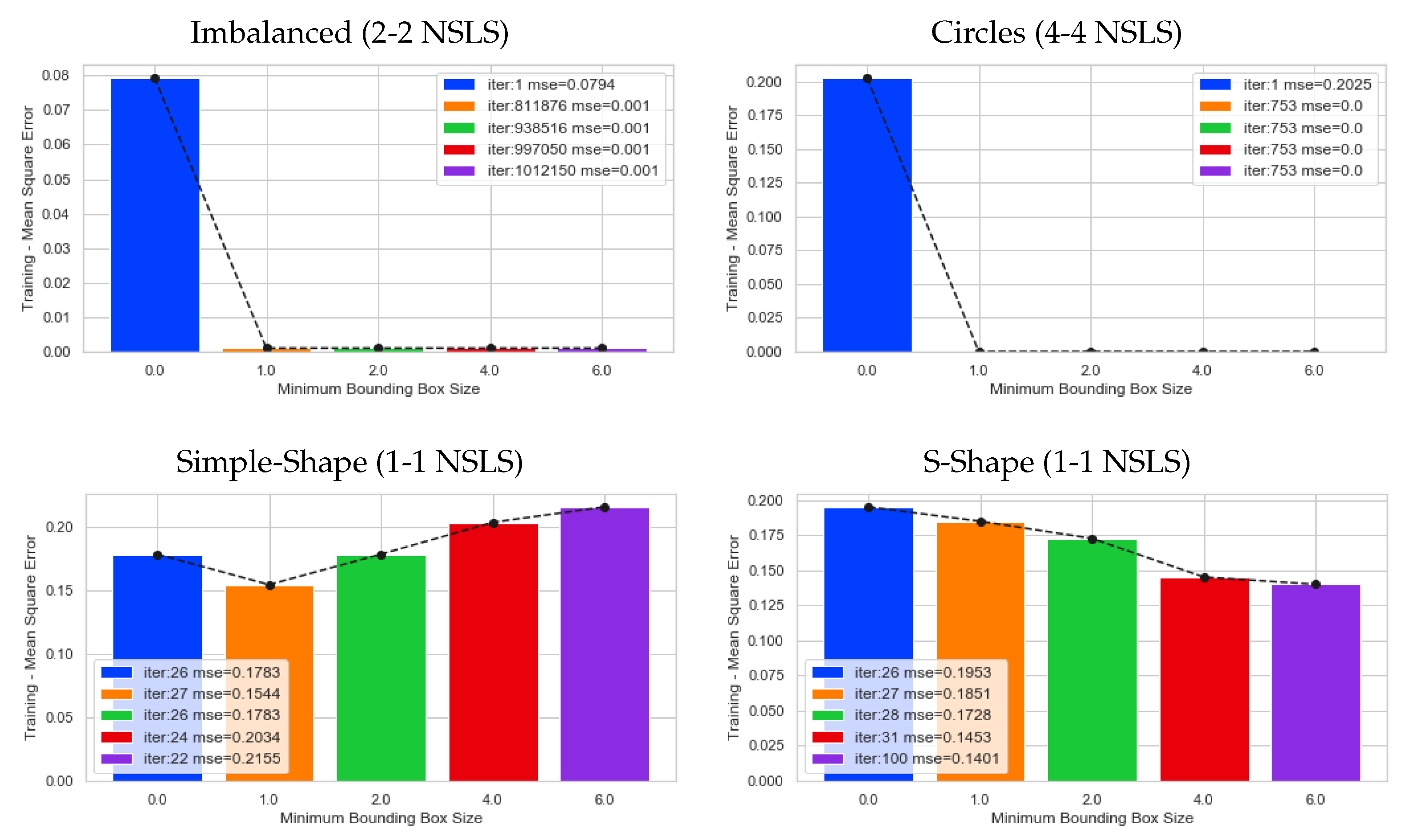
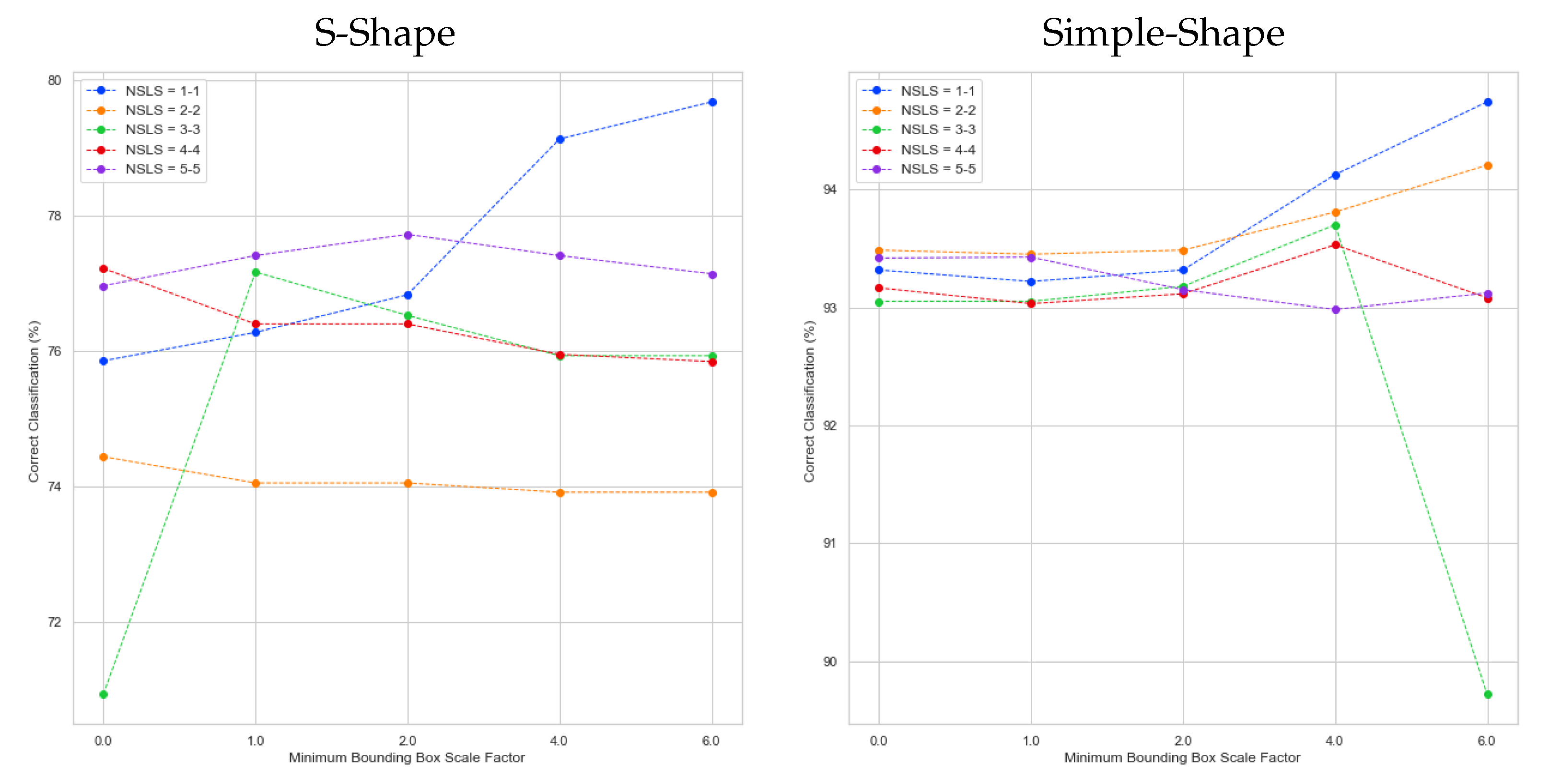
5. Results
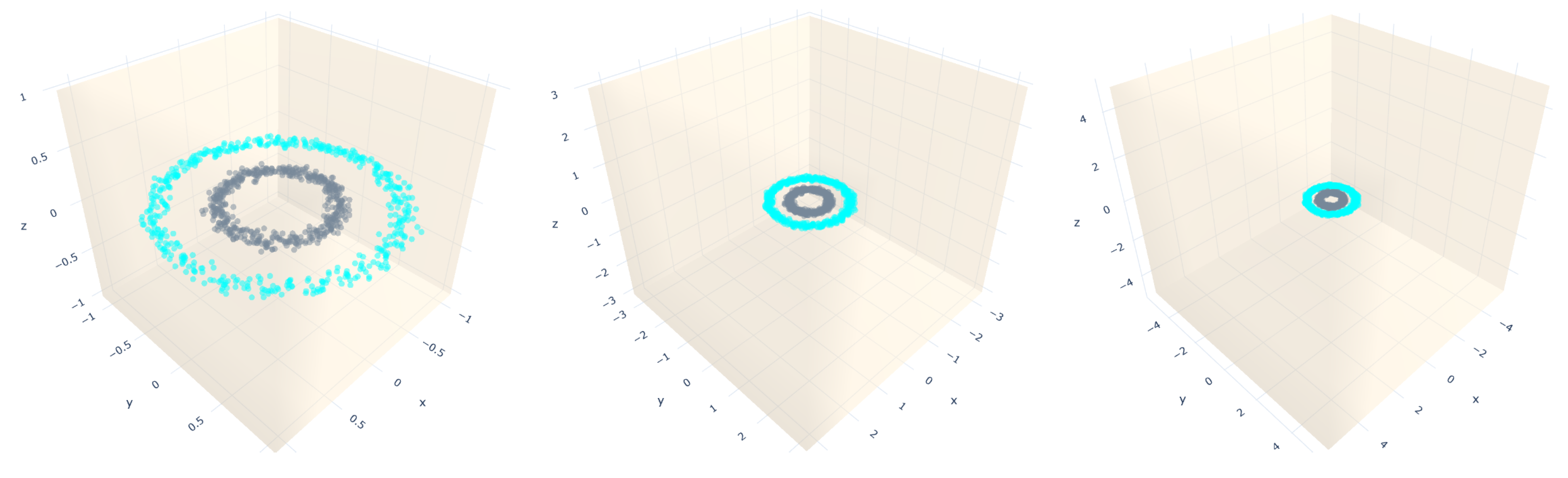
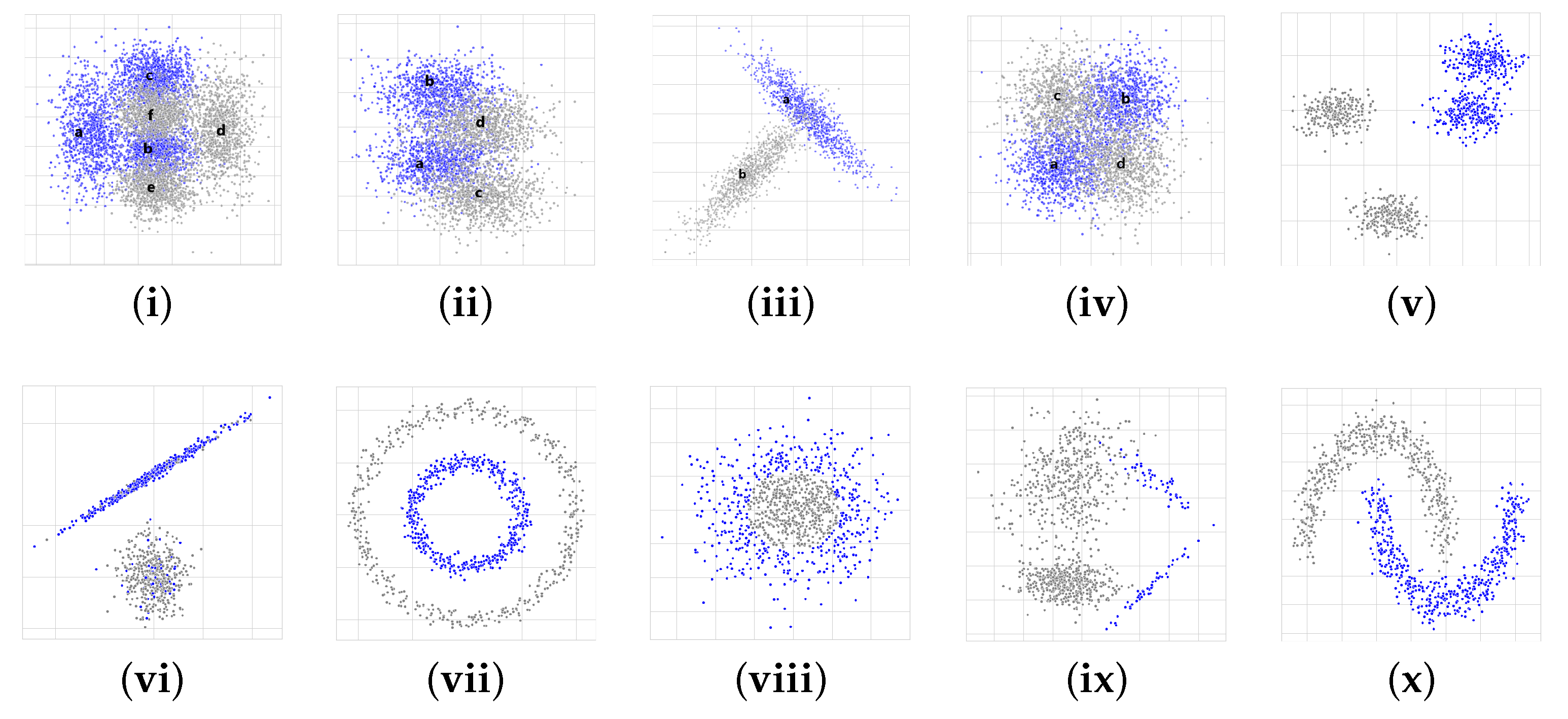
5.1. Artificial Datasets
- (v)
- Blobs: it consists of two blobs from a Gaussian distribution for each class (gray and blue colors, respectively).
- (vi)
- Blobs with noise: they are composed of one blob for each class. The label of 20% of the samples from each blob is randomly exchanged.
- (vii)
- Circles: they are a sample that falls into two concentric circles.
- (viii)
- Gaussian quantiles: they are constructed by taking a multi-dimensional standard normal distribution and defining classes separated by nested concentric multi-dimensional spheres. Each class has an equal number of instances.
- (ix)
- Imbalanced: it contains the number of gray instances that significantly outnumbers (nine times) the blue instances , which leads to class imbalance.
- (x)
- Moon: it is composed of two half moons, one for each class.
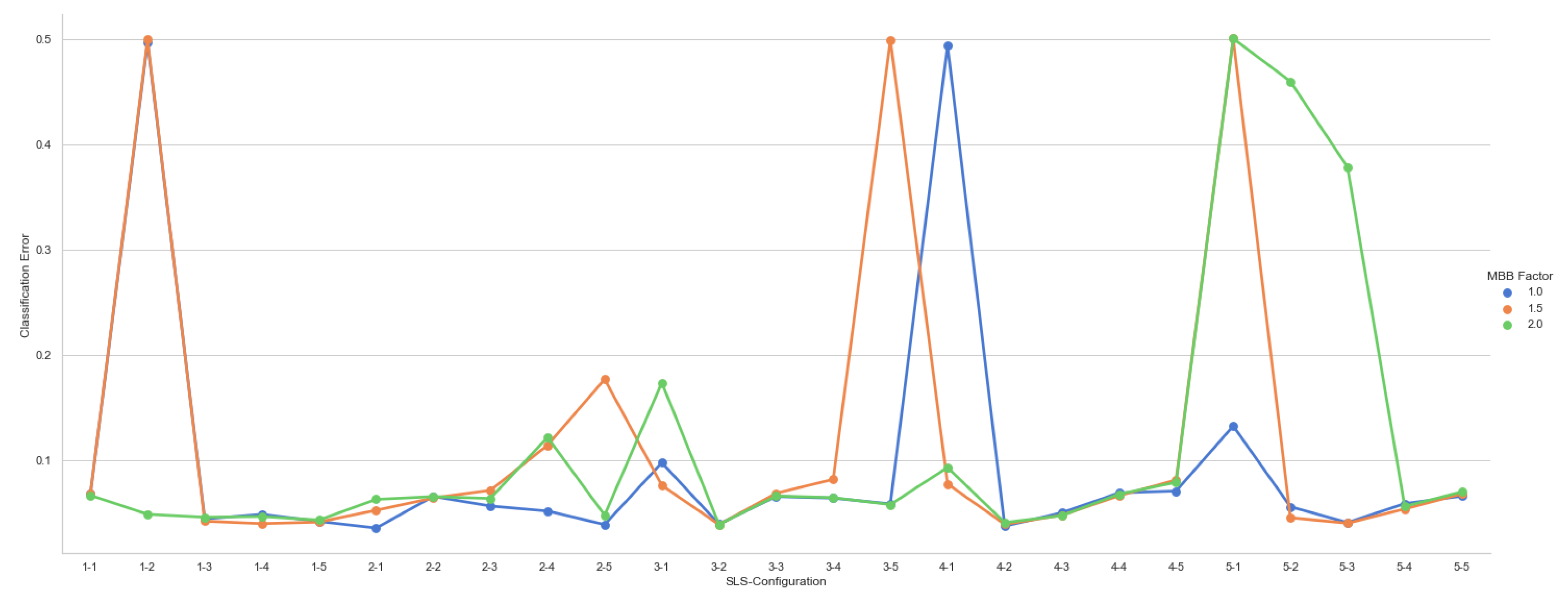
5.2. Experiments
5.3. Public Datasets
- Pre-processing: each dataset is normalized between −1 and 1, and the mean of the column replaces the missing values.
- Validation: stratified dataset split, preserving the percentage of samples for each class (train 75%, test 25%); and 10-fold cross validation where the training dataset is divided into 10 stratified folds.
- Number of straight-line segments: from 1 to 10 for each class, obtaining 100 models.
- Model selection: exhaustive search to find the straight-line segments’ best configuration.
- Gradient descent parameters: = 0.1; = 0.1; = 0.5 and = 0.001.
- All the experiments were performed in a 64-bit computer with 12 cores of 3.60 GHz under the Ubuntu/Linux operating system.
6. Discussion
7. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bishop, C. Pattern Recognition and Machine Learning; Springer: New York, NY, USA, 2006; Volume 4. [Google Scholar]
- Ali, S.; Smith, K.A. On learning algorithm selection for classification. Appl. Soft Comput. 2006, 6, 119–138. [Google Scholar] [CrossRef]
- Aly, M. Survey on multiclass classification methods. Neural Netw. 2005, 19, 1–9. [Google Scholar]
- Bengio, Y. Learning deep architectures for AI. Found. Trends Mach. Learn. 2009, 2, 1–127. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, C.; Zhang, X.; Almpanidis, G. An up-to-date comparison of state-of-the-art classification algorithms. Expert Syst. Appl. 2017, 82, 128–150. [Google Scholar] [CrossRef]
- Olson, M.L.; Wyner, A.J.; Berk, R.A. Modern Neural Networks Generalize on Small Data Sets; NeurIPS: Montrèal, QC, Canada, 2018. [Google Scholar]
- Brigato, L.; Iocchi, L. A Close Look at Deep Learning with Small Data. In Proceedings of the 2020 25th International Conference on Pattern Recognition (ICPR), Milan, Italy, 10–15 January 2021; pp. 2490–2497. [Google Scholar] [CrossRef]
- Ribeiro, J.; Hashimoto, R. A New Machine Learning Technique Based on Straight Line Segments. In Proceedings of the ICMLA 2006: 5th International Conference on Machine Learning and Applications, Orlando, FL, USA, 14–16 December 2006; pp. 10–16. [Google Scholar]
- Kohonen, T. Learning vector quantization. In Self-Organizing Maps; Springer: Berlin/Heidelberg, Germany, 1995; pp. 175–189. [Google Scholar]
- Pedreira, C. Learning Vector Quantization with Training Data Selection. IEEE Trans. Pattern Anal. Mach. Intell. 2006, 28, 157–162. [Google Scholar] [CrossRef] [PubMed]
- Hammer, B.; Villmann, T. Generalized Relevance Learning Vector Quantization. Neural Netw. 2002, 15, 1059–1068. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, C.; Wang, J. Extended nearest feature line classifier. In Pacific Rim International Conference on Artificial Intelligence; Springer: Berlin/Heidelberg, Germany, 2004; pp. 183–190. [Google Scholar]
- Li, S.Z.; Lu, J. Face recognition using the nearest feature line method. IEEE Trans. Neural Netw. 1999, 10, 439–443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ribeiro, J.; Hashimoto, R. A New Training Algorithm for Pattern Recognition Technique Based on Straight Line Segments. In Proceedings of the 2008 XXI Brazilian Symposium on Computer Graphics and Image Processing, Los Alamitos, CA, USA, 12–15 October 2008; pp. 19–26. [Google Scholar]
- Ribeiro, J.; Hashimoto, R. Pattern Recognition, Recent Advances; Chapter Pattern Recognition Based on Straight Line Segments; I-Tech: London, UK, 2010. [Google Scholar]
- Duda, R.O.; Hart, P.E.; Stork, D.G. Pattern Classification, 2nd ed.; John Wiley and Sons: Hoboken, NJ, USA, 2001. [Google Scholar]
- Kotsiantis, S. Supervised Machine Learning: A Review of Classification Techniques. In Proceeding of the 2007 Conference on Emerging Artificial Intelligence Applications in Computer Engineering: Real Word AI Systems with Applications in eHealth, HCI, Information Retrieval and Pervasive Technologies; IOS Press: Amsterdam, The Netherlands, 2007; pp. 3–24. [Google Scholar]
- Rodriguez, R.M.; Hashimoto, R. Combining dialectical optimization and gradient descent methods for improving the accuracy of straight line segment classifiers. In Proceedings of the 2011 24th SIBGRAPI Conference on Graphics, Patterns and Images, Alagoas, Brazil, 28–31 August 2011; pp. 321–328. [Google Scholar]
- Medina-Rodríguez, R.; Hashimoto, R.F. Evolutive Algorithms Applied To The Straight Line Segment Classifier. In Proceedings of the Workshop of Theses and Dissertations (WTD) in SIBGRAPI 2013 (XXVI Conference on Graphics, Patterns and Images), Arequipa, Peru, 5–8 August 2013. [Google Scholar]
- Medina-Rodríguez, R.; Castañón, C.B.; Hashimoto, R.F. Evaluation of the Impact of Initial Positions obtained by Clustering Algorithms on the Straight Line Segments Classifier. In Proceedings of the 2018 IEEE Latin American Conference on Computational Intelligence (LA-CCI), Gudalajara, Mexico, 7–9 November 2018; pp. 1–6. [Google Scholar]
- Fix, E.; Hodges, J.L., Jr. Discriminatory Analysis-Nonparametric Discrimination: Consistency Properties. Int. Stat. Rev. Int. Stat. 1989, 57, 238–247. [Google Scholar] [CrossRef]
- Cover, T.; Hart, P. Nearest neighbor pattern classification. Inf. Theory, IEEE Trans. 1967, 13, 21–27. [Google Scholar] [CrossRef]
- Biehl, M.; Hammer, B.; Villmann, T. Prototype-based models in machine learning. Wiley Interdiscip. Rev. Cogn. Sci. 2016, 7, 92–111. [Google Scholar] [CrossRef] [PubMed]
- Villmann, T.; Bohnsack, A.; Kaden, M. Can learning vector quantization be an alternative to SVM and deep learning?—recent trends and advanced variants of learning vector quantization for classification learning. J. Artif. Intell. Soft Comput. Res. 2017, 7, 65–81. [Google Scholar] [CrossRef] [Green Version]
- James, G.; Witten, D.; Hastie, T.; Tibshirani, R. Statistical Learning. In An Introduction to Statistical Learning: With Applications in R; Springer: New York, NY, USA, 2021; pp. 15–57. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Zhang, H. The optimality of naive Bayes. AA 2004, 1, 3. [Google Scholar]
- Asuncion, A.; Newman, D. UCI Machine Learning Repository. Available online: http://archive.ics.uci.edu/ml/ (accessed on 1 April 2021).
- Sanderson, C.; Curtin, R. Armadillo: A template-based C++ library for linear algebra. J. Open Source Softw. 2016, 1, 26. [Google Scholar] [CrossRef]
- Sanderson, C.; Curtin, R. A User-Friendly Hybrid Sparse Matrix Class in C++. arXiv 2018, arXiv:1805.03380. [Google Scholar]
- Quinlan, J.R. C4.5: Programs for Machine Learning; Morgan Kaufmann: Burlington, MA, USA, 1993. [Google Scholar]
- Ryszard, S.; Mozetic, I.; Hong, J.; Lavrac, N. The Multi-Purpose Incremental System AQ15 and Its Testing Application to Three Medical Domains. In Proceedings of the AAAI’86: Proceedings of the Fifth AAAI National Conference on Artificial Intelligence, Philadelphia, PA, USA, 11–15 August 1986; pp. 1041–1047. [Google Scholar]
- Smith, J.W.; Everhart, J.E.; Dickson, W.; Knowler, W.; Johannes, R. Using the ADAP Learning Algorithm to Forecast the Onset of Diabetes Mellitus. In Proceedings of the Annual Symposium on Computer Application in Medical Care, Washington, DC, USA, 6–9 November 1988; pp. 261–265. [Google Scholar]
- Eggermont, J.; Joost, N.K.; Kosters, W.A. Genetic Programming for Data Classification: Partitioning the Search Space. In Proceedings of the 2004 Symposium on Applied Computing (ACM SAC’04), Nicosia, Cyprus, 14–17 March 2004; pp. 1001–1005. [Google Scholar]
- Smirnov, E.; Sprinkhuizen-Kuyper, I.; Nalbantov, G. Unanimous Voting using Support Vector Machines. In Proceedings of the Sixteenth Belgium-Netherlands Conference on Artificial Intelligence (BNAIC-2004), Groningen, The Netherlands, 21–22 October 2004; pp. 43–50. [Google Scholar]
- Sigillito, V.G.; Wing, S.P.; Hutton, L.V.; Baker, K.B. Classification of radar returns from the ionosphere using neural networks. Johns Hopkins APL Tech. Dig. 1989, 10, 262–266. [Google Scholar]
- Bagirov, A.; Rubinov, A.; Soukhoroukova, N.; Yearwod, J. Unsupervised and Supervised Data Classification via Nonsmooth and Global Optimization. Top 2003, 11, 1–75. [Google Scholar] [CrossRef] [Green Version]
- Gorman, R.P.; Sejnowski, T.J. Analysis of hidden units in a layered network trained to classify sonar targets. Neural Netw. 1988, 1, 75–89. [Google Scholar] [CrossRef]
- Fernández-Delgado, M.; Cernadas, E.; Barro, S.; Amorim, D. Do we need hundreds of classifiers to solve real world classification problems? J. Mach. Learn. Res. 2014, 15, 3133–3181. [Google Scholar]
- Amancio, D.R.; Comin, C.H.; Casanova, D.; Travieso, G.; Bruno, O.M.; Rodrigues, F.A.; da Fontoura Costa, L. A systematic comparison of supervised classifiers. PLoS ONE 2014, 9, e94137. [Google Scholar]
| Dist. | Class (C) | Mean () | Covariance () | Prob(P) | Label |
|---|---|---|---|---|---|
| F-Shape M = 3 | 0 | (a) | |||
| (b) | |||||
| (c) | |||||
| 1 | (d) | ||||
| (e) | |||||
| (f) | |||||
| S-Shape M = 2 | 0 | (a) | |||
| (b) | |||||
| 1 | (c) | ||||
| (d) | |||||
| Simple-Shape M = 1 | 0 | 1 | (a) | ||
| 1 | 1 | (b) | |||
| X-Shape M = 2 | 0 | (a) | |||
| (b) | |||||
| 1 | (c) | ||||
| (d) |
| Distribution | Bayes Classifier (%) |
|---|---|
| F-Shape | 91.43 |
| S-Shape | 84.39 |
| Simple-Shape | 96.95 |
| X-Shape | 81.11 |
| Dataset Type | Parameters |
|---|---|
| Blobs | n_samples = 160,000, centers = 4, n_features = 2, random_state = 222 |
| Blobs with Noise | n_samples = 160,000, n_features = 2, n_informative = 2, n_redundant = 0, n_repeated = 0, n_classes = 2, n_clusters_per_class = 1, class_sep = 2, flip_y = 0.2, weights = [0.5, 0.5], random_state = 222 |
| Circles | n_samples = 160,000, factor = 0.5, noise = 0.05, random_state = 222 |
| Gaussian Quantiles | n_samples = 160,000, n_features = 2, n_classes = 2, random_state = 222 |
| Imbalanced | n_samples = 160,000, n_features = 2, n_informative = 2, n_redundant = 0, n_repeated = 0, n_classes = 2, n_clusters_per_class = 2, class_sep = 1.5, flip_y = 0, weights = [0.9, 0.1], random_state = 222 |
| Moon | n_samples = 160,000, noise = 0.1, random_state = 222 |
| Distribution | 1st | 2nd | 3rd | 4th | 5th | 6th | 7th | 8th | 9th | 10th | Mean | Stdev |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Blobs | 99.76 | 89.20 | 99.63 | 60.57 | 99.38 | 82.69 | 96.25 | 97.85 | 93.66 | 90.68 | 90.97 | 12.03 |
| Blobs_noise | 89.05 | 89.42 | 89.90 | 89.22 | 89.89 | 90.03 | 90.02 | 89.73 | 89.91 | 89.02 | 89.62 | 0.40 |
| Circles | 99.99 | 99.99 | 99.99 | 99.99 | 99.99 | 99.99 | 99.99 | 99.99 | 99.99 | 99.99 | 99.99 | 0.002 |
| F-Shape | 82.16 | 82.00 | 82.08 | 82.05 | 82.08 | 82.19 | 82.10 | 82.12 | 82.02 | 82.02 | 82.08 | 0.06 |
| Gaussian | 97.27 | 97.23 | 97.33 | 97.42 | 97.33 | 97.24 | 97.21 | 97.32 | 97.35 | 97.41 | 97.31 | 0.07 |
| Imbalanced | 98.37 | 99.66 | 99.74 | 98.89 | 98.77 | 98.26 | 98.75 | 97.97 | 95.56 | 99.25 | 98.52 | 1.19 |
| Moon | 87.92 | 88.03 | 87.95 | 88.02 | 87.93 | 88.02 | 87.99 | 88.02 | 88.03 | 87.98 | 87.99 | 0.04 |
| S-Shape | 79.27 | 79.57 | 79.26 | 79.52 | 79.53 | 79.59 | 79.60 | 79.60 | 79.44 | 79.52 | 79.49 | 0.13 |
| Simple-Shape | 92.37 | 92.36 | 92.47 | 92.39 | 92.35 | 92.41 | 92.33 | 92.33 | 92.35 | 92.40 | 92.38 | 0.05 |
| X-Shape | 50.35 | 47.44 | 47.36 | 47.43 | 49.73 | 49.91 | 47.52 | 50.36 | 55.72 | 47.29 | 49.31 | 2.62 |
| Parameter | Values |
|---|---|
| Training Samples | 10 samples of 1000 examples |
| Test Sample | 160,000 examples |
| Validation | 10-Fold-Cross-Validation |
| mbb_factor | 0.0, 1.0, 2.0, 4.0, 6.0 |
| Num. SLS per class | 1-1, 2-2, 3-3, 4-4, 5-5 |
| Gradient Descent- | 0.1 |
| Gradient Descent- | 0.1 |
| Gradient Descent- | 0.5 |
| Gradient Descent- | 0.001 |
| Dataset | # Examples | Dim | Train-Size | Test-Size |
|---|---|---|---|---|
| Australian Credit Approval [31] | 690 | 14 | 517 | 173 |
| Breast Cancer Wisconsin [32] | 683 | 10 | 512 | 171 |
| Pima Indians Diabetes [33] | 768 | 8 | 576 | 192 |
| German Credit Data [34] | 1000 | 24 | 750 | 250 |
| Heart [35] | 270 | 13 | 202 | 68 |
| Ionosphere [36] | 351 | 34 | 263 | 88 |
| Liver Disorders [37] | 345 | 6 | 258 | 87 |
| Sonar Mines vs Rocks [38] | 208 | 60 | 156 | 52 |
| Dataset | The SLS Best Configuration | MSE-Validation (%) (1.0, 1.5 and 2.0) | MSE-Test (%) (1.0, 1.5 and 2.0) |
|---|---|---|---|
| Australian | {1-1, 2-3, 1-1} | ||
| Breast Cancer | {8-10, 8-10, 8-10} | ||
| Diabetes | {3-2, 7-2, 7-2} | ||
| German | {2-2, 1-1, 1-2} | ||
| Heart | {2-2, 1-2, 1-1} | ||
| Ionosphere | {10-8, 10-8, 10-8} | ||
| Liver Disorders | {5-2, 5-2, 5-2} | ||
| Sonar | {5-7, 6-7, 6-7} |
| Dataset | All | NN | SVM | SLS Classifier | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| [39] | [39] | [39] | Original | Proposed w/o MBB | Proposed w/ MBB | |||||
| Australian | 69.1 | 68.8 | 68.2 | 87.28 | 8-8 | 86.00 (1.49) | 1-1 | 89.81 (4.2) | 1-1 | 2.0 |
| Breast-Cancer | 97.4 | 97.4 | 97.0 | 94.74 | 8-8 | 98.20 (1.46) | 8-10 | 95.65 (1.6) | 8-10 | 2.0 |
| Diabetes | 79.0 | 78.1 | 78.3 | 73.44 | 7-7 | 76.79 (2.38) | 3-2 | 83.22 (3.5) | 3-2 | 1.0 |
| German | 79.0 | 78.1 | 77.6 | 72.40 | 1-1 | 72.16 (0.48) | 1-2 | 85.33 (1.6) | 1-2 | 2.0 |
| Heart | 88.2 | 86.9 | 88.1 | 80.88 | 6-6 | 78.42 (1.38) | 2-2 | 87.39 (6.2) | 2-2 | 1.0 |
| Ionosphere | 95.5 | 95.2 | 95.5 | 95.45 | 7-7 | 95.56 (3.53) | 10-8 | 94.50 (2.0) | 10-8 | 2.0 |
| Liver-Disorders | - | - | - | 72.41 | 5-5 | 72.08 (2.08) | 5-2 | 76.59 (7.5) | 5-2 | 2.0 |
| Sonar | 97.1 | 97.1 | 90.4 | 88.46 | 7-7 | 88.57 (1.51) | 6-7 | 92.38 (6.0) | 6-7 | 1.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Medina-Rodríguez, R.; Beltrán-Castañón, C.; Hashimoto, R.F. An Approach to Growth Delimitation of Straight Line Segment Classifiers Based on a Minimum Bounding Box. Entropy 2021, 23, 1541. https://doi.org/10.3390/e23111541
Medina-Rodríguez R, Beltrán-Castañón C, Hashimoto RF. An Approach to Growth Delimitation of Straight Line Segment Classifiers Based on a Minimum Bounding Box. Entropy. 2021; 23(11):1541. https://doi.org/10.3390/e23111541
Chicago/Turabian StyleMedina-Rodríguez, Rosario, César Beltrán-Castañón, and Ronaldo Fumio Hashimoto. 2021. "An Approach to Growth Delimitation of Straight Line Segment Classifiers Based on a Minimum Bounding Box" Entropy 23, no. 11: 1541. https://doi.org/10.3390/e23111541
APA StyleMedina-Rodríguez, R., Beltrán-Castañón, C., & Hashimoto, R. F. (2021). An Approach to Growth Delimitation of Straight Line Segment Classifiers Based on a Minimum Bounding Box. Entropy, 23(11), 1541. https://doi.org/10.3390/e23111541







