Entropy-Based Strategies for Rapid Pre-Processing and Classification of Time Series Data from Single-Molecule Force Experiments
Abstract
1. Introduction
2. Materials and Methodology
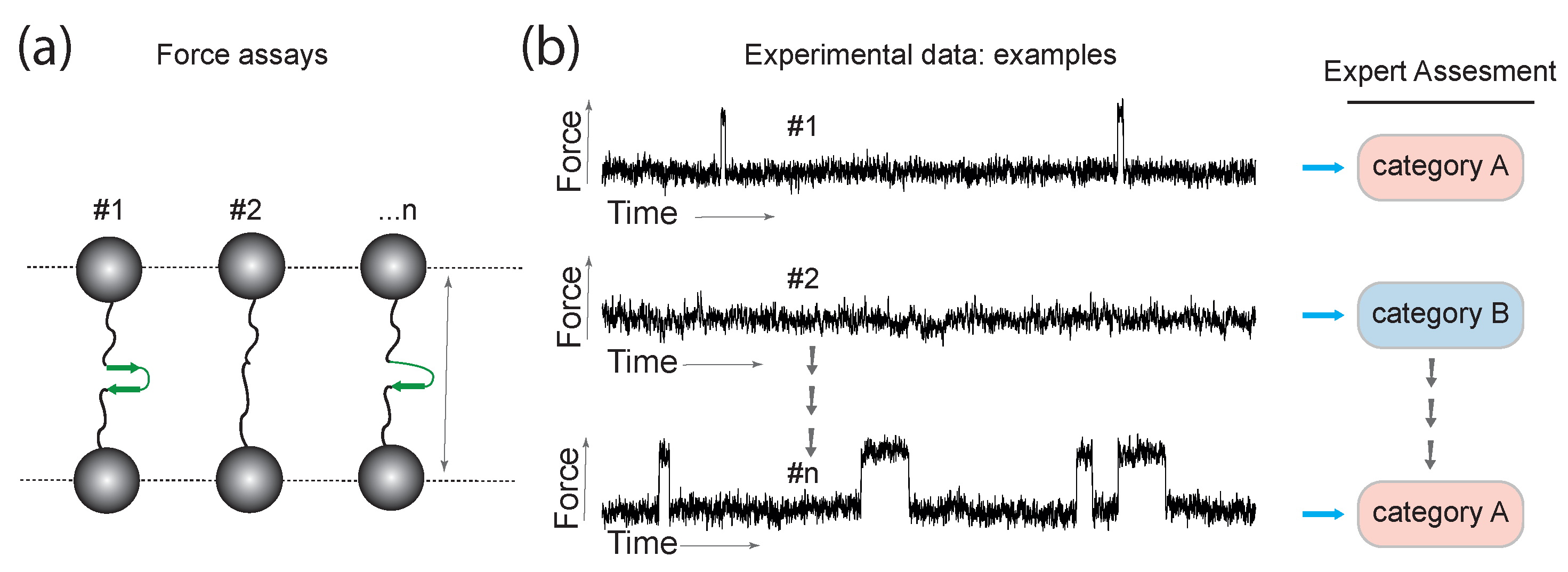
2.1. Experiments, Protocols, Signal Detection
Problem Formulation—Data Categories
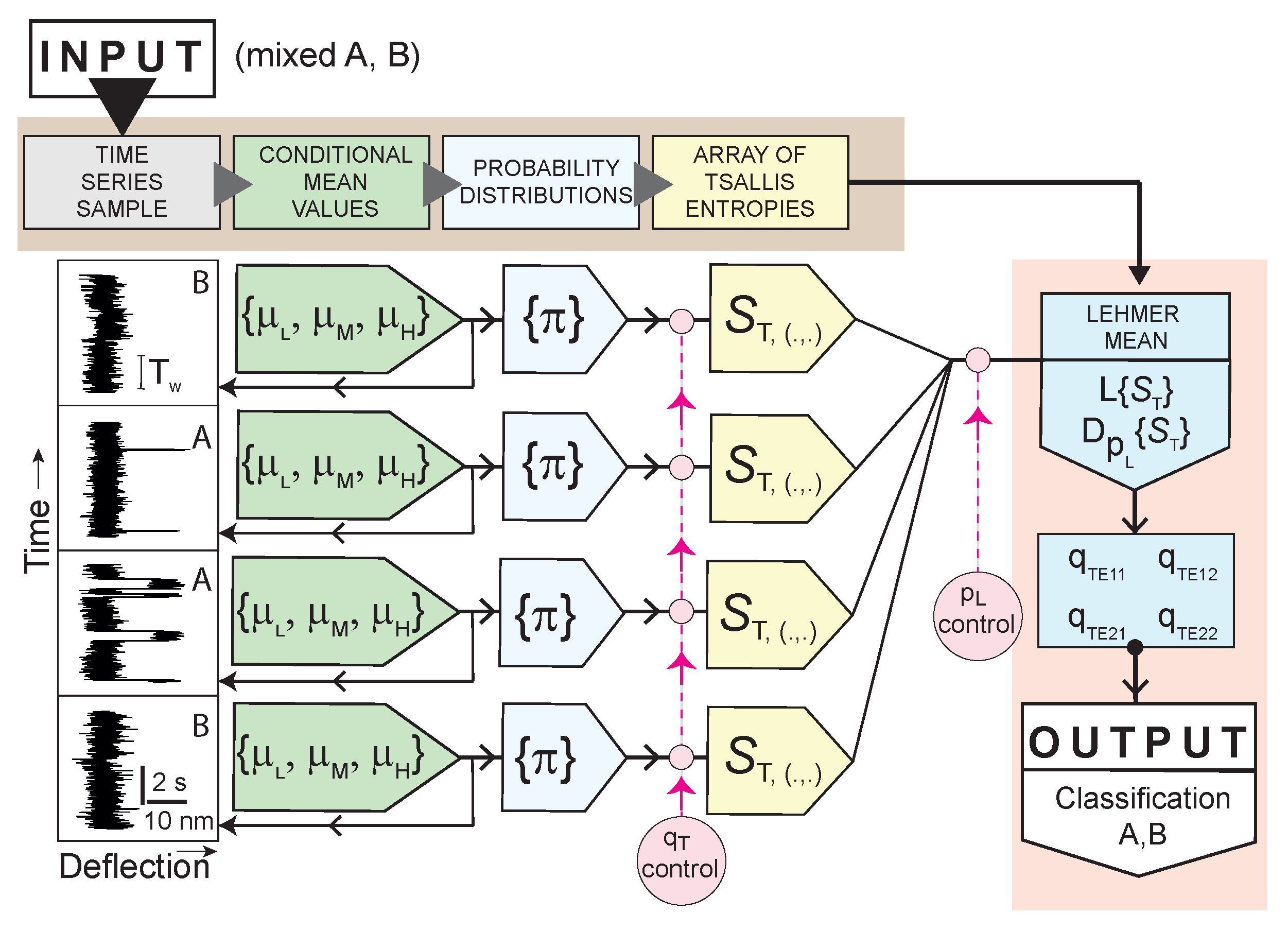
2.2. Measures and Methods of Supervised Classification
2.2.1. Time Series, Averages, Adaptive Histograms
| Algorithm 1: Conditional mean values for given time window. |
 |
2.2.2. Entropy of Histograms
| Algorithm 2: Lehmer mean of set of entropy values. |
 |
2.2.3. Long-Term Transformation into Entropic Systems with Related Lehmer Means
3. Numerical Results
3.1. Comparison of Methods for Specific Time-Series Classification
- the evaluation with the goals to emphasize the gains within the framework of applicability;
- the design of new potential classifiers with unified and specific mathematical structure;
- the comparison of new and previously established classification schemes;
- the identification of the proper parameters (meta-parameters) that are useful for the classification.
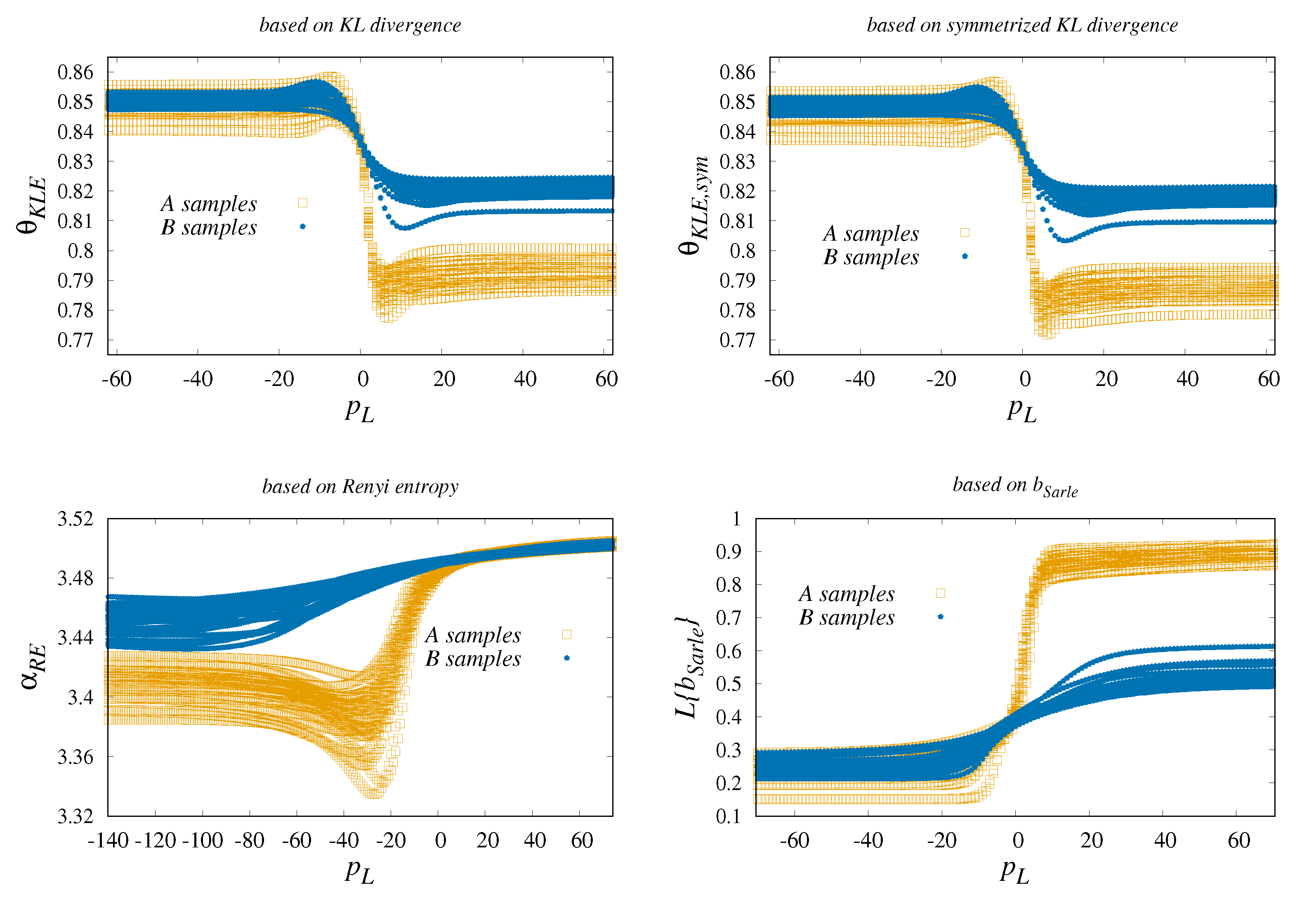
3.1.1. Classification Adapted from Kullback–Leibler Form
3.1.2. Classification which Converts the Original Time Series into Rényi Entropy Series
3.1.3. Problem of Sarle’s b Revisited
3.2. Integration over the Values-Option for t-Testing
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Neuman, K.C.; Nagy, A. Single-molecule force spectroscopy: Optical tweezers, magnetic tweezers and atomic force microscopy. Nat. Methods 2008, 5, 491–505. [Google Scholar] [CrossRef]
- Ramanathan, A.; Savol, A.J.; Langmead, C.J.; Agarwal, P.K.; Chennubhotla, C.S. Discovering Conformational Sub-States Relevant to Protein Function. PLoS ONE 2011, 6, e15827. [Google Scholar] [CrossRef]
- Krammer, C.; Schatzl, H.; Vorberg, I. Prion-like propagation of cytosolic protein aggregates Insights from cell culture models. Prion 2009, 3, 206–212. [Google Scholar] [CrossRef]
- Yu, H.; Liu, X.; Neupane, K.; Gupta, A.; Brigley, A.; Solanki, A.; Sosova, I.; Woodside, M. Direct observation of multiple misfolding pathways in a single prion protein molecule. Proc. Natl. Acad. Sci. USA 2012, 109, 5283–5288. [Google Scholar] [CrossRef] [PubMed]
- Kolmogorov, A. New Metric Invariant of Transitive Dynamical Systems and Endomorphisms of Lebesgue Spaces. Dokl. Russ. Acad. Sci. 1958, 119, 861–864. [Google Scholar]
- Sinai, Y. On the Notion of Entropy of a Dynamical System. Dokl. Russ. Acad. Sci. 1959, 124, 768–771. [Google Scholar]
- Richman, J.S.; Moorman, J.R. Physiological time-series analysis using approximate entropy and sample entropy. Am. J. Physiol. Heart Circ. Physiol. 2000, 278, H2039–H2049. [Google Scholar] [CrossRef]
- Xiong, J.; Liang, X.; Zhao, L.; Lo, B.; Li, J.; Liu, C. Improving Accuracy of Heart Failure Detection Using Data Refinement. Entropy 2020, 22, 520. [Google Scholar] [CrossRef]
- Costa, M.; Goldberger, A.; Peng, C. Multiscale entropy analysis of biological signals. Phys. Rev. E 2005, 71, 021906. [Google Scholar] [CrossRef]
- Tavakoli, M.; Taylor, J.N.; Li, C.B.; Komatsuzaki, T.; Pressé, S. Single Molecule Data Analysis: An Introduction. In Advances in Chemical Physics (Book 162); Rice, S.A., Dinner, A.R., Eds.; O’Reilly: Sebastopol, CA, USA, 2013; pp. 205–306. [Google Scholar]
- Tsallis, C. The nonadditive entropy Sq and its applications in physics and elsewhere: Some remarks. Entropy 2011, 13, 1765–1804. [Google Scholar] [CrossRef]
- Nielsen, F.; Nock, R. On Renyi and Tsallis entropies and divergences for exponential families. J. Phys. A 2011, 45, 032003. [Google Scholar] [CrossRef]
- Renyi, A. On measures of information and entropy. In Proceedings of the fourth Berkeley Symposium on Mathematics, Statistics and Probability 1960, Berkeley, CA, USA, 20 June–30 July 1960; Neyman, J., Ed.; Statistical Laboratory of the University of California, University of California Press: Berkeley, CA, USA, 1961; Volume 1, pp. 547–561. [Google Scholar]
- Moffitt, J.; Chemla, Y.; Izhaky, D.; Bustamante, C. Differential detection of dual traps improves the spatial resolution of optical tweezers. Proc. Natl. Acad. Sci. USA 2006, 103, 9006–9011. [Google Scholar] [CrossRef] [PubMed]
- Bauer, D.; Merz, D.; Pelz, B.; Theisen, K.; Yacyshyn, G.; Mokranjac, D.; Dima, R.; Rief, M.; Zoldak, G. Nucleotides regulate the mechanical hierarchy between subdomains of the nucleotide binding domain of the Hsp70 chaperone DnaK. Proc. Natl. Acad. Sci. USA 2015, 112, 10389–10394. [Google Scholar] [CrossRef] [PubMed]
- Bauer, D.; Meinhold, S.; Jakob, R.; Stigler, J.; Merkel, U.; Maier, T.; Rief, M.; Zoldak, G. A folding nucleus and minimal ATP binding domain of Hsp70 identified by single-molecule force spectroscopy. Proc. Natl. Acad. Sci. USA 2018, 115, 4666–4671. [Google Scholar] [CrossRef]
- Gebhardt, J.; Bornschlögl, T.; Rief, M. Full distance-resolved folding energy landscape of one single protein molecule. Proc. Natl. Acad. Sci. USA 2010, 107, 2013–2018. [Google Scholar] [CrossRef]
- Tolic-Norrelykke, S.; Schäffer, E.; Flyvbjerg, H. Calibration of optical tweezers with positional detection in the back focal plane. Rev. Sci. Instrum 2006, 77, 103101. [Google Scholar] [CrossRef]
- Baba, A.; Komatsuzaki, T. Construction of effective free energy landscape from single-molecule time series. Proc. Natl. Acad. Sci. USA 2007, 104, 19297–19302. [Google Scholar] [CrossRef]
- Schuetz, P.; Wuttke, R.; Schuler, B.; Caflisch, A. Free Energy Surfaces from Single-Distance Information. J. Phys. Chem. B 2010, 114, 15227–15235. [Google Scholar] [CrossRef]
- Gell-Mann, M.; Tsallis, C. Nonextensive Entropy: Interdisciplinary Applications; Oxford University Press: Oxford, UK, 2004. [Google Scholar]
- Shade, A.; Jones, S.; Caporaso, J.; Handelsman, J.; Knight, R.; Fierer, N.; Gilbert, J. Conditionally Rare Taxa Disproportionately Contribute to Temporal Changes in Microbial Diversity. mBio 2014, 5. [Google Scholar] [CrossRef]
- Bullen, P. Handbook of Means and Their Inequalities (Mathematics and Its Applications); Mathematics and Its Applications; Springer: Berlin/Heidelberg, Germany, 2003. [Google Scholar]
- Sluciak, O. On Inflection Points of the Lehmer Mean Function. arXiv, 2015; arXiv:1509.09277. [Google Scholar]
- Ito, M. Estimations of the Lehmer mean by the Heron mean and their generalizations involving refined Heinz operator inequalities. Adv. Oper. Theory 2018, 3, 763–780. [Google Scholar] [CrossRef]
- Amat, S.; Magrenan, A.; Ruiz, J.; Trillo, J.C.; Yanez, D.F. On the application of Lehmer means in signal and image processing. Int. J. Comput. Math. 2019, 97, 1–26. [Google Scholar] [CrossRef]
- Burlaga, L.; Vinas, A. Triangle for the entropic index q of non-extensive statistical mechanics observed by Voyager 1 in the distant heliosphere. Phys. A 2005, 356, 375–384. [Google Scholar] [CrossRef]
- Levner, I. Feature selection and nearest centroid classification for protein mass spectrometry. Bioinformatics 2005, 6, 68. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2014. [Google Scholar]
- Welch, B. The generalization of Student’s problem when several different population variances are involved. Biometrika 1947, 34, 28–35. [Google Scholar] [CrossRef]
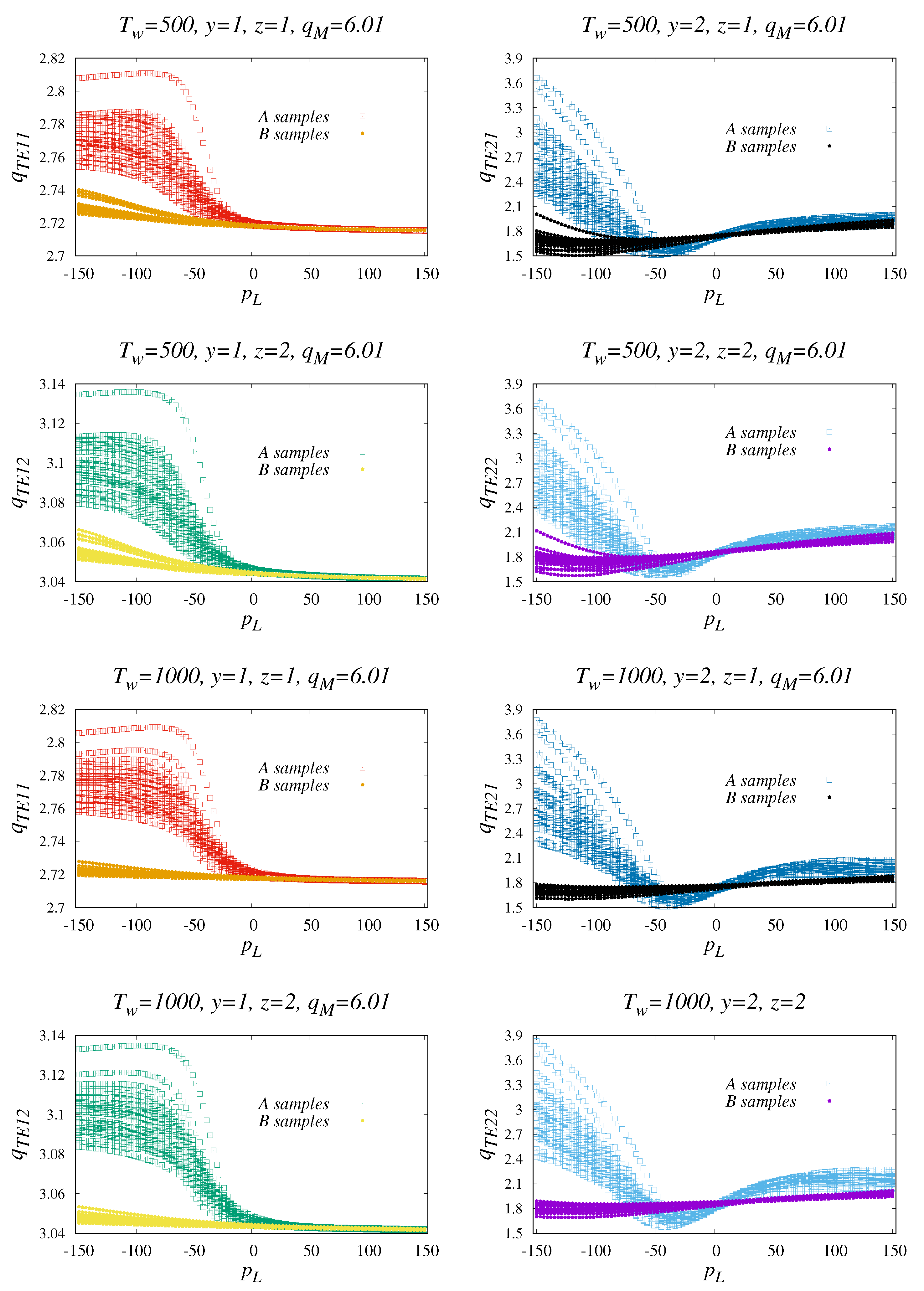
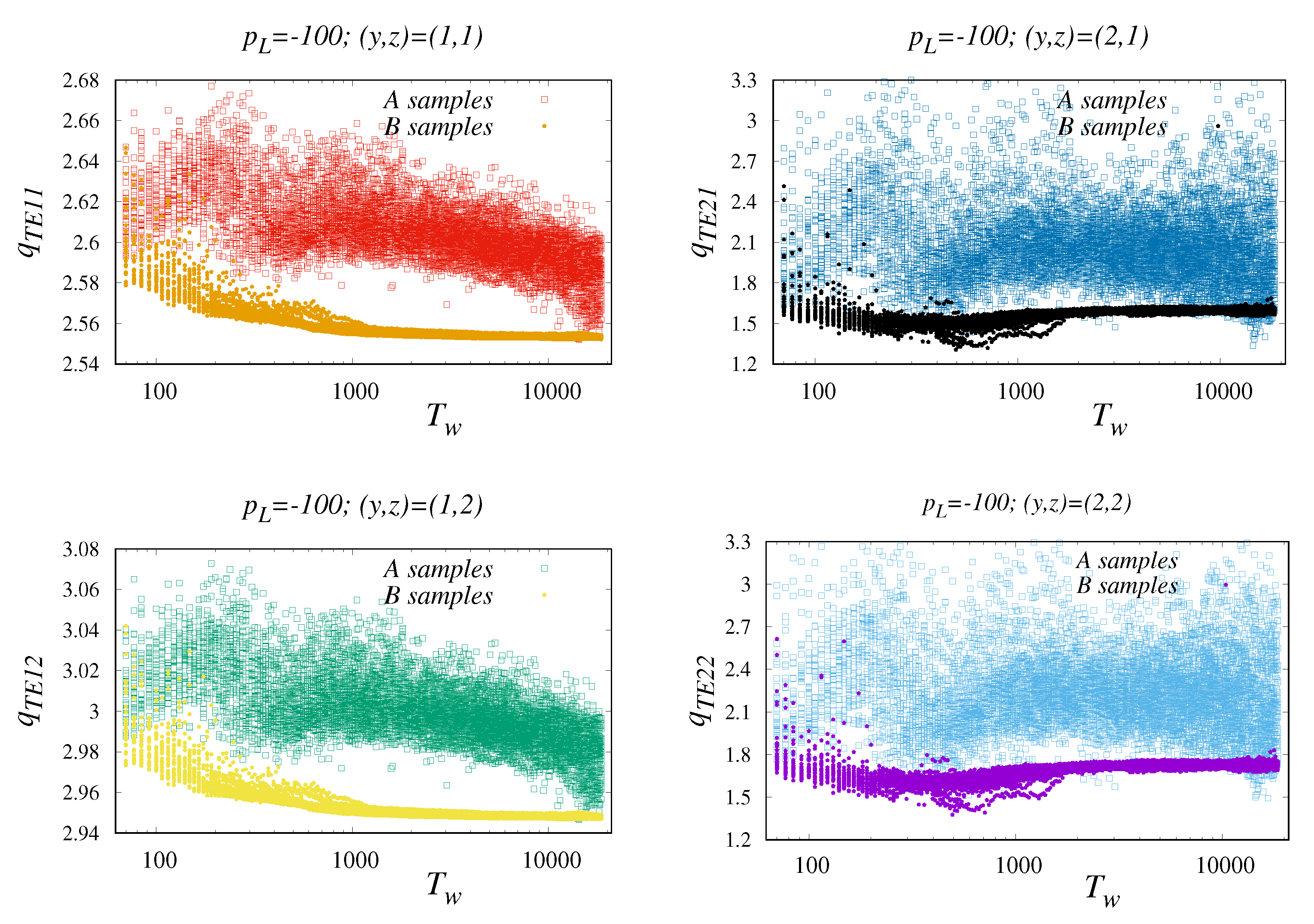
| Window | Category | Indicator | Relative: | Min | 1st Qu | Median | Mean | 3rd Qu | Max |
|---|---|---|---|---|---|---|---|---|---|
| Samples | Median for A to Median for B | ||||||||
| 500 | A | 2.577 | 2.586 | 2.590 | 2.592 | 2.599 | 2.627 | ||
| 500 | B | 2.558 | 2.559 | 2.559 | 2.560 | 2.560 | 2.565 | ||
| 500 | A | 2.971 | 2.980 | 2.984 | 2.986 | 2.994 | 3.021 | ||
| 500 | B | 2.953 | 2.954 | 2.954 | 2.954 | 2.954 | 2.959 | ||
| 500 | A | 1.607 | 1.718 | 1.801 | 1.828 | 1.894 | 2.386 | ||
| 500 | B | 1.427 | 1.511 | 1.522 | 1.520 | 1.541 | 1.629 | ||
| 500 | A | 1.714 | 1.835 | 1.928 | 1.947 | 2.013 | 2.466 | ||
| 500 | B | 1.524 | 1.623 | 1.636 | 1.633 | 1.658 | 1.749 | ||
| 1000 | A | 2.583 | 2.589 | 2.595 | 2.597 | 2.601 | 2.628 | ||
| 1000 | B | 2.554 | 2.555 | 2.556 | 2.556 | 2.556 | 2.559 | ||
| 1000 | A | 2.978 | 2.984 | 2.991 | 2.991 | 2.996 | 3.023 | ||
| 1000 | B | 2.949 | 2.950 | 2.950 | 2.950 | 2.951 | 2.954 | ||
| 1000 | A | 1.735 | 1.818 | 1.918 | 1.940 | 2.020 | 2.473 | ||
| 1000 | B | 1.476 | 1.542 | 1.567 | 1.558 | 1.578 | 1.589 | ||
| 1000 | A | 1.867 | 1.944 | 2.050 | 2.069 | 2.135 | 2.576 | ||
| 1000 | B | 1.584 | 1.660 | 1.689 | 1.679 | 1.704 | 1.716 | ||
| 2000 | A | 2.579 | 2.588 | 2.591 | 2.592 | 2.595 | 2.608 | ||
| 2000 | B | 2.553 | 2.554 | 2.554 | 2.554 | 2.555 | 2.556 | ||
| 2000 | A | 2.973 | 2.982 | 2.986 | 2.986 | 2.991 | 3.002 | ||
| 2000 | B | 2.948 | 2.949 | 2.949 | 2.949 | 2.949 | 2.951 | ||
| 2000 | A | 1.711 | 1.787 | 1.834 | 1.867 | 1.933 | 2.220 | ||
| 2000 | B | 1.564 | 1.577 | 1.583 | 1.584 | 1.590 | 1.601 | ||
| 2000 | A | 1.816 | 1.929 | 1.985 | 2.012 | 2.090 | 2.328 | ||
| 2000 | B | 1.686 | 1.703 | 1.710 | 1.711 | 1.719 | 1.733 |
| p-Value | 95% Confidence Interval | ||||
|---|---|---|---|---|---|
| 500 | × | ||||
| 500 | × | ||||
| 500 | × | ||||
| 500 | × | ||||
| 1000 | |||||
| 1000 | |||||
| 1000 | |||||
| 1000 | × | ||||
| 2000 | × | ||||
| 2000 | × | ||||
| 2000 | |||||
| 2000 | × |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Horvath, D.; Žoldák, G. Entropy-Based Strategies for Rapid Pre-Processing and Classification of Time Series Data from Single-Molecule Force Experiments. Entropy 2020, 22, 701. https://doi.org/10.3390/e22060701
Horvath D, Žoldák G. Entropy-Based Strategies for Rapid Pre-Processing and Classification of Time Series Data from Single-Molecule Force Experiments. Entropy. 2020; 22(6):701. https://doi.org/10.3390/e22060701
Chicago/Turabian StyleHorvath, Denis, and Gabriel Žoldák. 2020. "Entropy-Based Strategies for Rapid Pre-Processing and Classification of Time Series Data from Single-Molecule Force Experiments" Entropy 22, no. 6: 701. https://doi.org/10.3390/e22060701
APA StyleHorvath, D., & Žoldák, G. (2020). Entropy-Based Strategies for Rapid Pre-Processing and Classification of Time Series Data from Single-Molecule Force Experiments. Entropy, 22(6), 701. https://doi.org/10.3390/e22060701






