Evaluation of Survival Outcomes of Endovascular Versus Open Aortic Repair for Abdominal Aortic Aneurysms with a Big Data Approach
Abstract
1. Introduction
- It strives to compare the treatment effects of EVAR and OAR under the clinical trial framework, as opposed to the commonly adopted observational data analysis framework. The conclusion so generated may have important and direct clinical implications.
- It advances from the existing emulation analyses by investigating a new disease condition and treatments, which may further expand the paradigm of emulation analysis.
- Deep learning techniques, as opposed to “simple” regressions, are adopted. This study may assist in introducing deep learning to the emulation paradigm, as well as further fostering deep learning research. Specifically, this is the first application of deep learning to the emulation analysis and study of rAAA. Building on the existing deep learning components, we assemble an analysis pipeline that mimics the “propensity score + inverse probability treatment (IPT) weighting Cox regression” approach [18,19].
2. Methods
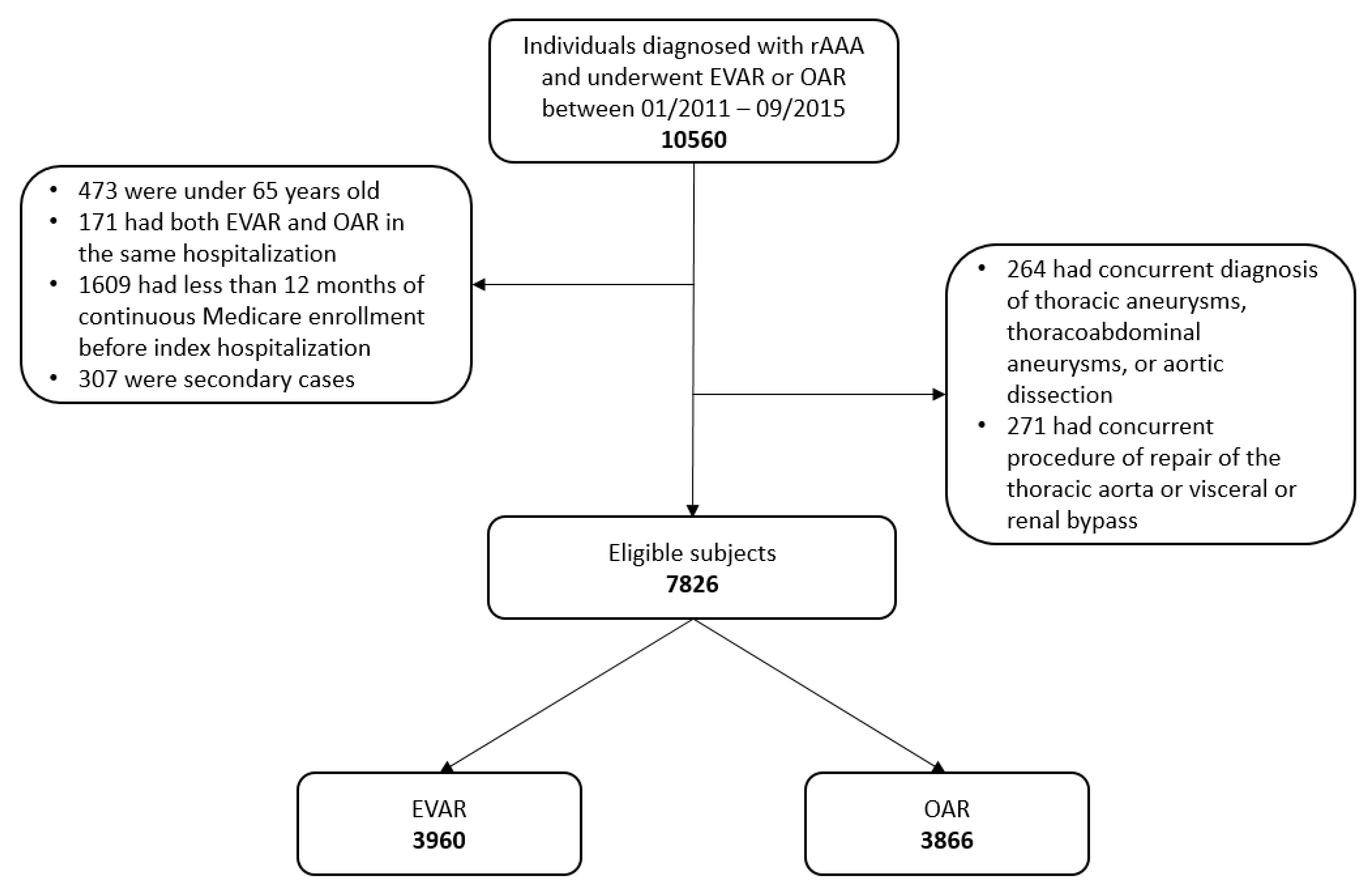
2.1. Data Source
2.2. The Target Randomized Clinical Trial
2.3. The Emulated Trial
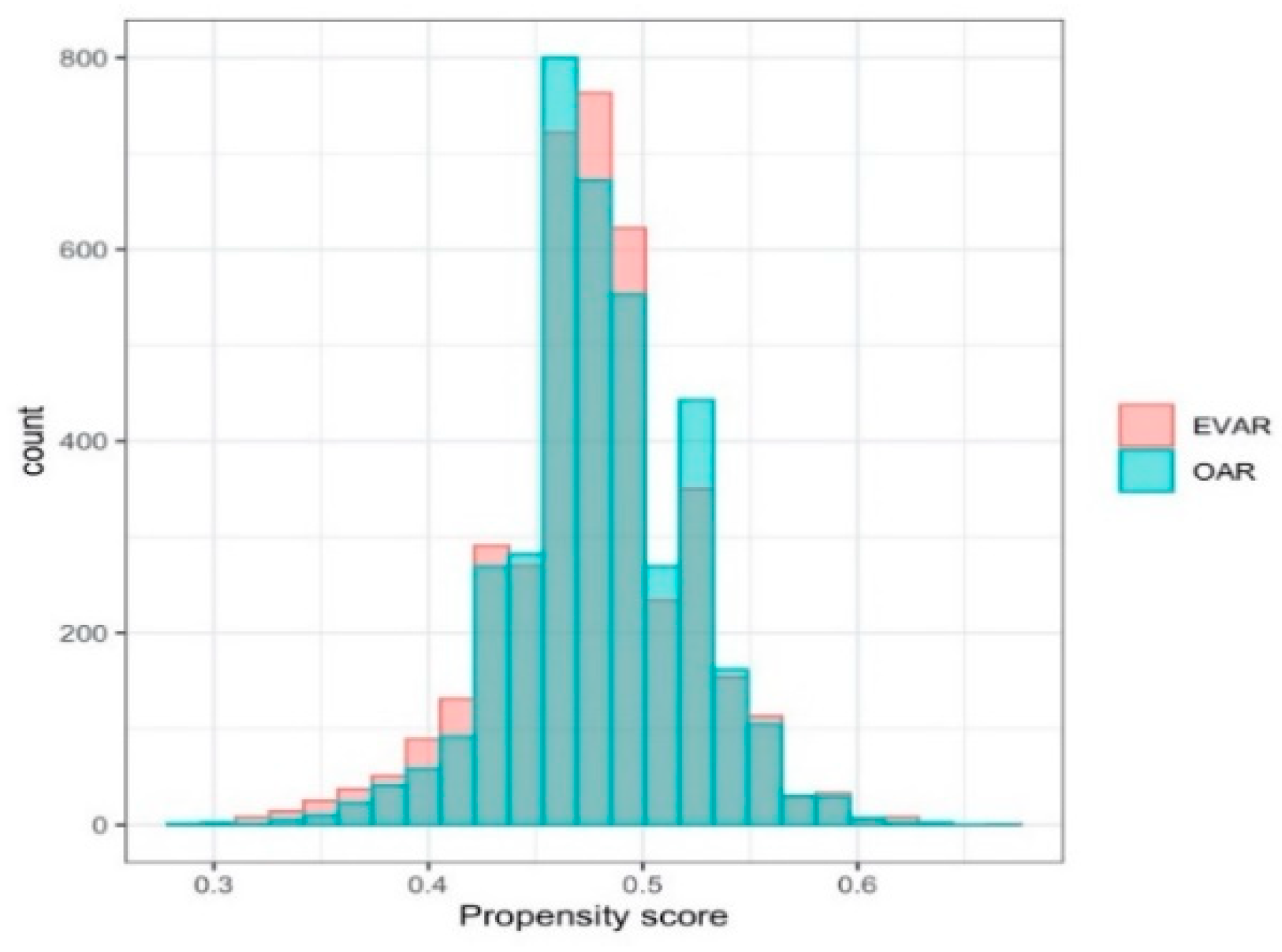
2.4. Data Analysis
Remarks
3. Results
3.1. Patient Characteristics and Unadjusted Incidences
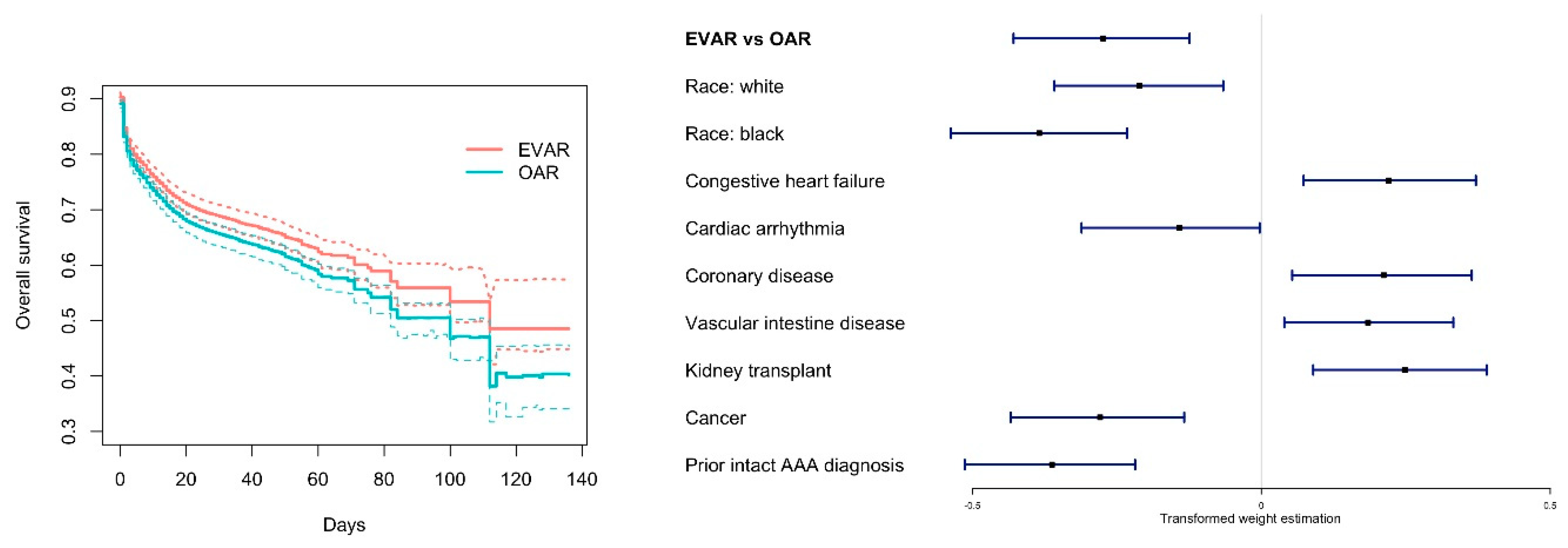
3.2. Analysis of the Emulated Trial
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Variable | ICD-9-CM Code * |
| Inclusion | |
| Ruptured abdominal aortic aneurysm | 441.3 |
| Endovascular aortic repair | 39.71 |
| Open aortic repair | 38.44, 39.25, 39.52, 38.34, 38.64, 38.40, 38.60 |
| Exclusion | |
| Thoracic aneurysms | 441.1, 441.2 |
| Thoracoabdominal aneurysms | 441.6, 441.7 |
| Aortic dissection | 441.00-441.03 |
| Repair of the thoracic aorta | 38.35, 38.45, 39.73 |
| Visceral or renal bypass | 38.46, 39.24, 39.26 |
| Medical history | |
| Congestive heart failure | 398.91, 402.01, 402.11, 402.91, 404.01, 404.03, 404.11, 404.91, 404.13, 404.93, 425.4, 425.5, 425.7, 425.8, 425.9, 428.0, 428.1, 428.20, 428.22, 428.30, 428.32, 428.40, 428.42, 428.9 |
| Cardiac arrhythmia | 426.0, 426.10, 426.11, 426.12, 426.13, 426.7, 426.9, 427.0, 427.1, 427.2, 427.3, 427.9, V45.0, V53.3 |
| Valvular disease | 093.2, 394, 395, 396, 397, 424, V42.2, V43.3 |
| Coronary disease | 412, 413, 414, 429.2 |
| Diabetes | 250 |
| Hypertension | 401, 402, 403, 404, 405 |
| Chronic Obstructive Pulmonary diseases | 416, 417.9, 490, 491, 492, 493, 494, 495.0, 495.1, 495.2, 495.3, 495.4, 495.5, 495.6, 495.8, 495.9, 496, 500, 501, 502, 503, 504, 505, 506.0, 506.2, 506.4, 506.9, 508.1, 508.8, 508.9 |
| Clinically significant lower extremity vascular diseases | 440.22, 440.23, 440.24, 440.3, 444.22, V43.4, |
| Renal atherosclerosis | 440.1 |
| Vascular intestine disease | 557.1 |
| Renal failure w dialysis | V45.1, V56.0, V56.1, V56.2, V56.3, V56.8, 585.6, 39.95 (w/o 586) |
| Renal failure without dialysis | 403.01, 403.11, 403.91, 404.02, 404.03, 404.12, 404.13, 404.92, 404.93, 585 (w/o 585.6), 588.0 |
| Other renal diseases | 582, 583.0, 583.1, 583.2, 583.4 |
| Kidney transplant | V420 |
| Liver disease | 070.22, 070.23, 070.32, 070.33, 070.44, 070.54, 070.9, 456.0, 456.1, 571, 572.1, 572.2, 572.3, 572.4, 572.8, 573.0, 573.1, 573.8, 573.9 |
| Cerebrovascular diseases and paralysis | 342, 344.1, 344.3, 344.4, 344.5, 344.9, 437.0, 438 |
| Other neurological diseases | 330, 331, 332, 333, 334.0, 334.1, 334.2, 334.4, 334.8, 335.0, 335.1, 335.2, 335.8, 335.9, 336.0, 336.2, 343, 344.0, 348.1, 348.3, 344.2, 344.6, 345, 437.3, 437.4, 437.5, 437.6, 437.7 |
| Hyperlipidemia | 272 |
| Cancer | 140, 141, 142, 143, 144,145, 146, 147, 148, 149, 150, 151, 152, 153, 154, 155, 156, 157, 158,159, 160, 161, 162, 163, 164, 165, 170, 171,172, 174, 175, 176, 179, 180, 181, 182, 183, 184, 185, 186, 187, 188, 189, 190, 191, 192, 193, 194, 195, 196, 197, 198, 199, 200, 201, 202, 203.0, 238.6 |
| Rheumatoid arthritis | 446, 701.0, 710.0, 710.1, 710.2, 710.3, 710.4, 710.8, 710.9, 711.2, 719.3, 714,720, 725, 728.5, 728.89 |
| Prior intact AAA diagnosis | 441.4, without mention 441.3 |
Appendix B
| Algorithm A1. Algorithm for the bootstrap type analysis (Step 3). |
| 1. Initialize . |
| 2. Update , randomly sample 0.632n subjects without replacement from the original data and conduct the following procedure. |
| Step 1. Estimate the propensity score. |
| • Construct a simple multilayer perceptron with one hidden layer, ReLU and sigmoid activations, and binary cross-entropy as the loss function. |
| • For a fixed learning rate from a vector of hyper-parameter values: ○ Compile the model using a stochastic gradient descent optimizer with Nesterov momentum. ○ Compute the classification accuracy as the metrics for model selection. |
| • Select the best model with the highest accuracy from the above grid search, and calculate the IPT weight by transforming the estimated propensity score. |
| Step 2. Assess the treatment effects using the IPT weighted survival analysis. |
| • Construct a multilayer perceptron with two hidden layers, ReLU activations, and the proposed loss function . |
| • For a fixed learning rate, the number of nodes in hidden layers from a grid of hyper-parameter values: ○ Compile the model using Adam optimizer. ○ Estimate the weights , and compute the concordance statistic for model selection. |
| • Select the best model with the largest concordance, and obtain the effects of treatment and other confounders by multiplying the estimated weights across layers. |
| 3. Repeat until is large enough (e.g., ). |
References
- Kent, K.C.; Zwolak, R.M.; Egorova, N.N.; Riles, T.S.; Manganaro, A.; Moskowitz, A.J.; Gelijns, A.C.; Greco, G. Analysis of risk factors for abdominal aortic aneurysm in a cohort of more than 3 million individuals. J. Vasc. Surg. 2010, 52, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Heikkinen, M.; Salenius, J.P.; Auvinen, O. Ruptured abdominal aortic aneurysm in a well- defined geographic area. J. Vasc. Surg. 2002, 36, 291–296. [Google Scholar] [CrossRef] [PubMed]
- Minino, A.M.; Murphy, S.H.; Xu, J.; Kochanek, K.D. Division of Vital Statistics. Deaths: Final Data for 2009. Centers for Disease Control and Prevention. Natl. Vital Stat. Rep. 2011, 59, 10. [Google Scholar]
- Sakalihasan, N.; Limet, R.; Defawe, O.D. Abdominal aortic aneurysm. Lancet 2005, 365, 1577–1589. [Google Scholar] [CrossRef]
- MacSweeney, S.T.R.; Ellis, M.; Greenhalgh, R.M.; Powell, J.T. Smoking and growth rate of small abdominal aortic aneurysms. Lancet 1994, 344, 651–652. [Google Scholar] [CrossRef]
- Schermerhorn, M.L.; Buck, D.B.; O’Malley, A.J.; Curran, T.; McCallum, J.C.; Darling, J.; Landon, B.E. Long-Term Outcomes of Abdominal Aortic Aneurysm in the Medicare Population. N. Engl. J. Med. 2015, 373, 328–338. [Google Scholar] [CrossRef]
- Lederle, F.A.; Freischlag, J.A.; Kyriakides, T.C.; Padberg, F.T.; Matsumura, J.S.; Kohler, T.R.; Lin, P.H.; Jean-Claude, J.M.; Cikrit, D.F.; Peduzzi, P.N.; et al. Outcomes Following Endovascular vs Open Repair of Abdominal Aortic Aneurysm: A Randomized Trial. JAMA 2009, 302, 1535–1542. [Google Scholar] [CrossRef]
- De Bruin, J.L.; Baas, A.F.; Buth, J.; Prinssen, M.; Verhoeven, E.L.G.; Cuypers, P.W.M.; van Sambeek, M.R.H.M.; Balm, R.; Grobbee, D.E.; Blankensteijn, J.D. Blankensteijn Long-Term Outcome of Open or Endovascular Repair of Abdominal Aortic Aneurysm. N. Engl. J. Med. 2010, 362, 1881–1889. [Google Scholar] [CrossRef]
- Patel, R. Endovascular versus open repair of abdominal aortic aneurysm in 15-years’ follow-up of the UK endovascular aneurysm repair trial 1 (EVAR trial 1): A randomised controlled trial. Lancet 2016, 388, 2366–2374. [Google Scholar] [CrossRef]
- Edwards, S.T.; Schermerhorn, M.L.; O’Malley, A.J.; Bensley, R.P.; Hurks, R.; Cotterill, P.; Landon, B.E. Landon Comparative effectiveness of endovascular versus open repair of ruptured abdominal aortic aneurysm in the Medicare population. J. Vasc. Surg. 2014, 59, 575–582. [Google Scholar] [CrossRef]
- Behrendt, C.-A.; Sedrakyan, A.; Rieß, H.C.; Heidemann, F.; Kölbel, T.; Debus, E.S. Short-term and long-term results of endovascular and open repair of abdominal aortic aneurysms in Germany. J. Vasc. Surg. 2017, 66, 1704–1711. [Google Scholar] [CrossRef] [PubMed]
- Egorova, N.N.; Vouyouka, A.G.; McKinsey, J.F.; Faries, P.L.; Kent, K.C.; Moskowitz, A.J.; Gelijns, A. Effect of gender on long-term survival after abdominal aortic aneurysm repair based on results from the Medicare national database. J. Vasc. Surg. 2011, 54, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Jackson, R.S.; Chang, D.C.; Freischlag, J.A. Comparison of Long-term Survival After Open vs Endovascular Repair of Intact Abdominal Aortic Aneurysm Among Medicare Beneficiaries. JAMA 2012, 307, 1621–1628. [Google Scholar] [CrossRef] [PubMed]
- Hernán, M.A.; Robins, J.M. Causal Inference: What If; Chapman Hill/Crc: Boca Raton, FL, USA, 2020. [Google Scholar]
- Van der Laan, M.J.; Rose, S. Targeted Learning: Causal Inference for Observational and Experimental Data; Springer Science & Business Media: New York, NY, USA, 2011; ISBN 978-1-4419-9782-1. [Google Scholar]
- Chipman, H.A.; George, E.I.; McCulloch, R.E. BART: Bayesian additive regression trees. Ann. Appl. Stat. 2010, 4, 266–298. [Google Scholar] [CrossRef]
- Hernán, M.A.; Alonso, A.; Logan, R.; Grodstein, F.; Stampfer, M.J.; Willett, W.C.; Manson, J.E.; Robins, M. Observational studies analyzed like randomized experiments: An application to postmenopausal hormone therapy and coronary heart disease. Epidemiology (Camb. Mass.) 2008, 19, 766–779. [Google Scholar] [CrossRef]
- Dickerman, B.A.; García-Albéniz, X.; Logan, R.W.; Denaxas, S.; Hernán, M.A. Avoidable flaws in observational analyses: An application to statins and cancer. Nat. Med. 2019, 25, 1601–1606. [Google Scholar] [CrossRef]
- Petito, L.C.; García-Albéniz, X.; Logan, R.W.; Howlader, N.; Mariotto, A.B.; Dahabreh, I.J.; Hernán, M.A. Estimates of Overall Survival in Patients With Cancer Receiving Different Treatment Regimens: Emulating Hypothetical Target Trials in the Surveillance, Epidemiology, and End Results (SEER)–Medicare Linked Database. JAMA Netw Open 2020, 3, e200452. [Google Scholar] [CrossRef]
- Zigler, C.M.; Kim, C.; Choirat, C.; Hansen, J.B.; Wang, Y.; Hund, L.; Samet, J.; King, G.; Dominici, F. Causal Inference Methods for Estimating Long-Term Health Effects of Air Quality Regulations. Europe PMC 2016, 187, 5–49. [Google Scholar]
- Deng, L.; Yu, D. Deep Learning: Methods and Applications. SIG 2014, 7, 197–387. [Google Scholar] [CrossRef]
- Lv, Y.; Duan, Y.; Kang, W.; Li, Z.; Wang, F.-Y. Traffic Flow Prediction With Big Data: A Deep Learning Approach—IEEE Journals & Magazine. IEEE Trans. Intell. Transp. Syst. 2015, 16, 865–873. [Google Scholar]
- Wang, D.; Khosla, A.; Gargeya, R.; Irshad, H.; Beck, A.H. Deep Learning for Identifying Metastatic Breast Cancer. arXiv 2016, arXiv:1606.05718. [Google Scholar]
- Badgeley, M.A.; Zech, J.R.; Oakden-Rayner, L.; Glicksberg, B.S.; Liu, M.; Gale, W.; McConnell, M.V.; Percha, B.; Snyder, T.M.; Dudley, J.T. Dudley Deep learning predicts hip fracture using confounding patient and healthcare variables. NPJ Digit. Med. 2019, 2, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Chung, K.; Yoo, H.; Choe, D. Ambient context-based modeling for health risk assessment using deep neural network. J. Ambient Intell. Humaniz. Comput. 2020, 11, 1387–1395. [Google Scholar] [CrossRef]
- Hsiao, H.C.; Chen, S.H.; Tsai, J.J. Deep Learning for Risk Analysis of Specific Cardiovascular Diseases Using Environmental Data and Outpatient Records. In Proceedings of the 2016 IEEE 16th International Conference on Bioinformatics and Bioengineering (BIBE), Taichung, Taiwan, 31 October–2 November 2016. [Google Scholar]
- Katzman, J.L.; Shaham, U.; Cloninger, A.; Bates, J.; Jiang, T.; Kluger, Y. DeepSurv: Personalized treatment recommender system using a Cox proportional hazards deep neural network. BMC Med. Res. Methodol. 2018, 18, 24. [Google Scholar] [CrossRef]
- Mues, K.E.; Liede, A.; Liu, J.; Wetmore, J.B.; Zaha, R.; Bradbury, B.D.; Collins, A.J.; Gilbertson, D.T. Use of the Medicare database in epidemiologic and health services research: A valuable source of real-world evidence on the older and disabled populations in the US. Clin. Epidemiol. 2017, 9, 267. [Google Scholar] [CrossRef]
- Brennan, N.; Oelschlaeger, A.; Cox, C.; Tavenner, M. Leveraging The Big-Data Revolution: CMS Is Expanding Capabilities To Spur Health System Transformation. Health Aff. 2014, 33, 1195–1202. [Google Scholar] [CrossRef]
- Hernán, M.A.; Robins, J.M. Using Big Data to Emulate a Target Trial When a Randomized Trial Is Not Available. Am. J. Epidemiol. 2016, 183, 758–764. [Google Scholar] [CrossRef]
- Westreich, D.; Cole, S.R.; Schisterman, E.F.; Platt, R.W. A simulation study of finite-sample properties of marginal structural Cox proportional hazards models. Stat. Med. 2012, 31, 2098–2109. [Google Scholar] [CrossRef] [PubMed]
- Robins, J.M. Association, causation, and marginal structural models. Synthese 1999, 121, 151–179. [Google Scholar] [CrossRef]
- Jiang, W.; Simon, R. A comparison of bootstrap methods and an adjusted bootstrap approach for estimating the prediction error in microarray classification. Stat. Med. 2007, 26, 5320–5334. [Google Scholar] [CrossRef] [PubMed]
- Ching, T.; Zhu, X.; Garmire, L.X. Cox-nnet: An artificial neural network method for prognosis prediction of high-throughput omics data. PLoS Comput. Biol. 2018, 14, e1006076. [Google Scholar] [CrossRef] [PubMed]
- Allen-Zhu, Z.; Li, Y.; Song, Z. A convergence theory for deep learning via over-parameterization. In Proceedings of the 36th International Conference on Machine Learning (PMLR 2019), Long Beach, CA, USA, 9–15 June 2019; Volume 97, pp. 242–252. [Google Scholar]
- Farrell, M.H.; Liang, T.; Misra, S. Deep neural networks for estimation and inference. arxiv 2018, arXiv:1809.09953. [Google Scholar]
- Ali, F.; El-Sappagh, S.; Islam, S.M.R.; Kwak, D.; Ali, A.; Imran, M.; Kwak, K.-S. A smart healthcare monitoring system for heart disease prediction based on ensemble deep learning and feature fusion. Inf. Fusion 2012, 63, 208–222. [Google Scholar] [CrossRef]
- Ali, F.; El-Sappagh, S.; Islam, S.M.R.; Ali, A.; Attique, M.; Imran, M.; Kwak, K.-S. An intelligent healthcare monitoring framework using wearable sensors and social networking data. Future Gener. Comput. Syst. 2020, 114, 23–43. [Google Scholar] [CrossRef]
- Jain, K.; Neelakantan, M.; Key, P. Limitations in the Analysis of Atherectomy Using Medicare Big Data. J. Endovasc. Ther. 2020. [Google Scholar] [CrossRef] [PubMed]
- Selden, T.M. Differences Between Public And Private Hospital Payment Rates Narrowed, 2012–2016: A data analysis comparing payment rate differences between private insurance and Medicare for inpatient hospital stays, emergency department visits, and outpatient hospital care. Health Aff. 2020, 39, 94–99. [Google Scholar]
- Dardik, A.; Burleyson, G.P.; Bowman, H.; Gordon, T.A.; Williams, G.M.; Webb, T.H.; Perler, B.A. Surgical repair of ruptured abdominal aortic aneurysms in the state of Maryland: Factors influencing outcome among 527 recent cases. J. Vasc. Surg. 1998, 28, 413–421. [Google Scholar] [CrossRef][Green Version]

| EVAR (N = 3930) | OAR (N = 3866) | p-Value * | |
|---|---|---|---|
| Demographic | |||
| Age, mean(sd) | 78.03 (7.52) | 76.59 (6.90) | <0.0001 |
| Male | 3023 (76.34) | 2786 (72.06) | <0.0001 |
| Race | 0.0030 | ||
| White | 3588 (90.77) | 3550 (92.02) | |
| Black | 249 (6.30) | 178 (4.61) | |
| Other | 116 (2.93) | 130 (3.37) | |
| Medical conditions | |||
| Congestive heart failure | 464 (11.72) | 299 (7.73) | <0.0001 |
| Cardiac arrhythmia | 596 (15.05) | 438 (11.33) | <0.0001 |
| Valvular disease | 199 (5.03) | 172 (4.45) | 0.2304 |
| Coronary disease | 758 (19.14) | 603 (15.60) | <0.0001 |
| Diabetes | 329 (8.31) | 256 (6.62) | 0.0045 |
| Hypertension | 1250 (31.25) | 1078 (27.88) | 0.0004 |
| Chronic obstructive pulmonary diseases | 707 (17.85) | 584 (15.11) | 0.0011 |
| Clinically significant lower extremity vascular diseases | 26 (0.66) | 27 (0.70) | 0.8215 |
| Renal atherosclerosis | 20 (0.51) | 27 (0.70) | 0.2684 |
| Vascular intestine disease | 7 (0.18) | 2 (0.05) | 0.1027 |
| Renal failure | 493 (12.45) | 358 (9.26) | <0.0001 |
| Other renal diseases | 3 (0.08) | 1 (0.03) | 0.3289 |
| Kidney transplant | 4 (0.10) | 3 (0.08) | 0.7291 |
| Liver disease | 33 (0.83) | 30 (0.78) | 0.7766 |
| Cerebrovascular diseases and paralysis | 93 (2.35) | 67 (1.73) | 0.0544 |
| Other neurological diseases | 153 (3.86) | 114 (2.95) | 0.0258 |
| Hyperlipidemia | 817 (20.63) | 687 (17.77) | 0.0013 |
| Cancer | 132 (3.33) | 87 (2.25) | 0.0037 |
| Rheumatoid arthritis | 76 (1.92) | 39 (1.01) | 0.0008 |
| Prior intact AAA diagnosis | 511 (12.90) | 440 (11.38) | 0.0393 |
| Other | |||
| Year in which repair was performed | <0.0001 | ||
| 2011 | 808 (20.40) | 1013 (26.20) | |
| 2012 | 869 (21.94) | 913 (23.62) | |
| 2013 | 819 (20.68) | 785 (20.31) | |
| 2014 | 837 (21.14) | 701 (18.13) | |
| 2015 | 627 (15.83) | 454 (11.74) | |
| Outcome (followed until death, loss to follow-up, or 06/30/2019) | |||
| All-cause mortality | 2430 (61.36) | 2542 (65.75) | <0.0001 |
| Perioperative mortality (in-hospital or 30 days after discharge) | 1107 (27.95) | 1704 (44.08) | <0.0001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mei, H.; Xu, Y.; Wang, J.; Ma, S. Evaluation of Survival Outcomes of Endovascular Versus Open Aortic Repair for Abdominal Aortic Aneurysms with a Big Data Approach. Entropy 2020, 22, 1349. https://doi.org/10.3390/e22121349
Mei H, Xu Y, Wang J, Ma S. Evaluation of Survival Outcomes of Endovascular Versus Open Aortic Repair for Abdominal Aortic Aneurysms with a Big Data Approach. Entropy. 2020; 22(12):1349. https://doi.org/10.3390/e22121349
Chicago/Turabian StyleMei, Hao, Yaqing Xu, Jiping Wang, and Shuangge Ma. 2020. "Evaluation of Survival Outcomes of Endovascular Versus Open Aortic Repair for Abdominal Aortic Aneurysms with a Big Data Approach" Entropy 22, no. 12: 1349. https://doi.org/10.3390/e22121349
APA StyleMei, H., Xu, Y., Wang, J., & Ma, S. (2020). Evaluation of Survival Outcomes of Endovascular Versus Open Aortic Repair for Abdominal Aortic Aneurysms with a Big Data Approach. Entropy, 22(12), 1349. https://doi.org/10.3390/e22121349





