A Nonparametric Model for Multi-Manifold Clustering with Mixture of Gaussians and Graph Consistency
Abstract
1. Introduction
- A constrained MoG distribution has been applied to model the non-Gaussian manifold distribution.
- We integrate the graph theory with DPM to capture the manifold geometrical information.
- The variational inference-based optimization framework is proposed to carry out the model inference and learning.
2. Materials and Methods
2.1. Dirichlet Process Mixture Model
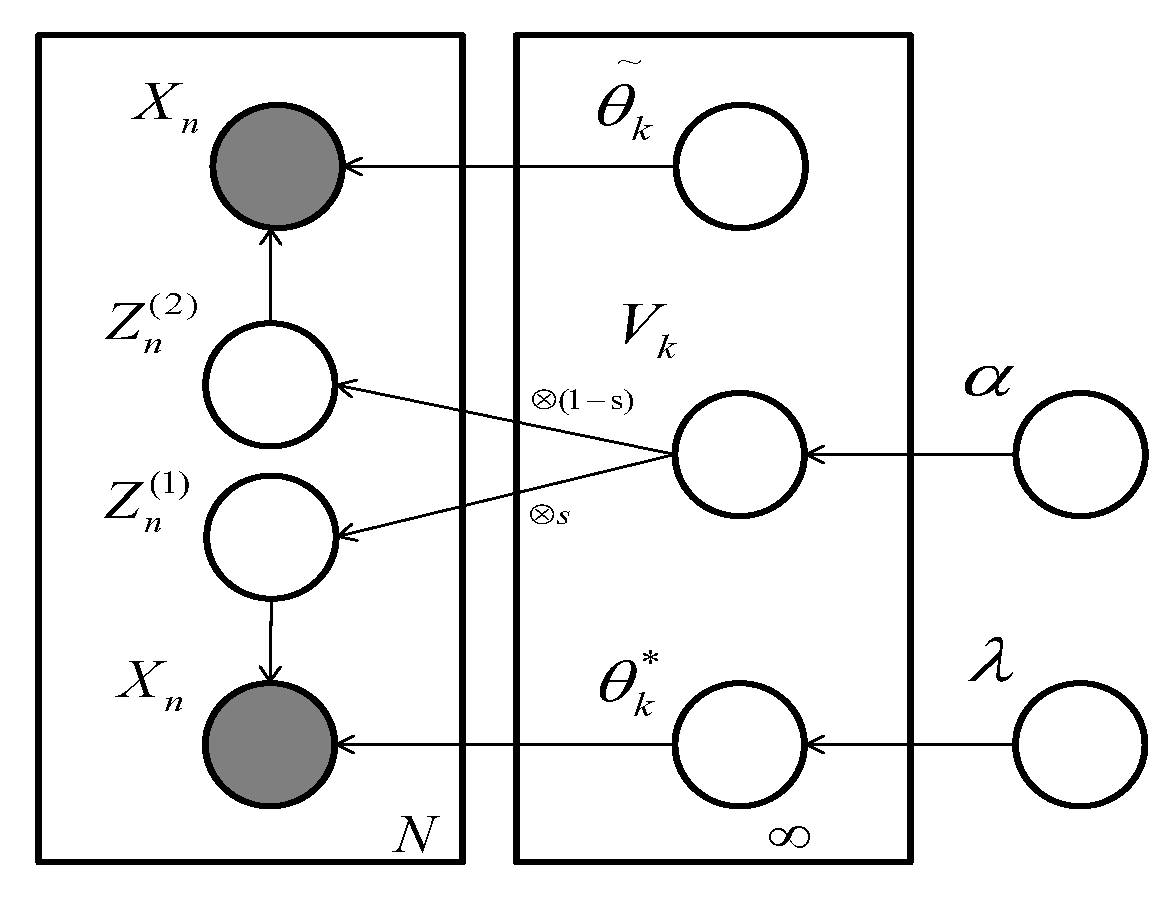
2.2. Our Proposed Method
- For draw
- For draw
- For every data point i:
- (a)
- Choose
- (b)
- Choose
- (c)
- Draw
- (d)
- Draw
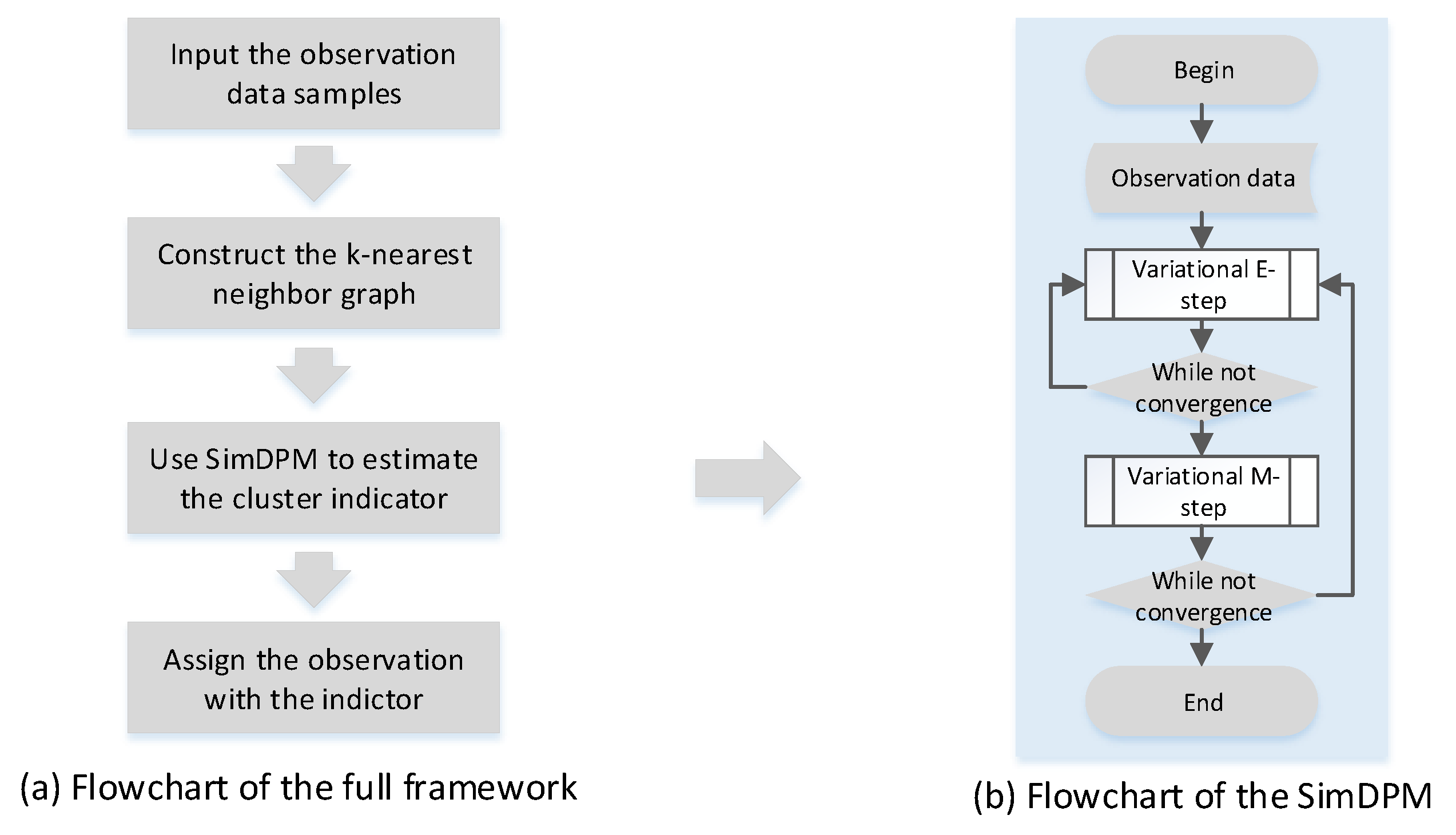
2.3. Variational Expectation Maximization Inference
2.4. Agorithm
| Algorithm 1 Semi-supervised DPM clustering algorithm. |
| Require: unlabeled dataset . |
| Ensure: variational parameters, , and model parameter, |
|
3. Results
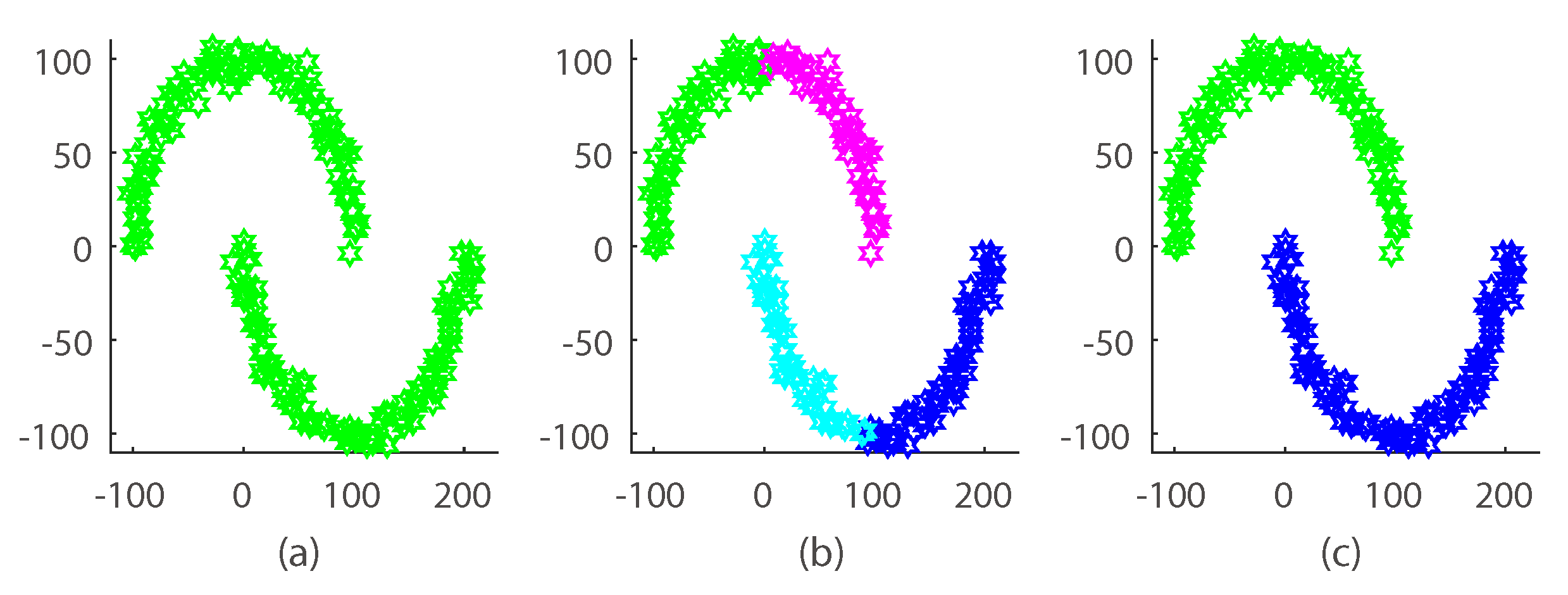
3.1. Synthetic Dataset
3.2. Real Dataset
- The proposed method obtained the highest clustering accuracy especially on the Non-L and coil20 dataset compared with the non-prespecified cluster number methods, which validates the effectiveness of our non-linear assumption.
- DP-space performed better than our method on the L and others dataset, the reason being that DP-space has a prior structure assumption, which introduces additional manifold geometric information.
- LRR, LatLRR and K-means outperformed our algorithm on some coil20 and leaf subdatasets, the reason being that our method needed to estimate the cluster number along with clustering. This made our algorithm hard to optimize.
- Compared to the coil20 and leaf dataset, our method achieved an all-around performance boosting on the motion segmentation dataset; this is because the simple clustering task (the linear manifold has only three classes) was easy for our algorithm to optimize and model.
- Compared to the leaf dataset, our method achieved a better clustering performance boosting on the coil20 dataset. The reason is that coil20 is a well-defined manifold dataset, in which the structure among samples is easy to capture by the graph Laplacian.
- Our manifold model consistently produced the suitable cluster number with the increasing of the data cluster size, which indicates that our model could provide a flexible model size when fitting different datasets.
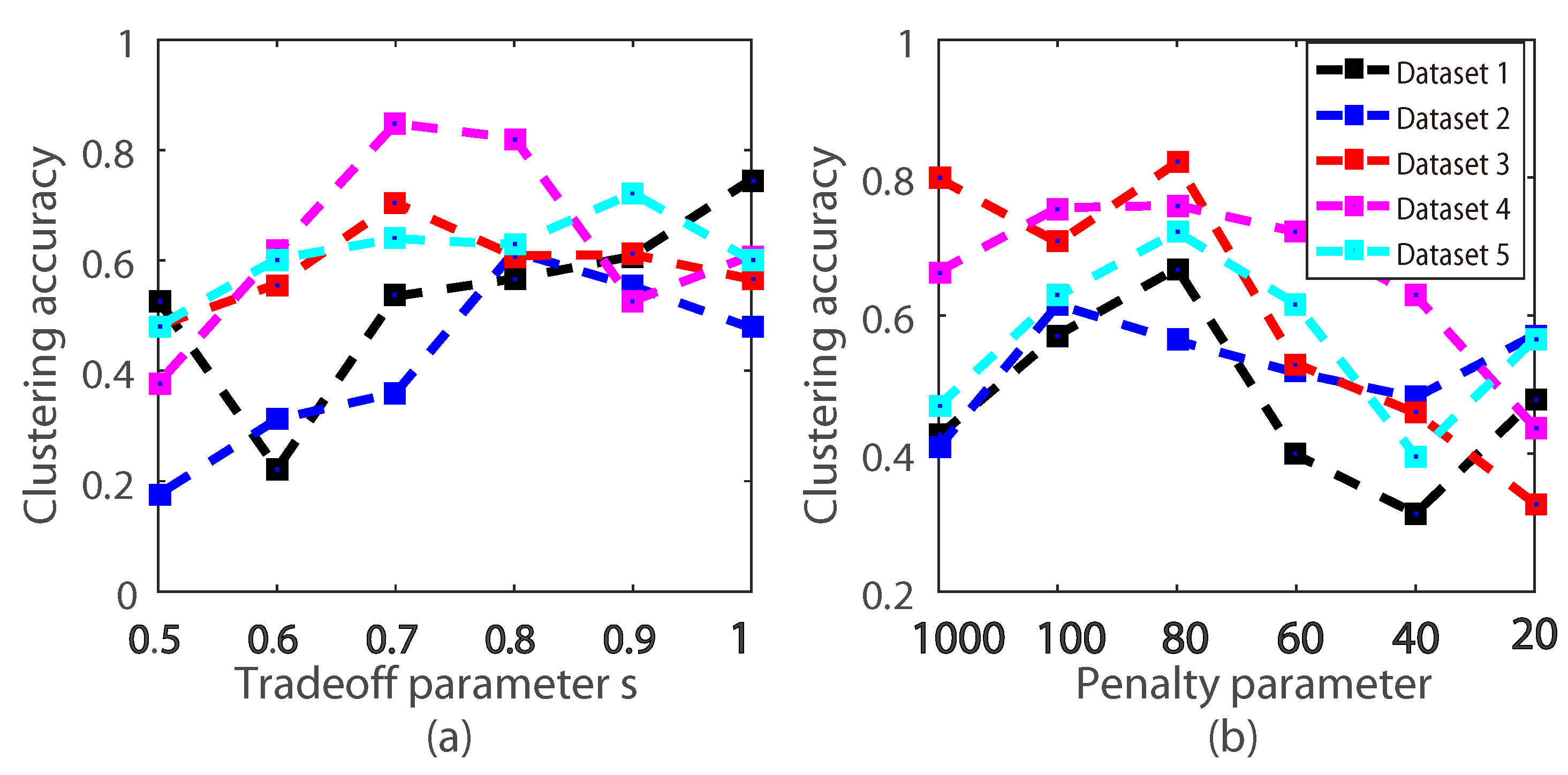
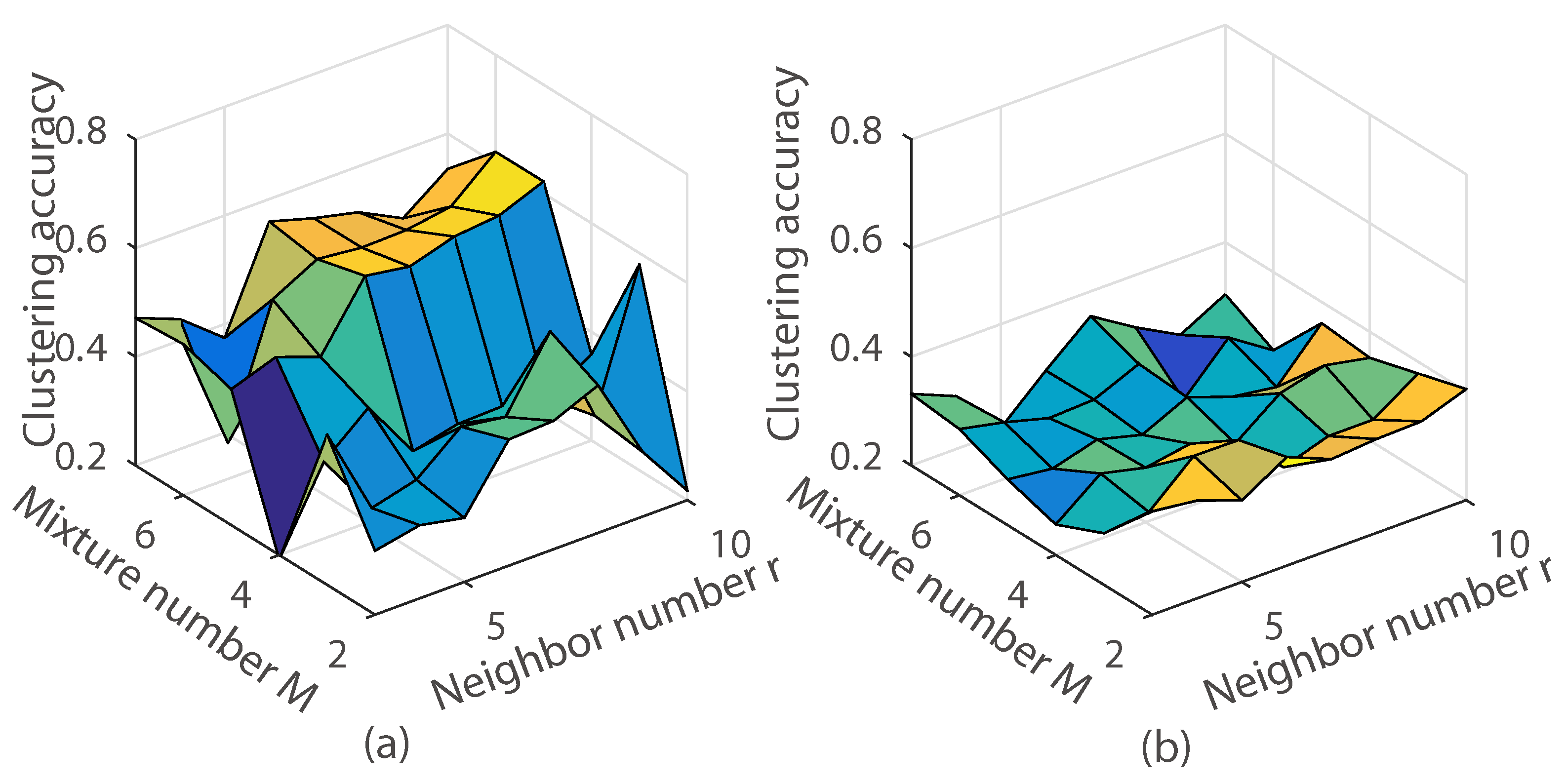
3.3. The Effect of the Algorithm Parameters
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Akogul, S.; Erisoglu, M. An Approach for Determining the Number of Clusters in a Model-Based Cluster Analysis. Entropy 2017, 19, 452. [Google Scholar] [CrossRef]
- Zang, W.; Zhang, W.; Zhang, W.; Liu, X. A Kernel-Based Intuitionistic Fuzzy C-Means Clustering Using a DNA Genetic Algorithm for Magnetic Resonance Image Segmentation. Entropy 2017, 19, 578. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, Y.; Blasch, E.; Ling, H. Simultaneous Trajectory Association and Clustering for Motion Segmentation. IEEE Signal Process. Lett. 2017, 25, 145–149. [Google Scholar] [CrossRef]
- Bansal, S.; Aggarwal, D. Color Image Segmentation Using CIELab Color Space Using Ant Colony Optimization. Int. J. Comput. Appl. 2011, 29, 28–34. [Google Scholar] [CrossRef]
- Chen, P.Y.; Hero, A.O. Phase Transitions in Spectral Community Detection. IEEE Trans. Signal Process. 2015, 63, 4339–4347. [Google Scholar] [CrossRef]
- Sun, J.; Zhou, A.; Keates, S.; Liao, S. Simultaneous Bayesian Clustering and Feature Selection through Student’s t Mixtures Model. IEEE Trans. Neural Netw. Learn. Syst. 2018, 29, 1187–1199. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Chen, L.; Guo, L. KL Divergence-Based Fuzzy Cluster Ensemble for Image Segmentation. Entropy 2018, 20, 273. [Google Scholar]
- Mo, Y.; Cao, Z.; Wang, B. Occurrence-Based Fingerprint Clustering for Fast Pattern-Matching Location Determination. IEEE Commun. Lett. 2012, 16, 2012–2015. [Google Scholar] [CrossRef]
- Cai, D.; Mei, Q.; Han, J.; Zhai, C. Modeling hidden topics on document manifold. In Proceedings of the 17th ACM Conference on Information and Knowledge Management, Napa Valley, CA, USA, 26–30 October 2008; pp. 911–920. [Google Scholar]
- Li, B.; Lu, H.; Zhang, Y.; Lin, Z.; Wu, W. Subspace Clustering under Complex Noise. IEEE Trans. Circuits Syst. Video Technol. 2018, 1. [Google Scholar] [CrossRef]
- Rahkar Farshi, T.; Demirci, R.; Feiziderakhshi, M.R. Image Clustering with Optimization Algorithms and Color Space. Entropy 2018, 20, 296. [Google Scholar] [CrossRef]
- Men, B.; Long, R.; Li, Y.; Liu, H.; Tian, W.; Wu, Z. Combined Forecasting of Rainfall Based on Fuzzy Clustering and Cross Entropy. Entropy 2017, 19, 694. [Google Scholar] [CrossRef]
- Wang, Y.; Jiang, Y.; Wu, Y.; Zhou, Z.H. Spectral clustering on multiple manifolds. IEEE Trans. Neural Netw. 2011, 22, 1149–1161. [Google Scholar] [CrossRef] [PubMed]
- Gholami, B.; Pavlovic, V. Probabilistic Temporal Subspace Clustering. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 3066–3075. [Google Scholar]
- Vidal, R.; Ma, Y.; Sastry, S. Generalized Principal Component Analysis (GPCA). IEEE Trans. Pattern Anal. Mach. Intell. 2005, 27, 1945–1959. [Google Scholar] [CrossRef] [PubMed]
- Elhamifar, E.; Vidal, R. Sparse Subspace Clustering: Algorithm, Theory, and Applications. IEEE Trans. Pattern Anal. Mach. Intell. 2013, 35, 2765–2781. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Lin, Z.; Yan, S.; Sun, J.; Yu, Y.; Ma, Y. Robust Recovery of Subspace Structures by Low-Rank Representation. IEEE Trans. Pattern Anal. Mach. Intell. 2013, 35, 171–184. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Min, H.; Zhao, Z.; Zhu, L.; Huang, D.; Yan, S. Robust and Efficient Subspace Segmentation via Least Squares Regression. In Proceedings of the European Conference on Computer Vision, Florence, Italy, 7–13 October 2012; pp. 347–360. [Google Scholar]
- Souvenir, R.; Pless, R. Manifold Clustering. In Proceedings of the Tenth IEEE International Conference on Computer Vision (ICCV 2005), Beijing, China, 17–20 October 2005; pp. 648–653. [Google Scholar]
- Allab, K.; Labiod, L.; Nadif, M. Multi-Manifold Matrix Decomposition for Data Co-Clustering. Pattern Recognit. 2017, 64, 386–398. [Google Scholar] [CrossRef]
- Peng, X.; Xiao, S.; Feng, J.; Yau, W.Y.; Yi, Z. Deep Subspace Clustering with Sparsity Prior. In Proceedings of the 25th International Joint Conference on Artificial Intelligence (IJCAI 2016), New York, NY, USA, 9–15 July 2016; pp. 1925–1931. [Google Scholar]
- Ji, P.; Zhang, T.; Li, H.; Salzmann, M.; Reid, I. Deep Subspace Clustering Networks. In Proceedings of the Advances in Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017; pp. 24–33. [Google Scholar]
- He, X.; Cai, D.; Shao, Y.; Bao, H.; Han, J. Laplacian Regularized Gaussian Mixture Model for Data Clustering. IEEE Trans. Knowl. Data Eng. 2011, 23, 1406–1418. [Google Scholar] [CrossRef]
- Neal, R.M. Markov Chain Sampling Methods for Dirichlet Process Mixture Models. J. Comput. Graph. Stat. 2000, 9, 249–265. [Google Scholar]
- Wei, X.; Yang, Z. The Infinite Student’s T-Factor Mixture Analyzer for Robust Clustering and Classification. Pattern Recognit. 2012, 45, 4346–4357. [Google Scholar] [CrossRef]
- Nguyen, T.V.; Phung, D.; Nguyen, X.; Venkatesh, S.; Bui, H. Bayesian Nonparametric Multilevel Clustering with Group-Level Contexts. In Proceedings of the 31st International Conference on Machine Learning, Beijing, China, 21–26 June 2014; pp. 288–296. [Google Scholar]
- Palla, K.; Ghahramani, Z.; Knowles, D.A. A Nonparametric Variable Clustering Model. In Proceedings of the Advances in Neural Information Processing Systems, Lake Tahoe, NV, USA, 3–6 December 2012; pp. 2987–2995. [Google Scholar]
- Wang, Y.; Zhu, J. DP-Space: Bayesian Nonparametric Subspace Clustering with Small-Variance Asymptotics. In Proceedings of the 32nd International Conference on Machine Learning (ICML 2015), Lille, France, 6–11 July 2015; pp. 862–870. [Google Scholar]
- Straub, J.; Campbell, T.; How, J.P.; Fisher III, J.W. Efficient Global Point Cloud Alignment using Bayesian Nonparametric Mixtures. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR 2017), Honolulu, HI, USA, 21–26 July 2017; pp. 2941–2950. [Google Scholar]
- Straub, J.; Freifeld, O.; Rosman, G.; Leonard, J.J.; Fisher, J.W., III. The Manhattan Frame Model—Manhattan World Inference in the Space of Surface Normals. IEEE Trans. Pattern Anal. Mach. Intell. 2018, 40, 235–249. [Google Scholar] [CrossRef] [PubMed]
- Simo-Serra, E.; Torras, C.; Moreno-Noguer, F. 3D human pose tracking priors using geodesic mixture models. Int. J. Comput. Vis. 2017, 122, 388–408. [Google Scholar] [CrossRef]
- Sommer, S.; Lauze, F.; Hauberg, S.; Nielsen, M. Manifold valued statistics, exact principal geodesic analysis and the effect of linear approximations. In Proceedings of the 11th European Conference on Computer Vision (ECCV 2010), Crete, Greece, 5–11 September 2010; pp. 43–56. [Google Scholar]
- Huckemann, S.; Hotz, T.; Munk, A. Intrinsic shape analysis: Geodesic PCA for Riemannian manifolds modulo isometric lie group actions. Stat. Sin. 2010, 20, 1–58. [Google Scholar]
- Cao, X.; Chen, Y.; Zhao, Q.; Meng, D.; Wang, Y.; Wang, D.; Xu, Z. Low-Rank Matrix Factorization under General Mixture Noise Distributions. In Proceedings of the IEEE International Conference on Computer Vision, Santiago, Chile, 13–16 December 2015; pp. 1493–1501. [Google Scholar]
- Zhao, Q.; Meng, D.; Xu, Z.; Zuo, W.; Zhang, L. Robust Principal Component Analysis with Complex Noise. In Proceedings of the 31st International Conference on Machine Learning, Beijing, China, 21–26 June 2014; pp. 55–63. [Google Scholar]
- Liu, J.; Cai, D.; He, X. Gaussian Mixture Model with Local Consistency. In Proceedings of the 24th AAAI Conference on Artificial Intelligence, Atlanta, GA, USA, 11–15 July 2010; pp. 512–517. [Google Scholar]
- Bishop, C.M. Pattern Recognition and Machine Learning (Information Science and Statistics); Springer: New York, NY, USA, 2006; p. 049901. [Google Scholar]
- Blei, D.M.; Jordan, M.I. Variational inference for Dirichlet process mixtures. Bayesian Anal. 2006, 1, 121–143. [Google Scholar] [CrossRef]
- Frey, B.J.; Dueck, D. Clustering by passing messages between data points. Science 2007, 315, 972–976. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, A.; Laio, A. Machine learning. Clustering by fast search and find of density peaks. Science 2014, 344, 1492. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Yan, S. Latent Low-Rank Representation for subspace segmentation and feature extraction. In Proceedings of the 13th International Conference on Computer Vision, Barcelona, Spain, 6–13 November 2011; pp. 1615–1622. [Google Scholar]
- Zhang, Y.; Luo, B.; Zhang, L. Permutation Preference based Alternate Sampling and Clustering for Motion Segmentation. IEEE Signal Process. Lett. 2017, 25, 432–436. [Google Scholar] [CrossRef]
- Tron, R.; Vidal, R. A benchmark for the comparison of 3D motion segmentation algorithms. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Minneapolis, MN, USA, 17–22 June 2007; pp. 1–8. [Google Scholar]
- Nene, S.A.; Nayar, S.K.; Murase, H. Columbia Object Image Library (Coil-20); Technical Report; Columbia University: New York, NY, USA, 1996. [Google Scholar]
- Söderkvist, O. Computer Vision Classification of Leaves from Swedish Trees. Master’s Thesis, Linköping University, Linköping, Sweden, September 2001. Available online: http://www.diva-portal.org/smash/record.jsf?pid=diva2%3A303038dswid=-9927 (accessed on 26 October 2018).
- Zahn, C.T.; Roskies, R.Z. Fourier descriptors for plane closed curves. IEEE Trans. Comput. 1972, 100, 269–281. [Google Scholar] [CrossRef]





| Notations | Descriptions |
|---|---|
| Hyper parameter of normal-Wishart | |
| M | The mixture number |
| N | Number of the observation samples |
| Gaussian parameter | |
| MoG parameter | |
| s | Tradeoff parameter |
| K | The maximum cluster number |
| Parameter of the Beta distribution | |
| Class indicator of a Gaussian distribution | |
| Class indicator of the MoG distribution | |
| X | Unlabeled dataset |
| Variational parameter | |
| Variational parameter of normal-Wishart | |
| Variational parameter of categorical distribution | |
| A | An auxiliary parameter that equals |
| Penalty parameter used in the graph Laplacian | |
| L | Graph Laplacian |
| k-nearest neighbor graph | |
| Diagonal matrix whose entries are column sums of | |
| R | Posterior penalty term with graph Laplacian |
| r | Neighbor number used in the graph Laplacian |
| Method | Checkerboard | Others | ||
|---|---|---|---|---|
| L | Non-L | Average | ||
| SimDPM | 0.80 | 0.73 | 0.79 | 0.83 |
| DPM [38] | 0.42 | 0.37 | 0.41 | 0.45 |
| DP-space [28] | 0.84 | 0.48 | 0.78 | 0.94 |
| AP [39] | 0.29 | 0.32 | 0.29 | 0.31 |
| CFSFDP [40] | 0.40 | 0.19 | 0.36 | 0.47 |
| K-means | 0.48 | 0.48 | 0.49 | 0.47 |
| LRR [17] | 0.51 | 0.33 | 0.48 | 0.33 |
| LatLRR [41] | 0.52 | 0.31 | 0.47 | 0.34 |
| Method | Checkerboard | Others | ||
|---|---|---|---|---|
| L | Non-L | Average | ||
| Ground truth | 3.00 | 3.00 | 3.00 | 3.00 |
| The estimated cluster number | 3.33 | 3.60 | 3.10 | 3.09 |
| Method | Subdataset | 20 | ||||
|---|---|---|---|---|---|---|
| 2 | 4 | 6 | 8 | 10 | ||
| SimDPM | 0.30 | 0.55 | 0.56 | 0.60 | 0.69 | 0.72 |
| DPM [38] | 0.29 | 0.42 | 0.50 | 0.53 | 0.57 | 0.69 |
| DP-space citewang2015dp | 0.01 | 0.34 | 0.10 | 0.19 | 0.10 | 0.26 |
| AP [39] | 0.12 | 0.22 | 0.18 | 0.11 | 0.25 | 0.36 |
| CFSFDP [40] | 0 | 0.57 | 0.57 | 0.53 | 0.46 | 0.42 |
| K-means | 0 | 0.52 | 0.46 | 0.57 | 0.59 | 0.73 |
| LRR [17] | 0.11 | 0.62 | 0.56 | 0.47 | 0.52 | 0.70 |
| LatLRR [41] | 0 | 0.57 | 0.57 | 0.48 | 0.50 | 0.58 |
| Method | Subdataset | 20 | ||||
|---|---|---|---|---|---|---|
| 2 | 4 | 6 | 8 | 10 | ||
| Ground truth | 2.0 | 4.0 | 6.0 | 8.0 | 10.0 | 20.0 |
| The estimated cluster number | 3.1 | 5.0 | 6.3 | 6.7 | 11.3 | 21.7 |
| Method | Subdataset | 15 | ||||
|---|---|---|---|---|---|---|
| 2 | 4 | 6 | 8 | 10 | ||
| SimDPM | 0.29 | 0.62 | 0.46 | 0.50 | 0.54 | 0.38 |
| DPM [38] | 0.26 | 0.43 | 0.45 | 0.49 | 0.51 | 0.34 |
| DP-space [28] | 0.49 | 0.33 | 0 | 0 | 0 | 0.03 |
| AP [39] | 0 | 0.03 | 0.11 | 0 | 0.02 | 0 |
| CFSFDP [40] | 0.17 | 0.56 | 0.24 | 0.28 | 0.42 | 0 |
| K-means | 0.45 | 0.39 | 0.61 | 0.50 | 0.44 | 0.22 |
| LRR [17] | 0.76 | 0.45 | 0.48 | 0.33 | 0.40 | 0.22 |
| LatLRR [41] | 0.65 | 0.44 | 0.32 | 0.20 | 0.41 | 0.23 |
| Method | Subdataset | 15 | ||||
|---|---|---|---|---|---|---|
| 2 | 4 | 6 | 8 | 10 | ||
| Ground truth | 2.0 | 4.0 | 6.0 | 8.0 | 10.0 | 15.0 |
| The estimated class number | 4.7 | 6.3 | 10.2 | 14.5 | 16.2 | 20.2 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ye, X.; Zhao, J.; Chen, Y. A Nonparametric Model for Multi-Manifold Clustering with Mixture of Gaussians and Graph Consistency. Entropy 2018, 20, 830. https://doi.org/10.3390/e20110830
Ye X, Zhao J, Chen Y. A Nonparametric Model for Multi-Manifold Clustering with Mixture of Gaussians and Graph Consistency. Entropy. 2018; 20(11):830. https://doi.org/10.3390/e20110830
Chicago/Turabian StyleYe, Xulun, Jieyu Zhao, and Yu Chen. 2018. "A Nonparametric Model for Multi-Manifold Clustering with Mixture of Gaussians and Graph Consistency" Entropy 20, no. 11: 830. https://doi.org/10.3390/e20110830
APA StyleYe, X., Zhao, J., & Chen, Y. (2018). A Nonparametric Model for Multi-Manifold Clustering with Mixture of Gaussians and Graph Consistency. Entropy, 20(11), 830. https://doi.org/10.3390/e20110830




