Characterization of the Resistance to Powdery Mildew and Leaf Rust Carried by the Bread Wheat Cultivar Victo
Abstract
1. Introduction
2. Results
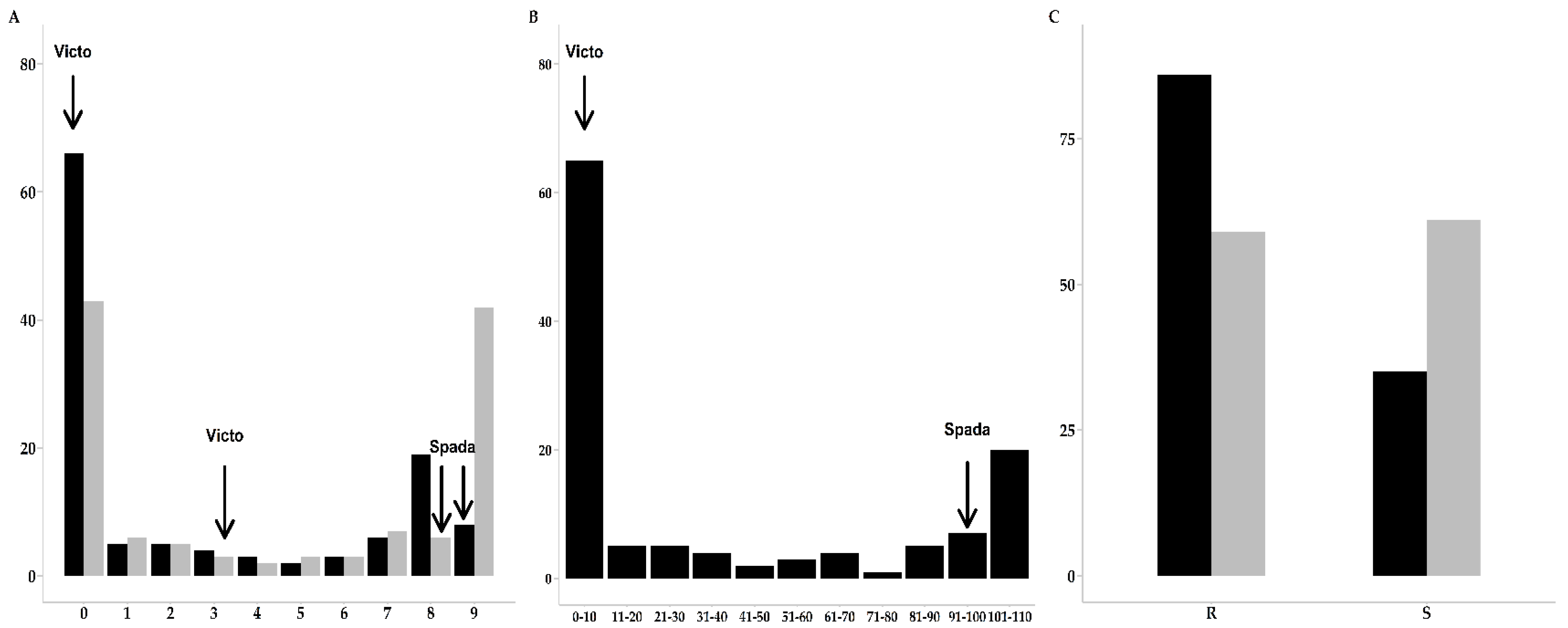
2.1. Phenotypic Evaluations of Leaf Rust and Powdery Mildew Resistance
2.2. Construction of the Victo x Spada Genetic Linkage Map
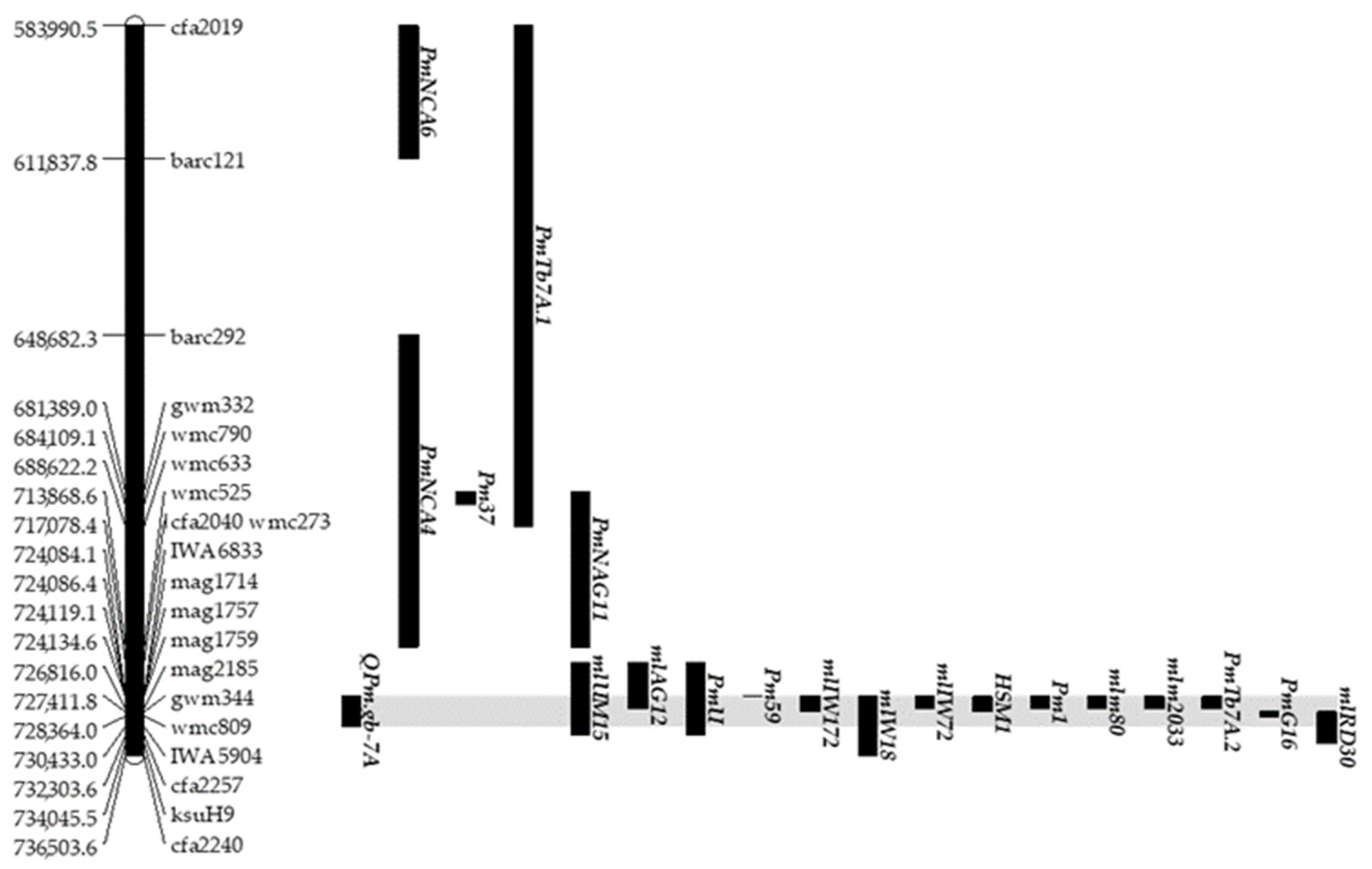
2.3. Identification of QPm.gb-7A, a Powdery Mildew Resistance QTL on Chromosome 7AL
- i.
- ii.
- iii.
- iv.
- Nine additional resistance loci showed a co-positional relationship of physical region with QPm.gb-7A: Pm59 (from 724.0 to 724.1Mbp), mlIW172 (from 724.1 to 727.4 Mbp), Pm1, mlIW72, mlm80, PmTb7A.2 and mlm2033.(from 724.1 to 726.8), HSM1 (from 724.1 to 727.4 Mbp), and PmG16 (from 727.4 to 728.3 Mbp) [40,48,49,50,51,52,53,54,55].
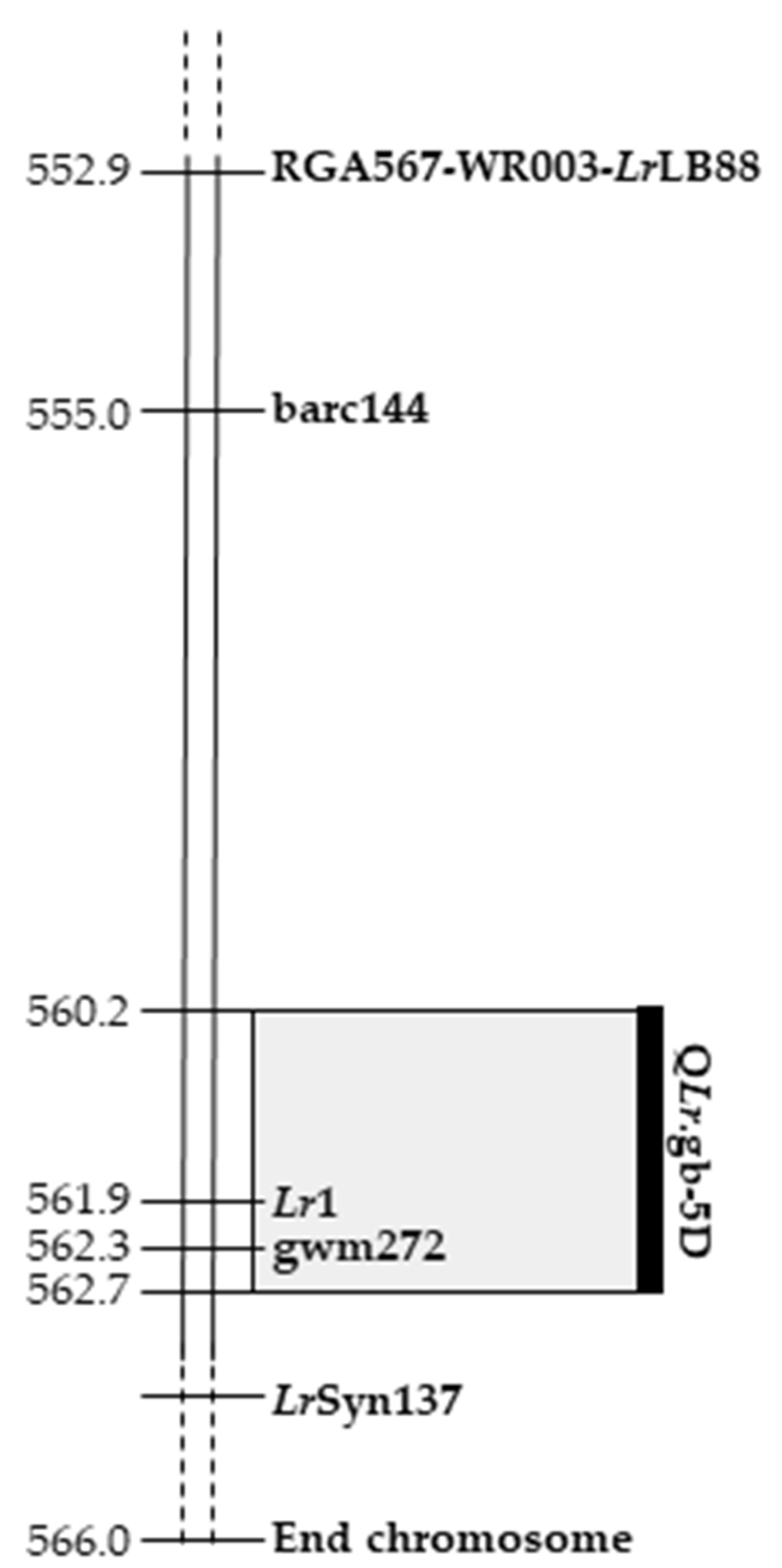
2.4. Identification of QLr.gb-5D, a with Leaf Rust Resistance QTL on Chromosome 5DL
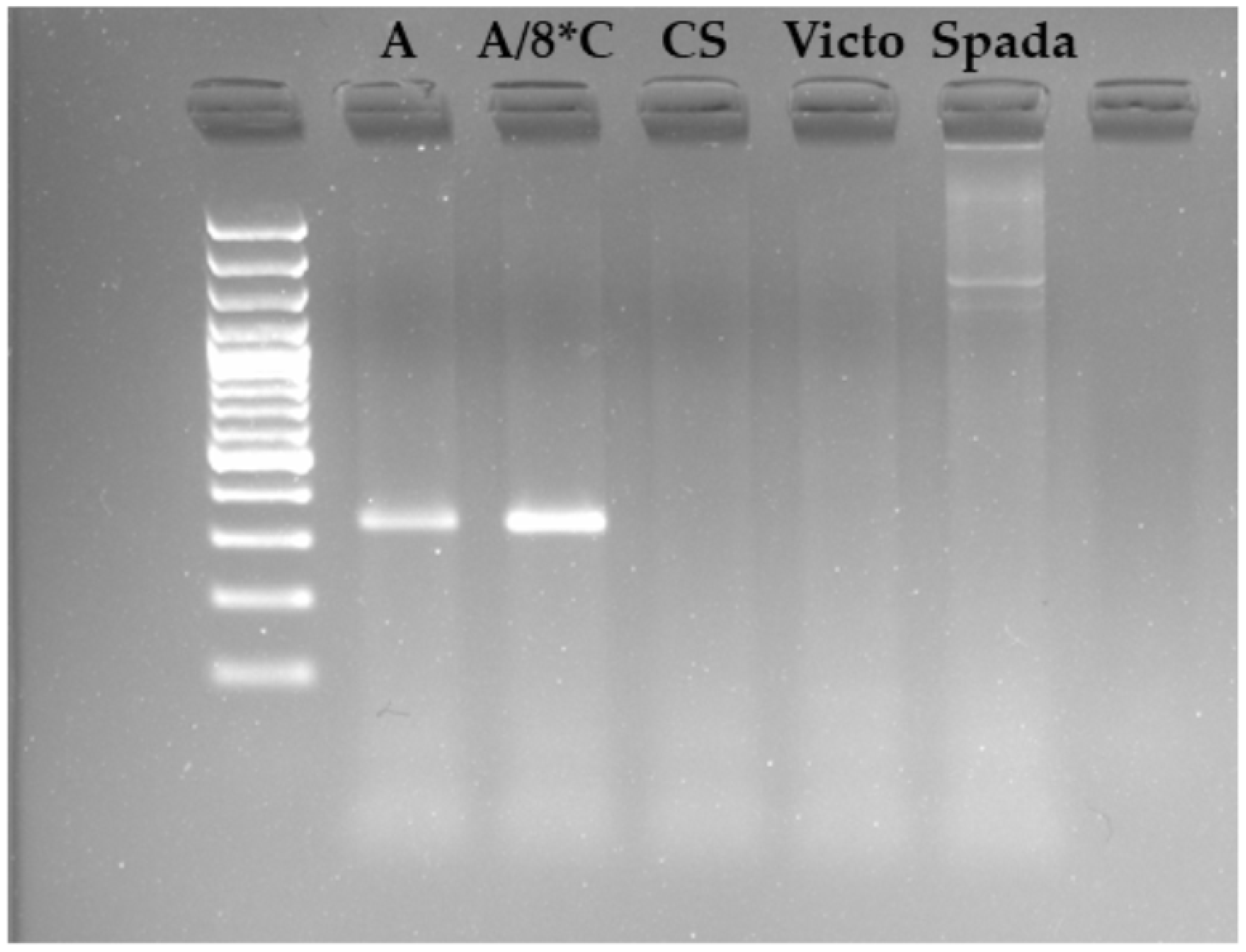
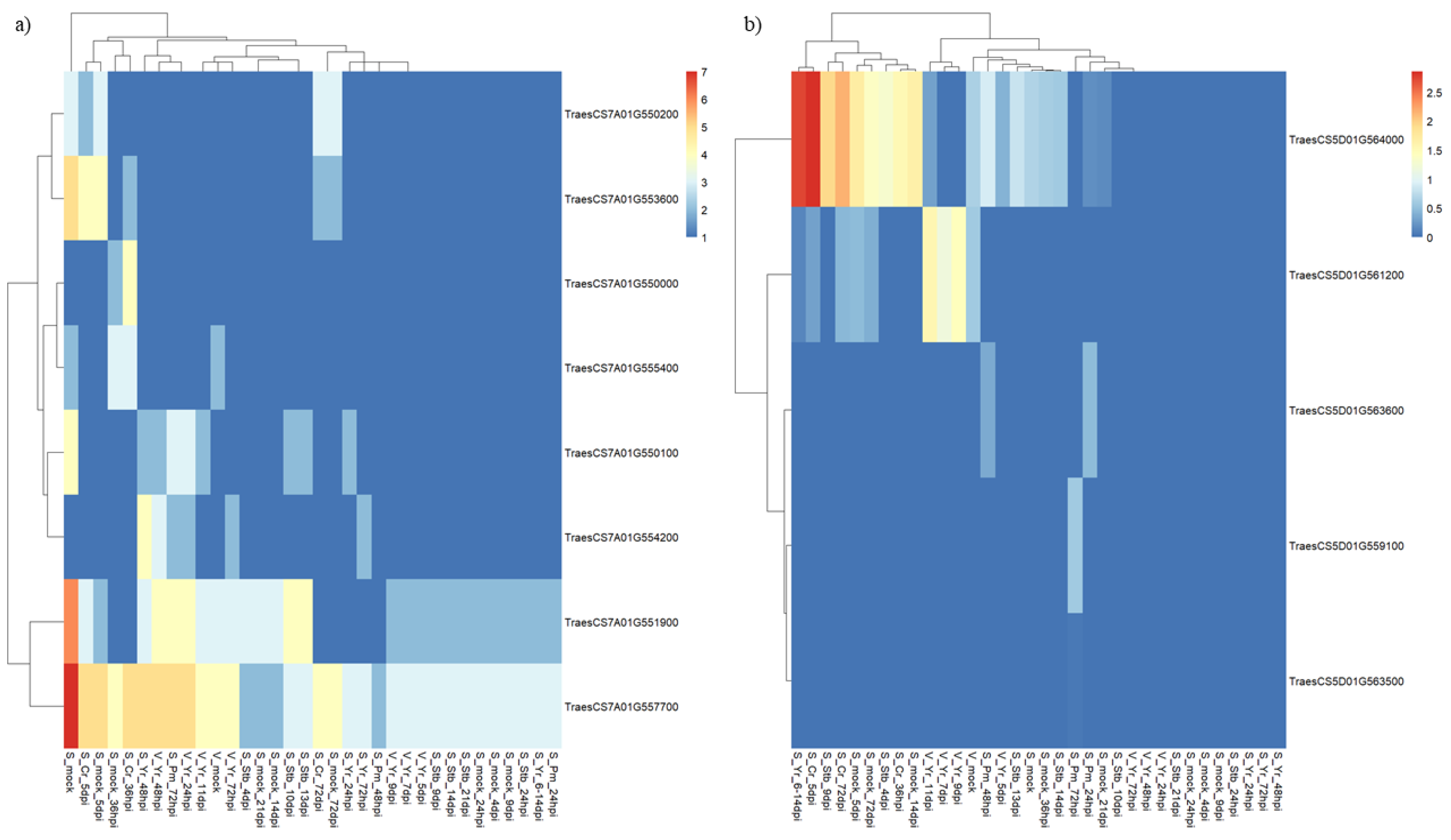
2.5. In Silico Functional Analysis of QPm.gb-7A Candidate Resistance Genes
2.6. In Silico Functional Analysis of QLr.gb-5D Candidate Resistance Genes
3. Discussion
3.1. A New Genetic Map for Triticum Aestivum
3.2. Identification of Powdery Mildew Resistance QTL in Hexaploid Wheat
3.3. Identification of Leaf Rust QTLs in Hexaploid Wheat
4. Materials and Methods
4.1. Genetic Materials
4.2. Phenotypic Evaluation
4.3. Statistical Analysis
4.4. Linkage Analysis
4.5. QTL Analysis
4.6. Analysis of Physical Regions Carrying QTLs Related to Disease
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Khan, M.H.; Bukhari, A.; Dar, Z.A.; Rizvi, S.M. Status and strategies in breeding for rust resistance in wheat. Agric. Sci. 2013, 4, 292–301. [Google Scholar] [CrossRef]
- Conner, R.L.; Kuzyk, A.D.; Su, H. Impact of powdery mildew on the yield of soft white spring wheat cultivars. Can. J. Plant Sci. 2003, 83, 725–728. [Google Scholar] [CrossRef]
- Bourras, S.; McNally, K.E.; Müller, M.C.; Wicker, T.; Keller, B. Avirulence genes in cereal powdery mildews: The gene-for-gene hypothesis 2.0. Front. Plant Sci. 2016, 7, 241. [Google Scholar] [CrossRef]
- McDonald, B.A.; Linde, C. The population genetics of plant pathogens and breeding strategies for durable resistance. Euphytica 2002, 124, 163–180. [Google Scholar] [CrossRef]
- Keller, B.; Krattinger, S.G. A new player in race-specific resistance. Nat. Plants 2018, 4, 197–198. [Google Scholar] [CrossRef] [PubMed]
- Mundt, C.C. Pyramiding for resistance durability: Theory and practice. Phytopathology 2018, 108, 792–802. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Lan, C.; He, Z.; Singh, R.P.; Rosewarne, G.M.; Chen, X.; Xia, X. Overview and application of QTL for adult plant resistance to leaf rust and powdery mildew in wheat. Crop. Sci. 2014, 54, 1907–1925. [Google Scholar] [CrossRef]
- Purcell, S.; Cherny, S.S.; Sham, P.C. Genetic Power Calculator: Design of linkage and association genetic mapping studies of complex traits. Bioinformatics 2003, 19, 149–150. [Google Scholar] [CrossRef]
- Zhu, C.; Gore, M.; Buckler, E.S.; Yu, J. Status and prospects of association mapping in plants. Plant Genome 2008, 1, 5–20. [Google Scholar] [CrossRef]
- McIntosh, R.A.; Dubcovsky, J.; Rogers, J.W.; Morris, C.; Appels, R.; Xia, C.X. Catalogue of gene Symbols for Wheat: 2013–2014 Supplement. KOMUGI Integr. Wheat Sci. 2013. Available online: http://www.shigen.nig.ac.jp/wheat/komugi/genes/symbolClassList.jsp (accessed on 22 December 2020).
- McIntosh, R.; Dubcovsky, J.; Rogers, W.; Morris, C.; Xia, X. Catalogue of gene symbols for wheat. 2017. Supplement. Annu. Wheat Newsl. 2017, 53, 1–20. [Google Scholar]
- Lillemo, M.; Asalf, B.; Singh, R.P.; Huerta-Espino, J.; Chen, X.M.; He, Z.H.; Bjørnstad, Å. The adult plant rust resistance loci Lr34/Yr18 and Lr46/Yr29 are important determinants of partial resistance to powdery mildew in bread wheat line Saar. Theor. Appl. Genet. 2008, 116, 1155–1166. [Google Scholar] [CrossRef] [PubMed]
- Lagudah, E.S.; Krattinger, S.G.; Herrera-Foessel, S.; Singh, R.P.; Huerta-Espino, J.; Spielmeyer, W.; Brown-Guedira, G.; Selter, L.L.; Keller, B. Gene-specific markers for the wheat gene Lr34/Yr18/Pm38 which confers resistance to multiple fungal pathogens. Theor. Appl. Genet. 2009, 119, 889–898. [Google Scholar] [CrossRef]
- Hare, R.; McIntosh, R. Genetic and cytogenetic studies of durable adult-plant resistance in ‘Hope’ and related cultivars to wheat rust. Plant Breed. 1979, 83, 356–367. [Google Scholar]
- Singh, R.P.; Nelson, J.C.; Sorrells, M.E. Mapping Yr28 and other genes for resistance to stripe rust in wheat. Crop. Sci. 2000, 40, 1148–1155. [Google Scholar] [CrossRef]
- Herrera-Foessel, S.A.; Singh, R.P.; Lillemo, M.; Huerta-Espino, J.; Bhavani, S.; Singh, S.; Lan, C.; Calvo-Salazar, V.; Lagudah, E.S. Lr67/Yr46 confers adult plant resistance to stem rust and powdery mildew in wheat. Theor. Appl. Genet. 2014, 127, 781–789. [Google Scholar] [CrossRef]
- Cloutier, S.; McCallum, B.D.; Loutre, C.; Banks, T.W.; Wicker, T.; Feuillet, C.; Keller, B.; Jordan, M.C. Leaf rust resistance gene Lr1, isolated from bread wheat (Triticum aestivum L.) is a member of the large psr567 gene family. Plant Mol. Biol. 2007, 65, 93–106. [Google Scholar] [CrossRef] [PubMed]
- Feuillet, C.; Travella, S.; Stein, N.; Albar, L.; Nublat, A.; Keller, B. Map-based isolation of the leaf rust disease resistance gene Lr10 from the hexaploid wheat (Triticum aestivum L.) genome. Proc. Natl. Acad. Sci. USA 2003, 100, 15253–15258. [Google Scholar] [CrossRef]
- Huang, L.; A Brooks, S.; Li, W.; Fellers, J.P.; Trick, H.N.; Gill, B.S. Map-based cloning of leaf rust resistance gene Lr21 from the large and polyploid genome of bread wheat. Genetics 2003, 164, 655–664. [Google Scholar] [PubMed]
- Thind, A.K.; Wicker, T.; Šimková, H.; Fossati, D.; Moullet, O.; Brabant, C.; Vrána, J.; Doležel, J.; Krattinger, S.G. Rapid cloning of genes in hexaploid wheat using cultivar-specific long-range chromosome assembly. Nat. Biotechnol. 2017, 35, 793–796. [Google Scholar] [CrossRef]
- Krattinger, S.G.; Lagudah, E.S.; Spielmeyer, W.; Singh, R.P.; Huerta-Espino, J.; McFadden, H.; Bossolini, E.; Selter, L.L.; Keller, B. A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 2009, 323, 1360–1363. [Google Scholar] [CrossRef] [PubMed]
- Milne, R.J.; Dibley, K.E.; Schnippenkoetter, W.H.; Mascher, M.; Lui, A.C.; Wang, L.; Lo, C.; Ashton, A.R.; Ryan, P.R.; Lagudah, E.S. The wheat Lr67 gene from the sugar transport protein 13 family confers multipathogen resistance in Barley. Plant Physiol. 2018, 179, 1285–1297. [Google Scholar] [CrossRef] [PubMed]
- Yahiaoui, N.; Srichumpa, P.; Dudler, R.; Keller, B. Genome analysis at different ploidy levels allows cloning of the powdery mildew resistance genePm3bfrom hexaploid wheat. Plant J. 2004, 37, 528–538. [Google Scholar] [CrossRef]
- Bhullar, N.K.; Street, K.; Mackay, M.; Yahiaoui, N.; Keller, B. Unlocking wheat genetic resources for the molecular identification of previously undescribed functional alleles at the Pm3 resistance locus. Proc. Natl. Acad. Sci. USA 2009, 106, 9519–9524. [Google Scholar] [CrossRef]
- Hurni, S.; Brunner, S.; Buchmann, G.; Herren, G.; Jordan, T.; Krukowski, P.; Wicker, T.; Yahiaoui, N.; Mago, R.; Keller, B. RyePm8and wheatPm3are orthologous genes and show evolutionary conservation of resistance function against powdery mildew. Plant J. 2013, 76, 957–969. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.P.; Hurni, S.; Ruinelli, M.; Brunner, S.; Sanchez-Martin, J.; Krukowski, P.; Peditto, D.; Buchmann, G.; Zbinden, H.; Keller, B. Evolutionary divergence of the rye Pm17 and Pm8 resistance genes reveals ancient diversity. Plant Mol. Biol. 2018, 98, 249–260. [Google Scholar] [CrossRef]
- Li, M.; Dong, L.; Li, B.; Wang, Z.; Xie, J.; Qiu, D.; Li, Y.; Shi, W.; Yang, L.; Wu, Q.; et al. A CNL protein in wild emmer wheat confers powdery mildew resistance. New Phytol. 2020, 228, 1027–1037. [Google Scholar] [CrossRef]
- Lu, P.; Guo, L.; Wang, Z.; Li, B.; Li, J.; Li, Y.; Qiu, D.; Shi, W.; Yang, L.; Wang, N.; et al. A rare gain of function mutation in a wheat tandem kinase confers resistance to powdery mildew. Nat. Commun. 2020, 11, 1–11. [Google Scholar] [CrossRef]
- Xing, L.; Hu, P.; Liu, J.; Witek, K.; Zhou, S.; Xu, J.; Zhou, W.; Gao, L.; Huang, Z.; Zhang, R.; et al. Pm21 from haynaldia villosa encodes a CC-NBS-LRR protein conferring powdery mildew resistance in wheat. Mol. Plant 2018, 11, 874–878. [Google Scholar] [CrossRef]
- He, H.; Zhu, S.; Zhao, R.; Jiang, Z.; Ji, Y.; Ji, J.; Qiu, D.; Li, H.; Bie, T. Pm21, encoding a typical CC-NBS-LRR protein, confers broad-spectrum resistance to wheat powdery mildew disease. Mol. Plant 2018, 11, 879–882. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Martín, J.; Steuernagel, B.; Ghosh, S.; Herren, G.; Hurni, S.; Adamski, N.; Vrána, J.; Kubaláková, M.; Krattinger, S.G.; Wicker, T.; et al. Rapid gene isolation in barley and wheat by mutant chromosome sequencing. Genome Biol. 2016, 17, 1–7. [Google Scholar] [CrossRef]
- Zou, S.; Wang, H.; Li, Y.; Kong, Z.; Tang, D. The NB-LRR gene Pm60 confers powdery mildew resistance in wheat. New Phytol. 2017, 218, 298–309. [Google Scholar] [CrossRef]
- Hewitt, T.; Mueller, M.C.; Molnár, I.; Mascher, M.; Holušová, K.; Šimková, H.; Kunz, L.; Zhang, J.; Li, J.; Bhatt, D.; et al. A highly differentiated region of wheat chromosome 7AL encodes a Pm1a immune receptor that recognizes its corresponding AvrPm1a effector from Blumeria graminis. New Phytol. 2021, 229, 2812–2826. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Guo, G.; Wang, Y.; Hu, T.; Wang, L.; Li, J.; Qiu, D.; Li, Y.; Wu, Q.; Lu, P.; et al. A rare single nucleotide variant in Pm5e confers powdery mildew resistance in common wheat. New Phytol. 2020, 228, 1011–1026. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, R.; Hu, Y.-G.; Chen, J. Genome-wide linkage mapping of quantitative trait loci for late-season physiological and agronomic traits in spring wheat under irrigated conditions. Agronomy 2018, 8, 60. [Google Scholar] [CrossRef]
- Gao, F.; Wen, W.; Liu, J.; Rasheed, A.; Yin, G.; Xia, X.; Wu, X.; He, Z. Genome-wide linkage mapping of QTL for yield components, plant height and yield-related physiological traits in the chinese wheat cross zhou 8425B/Chinese spring. Front. Plant Sci. 2015, 6, 1099. [Google Scholar] [CrossRef] [PubMed]
- Su, Q.; Zhang, X.; Zhang, W.; Zhang, N.; Song, L.; Liu, L.; Xue, X.; Liu, G.; Liu, J.; Meng, D.; et al. QTL detection for kernel size and weight in bread wheat (Triticum aestivum L.) using a high-density SNP and SSR-based linkage map. Front. Plant Sci. 2018, 9, 1484. [Google Scholar] [CrossRef]
- Li, F.; Wen, W.; Liu, J.; Zhang, Y.; Cao, S.; He, Z.; Rasheed, A.; Jin, H.; Zhang, C.; Yan, J.; et al. Genetic architecture of grain yield in bread wheat based on genome-wide association studies. BMC Plant Biol. 2019, 19, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Miranda, L.M.; Perugini, L.; Srnic, G.; Brown-Guedira, G.; Marshall, D.; Leath, S.; Murphy, J.P. Genetic mapping of a triticum monococcum -derived powdery mildew resistance gene in common wheat. Crop. Sci. 2007, 47, 2323–2329. [Google Scholar] [CrossRef]
- Chhuneja, P.; Kumar, K.; Stirnweis, D.; Hurni, S.; Keller, B.; Dhaliwal, H.S.; Singh, K. Identification and mapping of two powdery mildew resistance genes in Triticum boeoticum L. Theor. Appl. Genet. 2011, 124, 1051–1058. [Google Scholar] [CrossRef][Green Version]
- Perugini, L.D.; Murphy, J.P.; Marshall, D.; Brown-Guedira, G. Pm37, a new broadly effective powdery mildew resistance gene from Triticum timopheevii. Theor. Appl. Genet. 2008, 116, 417–425. [Google Scholar] [CrossRef]
- Srnic, G.; Murphy, J.P.; Lyerly, J.H.; Leath, S.; Marshall, D.S. Inheritance and chromosomal assignment of powdery mildew resistance genes in two winter wheat germplasm lines. Crop. Sci. 2005, 45, 1578–1586. [Google Scholar] [CrossRef]
- Maxwell, J.J.; Lyerly, J.H.; Cowger, C.; Marshall, D.; Brown-Guedira, G.; Murphy, J.P. MlAG12: A Triticum timopheevii-derived powdery mildew resistance gene in common wheat on chromosome 7AL. Theor. Appl. Genet. 2009, 119, 1489–1495. [Google Scholar] [CrossRef]
- Singrün, C.; Hsam, S.L.K.; Zeller, F.J.; Wenzel, G.; Mohler, V. Localization of a novel recessive powdery mildew resistance gene from common wheat line RD30 in the terminal region of chromosome 7AL. Theor. Appl. Genet. 2004, 109, 210–214. [Google Scholar] [CrossRef]
- Qiu, Y.C.; Zhou, R.H.; Kong, X.Y.; Zhang, S.S.; Jia, J.Z. Microsatellite mapping of a Triticum urartu Tum. derived powdery mildew resistance gene transferred to common wheat (Triticum aestivum L.). Theor. Appl. Genet. 2005, 111, 1524–1531. [Google Scholar] [CrossRef]
- Han, J.; Zhang, L.; Li, G.; Zhang, H.; Xie, C.; Yang, Z.; Sun, Q.; Liu, Z. Molecular mapping of powdery mildew resistance gene MlWE18 in wheat originated from wild emmer (Triticum turgidum var. dicoccoides). Acta Agronomica Sinica 2009, 35, 1791–1797. [Google Scholar] [CrossRef]
- Worthington, M.; Lyerly, J.; Petersen, S.; Brown-Guedira, G.; Marshall, D.; Cowger, C.; Parks, R.; Murphy, J.P. MlUM15: An Aegilops neglecta -derived powdery mildew resistance gene in common wheat. Crop. Sci. 2014, 54, 1397–1406. [Google Scholar] [CrossRef]
- Tan, C.; Li, G.; Cowger, C.; Carver, B.F.; Xu, X. Characterization of Pm59, a novel powdery mildew resistance gene in Afghanistan wheat landrace PI 181356. Theor. Appl. Genet. 2018, 131, 1145–1152. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.Q.; Sorrells, M.E.; Tanksley, S.D. RFLP markers linked to powdery mildew resistance genes Pm1, Pm2, Pm3, and Pm4 in wheat. Genome 1994, 37, 871–875. [Google Scholar] [CrossRef] [PubMed]
- Neu, C.; Stein, N.; Keller, B. Genetic mapping of the Lr20–Pm1 resistance locus reveals suppressed recombination on chromosome arm 7AL in hexaploid wheat. Genome 2002, 45, 737–744. [Google Scholar] [CrossRef] [PubMed]
- Yao, G.; Zhang, J.; Yang, L.; Xu, H.; Jiang, Y.; Xiong, L.; Zhang, C.; Zhang, Z.; Ma, Z.; Sorrells, M.E. Genetic mapping of two powdery mildew resistance genes in einkorn (Triticum monococcum L.) accessions. Theor. Appl. Genet. 2006, 114, 351–358. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.; Xie, C.; Ni, Z.; Yang, T.; Nevo, E.; Fahima, T.; Liu, Z.; Sun, Q. Identification and genetic mapping of a powdery mildew resistance gene in wild emmer (Triticum dicoccoides) accession IW72 from Israel. Euphytica 2007, 159, 385–390. [Google Scholar] [CrossRef]
- Ouyang, S.; Zhang, N.; Han, J.; Zhao, X.; Cui, Y.; Song, W.; Huo, N.; Liang, Y.; Xie, J.; Wang, Z.; et al. Fine physical and genetic mapping of powdery mildew resistance gene MlIW172 originating from wild emmer (Triticum dicoccoides). PLoS ONE 2014, 9, e100160. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Wen, Z.; Wang, J.; Fu, B.; Liu, J.; Xu, H.; Kong, Z.; Zhang, L.; Jia, H.; Ma, Z. Transfer and mapping of a gene conferring later-growth-stage powdery mildew resistance in a tetraploid wheat accession. Mol. Breed. 2013, 33, 669–677. [Google Scholar] [CrossRef]
- Ben-David, R.; Xie, W.; Peleg, Z.; Saranga, Y.; Dinoor, A.; Fahima, T. Identification and mapping of PmG16, a powdery mildew resistance gene derived from wild emmer wheat. Theor. Appl. Genet. 2010, 121, 499–510. [Google Scholar] [CrossRef] [PubMed]
- Lillemo, M.; Bjørnstad, Å.; Skinnes, H. Molecular mapping of partial resistance to powdery mildew in winter wheat cultivar Folke. Euphytica 2012, 185, 47–59. [Google Scholar] [CrossRef]
- Mohler, V.; Schmolke, M.; Zeller, F.J.; Hsam, S.L.K. Genetic analysis of Aegilops tauschii-derived seedling resistance to leaf rust in synthetic hexaploid wheat. J. Appl. Genet. 2020, 61, 163–168. [Google Scholar] [CrossRef] [PubMed]
- Qi, A.; Li, X.; Shi, L.; Liu, D.; Li, Z. Identification of a leaf rust resistance gene in the Chinese wheat line LB0288. Czech J. Genet. Plant Breed. 2016, 51, 43–49. [Google Scholar] [CrossRef]
- Moore, J.W.; Herrera-Foessel, S.; Lan, C.; Schnippenkoetter, W.; Ayliffe, M.; Huerta-Espino, J.; Lillemo, M.; Viccars, L.; Milne, R.J.; Periyannan, S.; et al. A recently evolved hexose transporter variant confers resistance to multiple pathogens in wheat. Nat. Genet. 2015, 47, 1494–1498. [Google Scholar] [CrossRef] [PubMed]
- Zhai, H.; Feng, Z.; Li, J.; Liu, X.; Xiao, S.; Ni, Z.; Sun, Q. QTL analysis of spike morphological traits and plant height in winter wheat (Triticum aestivum L.) using a high-density SNP and SSR-based linkage map. Front. Plant Sci. 2016, 7, 1617. [Google Scholar] [CrossRef]
- Dubcovsky, J.; Dvorak, J. Genome plasticity a key factor in the success of polyploid wheat under domestication. Science 2007, 316, 1862–1866. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wong, D.; Forrest, K.; Allen, A.; Chao, S.; Huang, B.E.; Maccaferri, M.; Salvi, S.; Milner, S.G.; Cattivelli, L.; et al. Characterization of polyploid wheat genomic diversity using a high-density 90 000 single nucleotide polymorphism array. Plant Biotechnol. J. 2014, 12, 787–796. [Google Scholar] [CrossRef] [PubMed]
- Maccaferri, M.; Zhang, J.; Bulli, P.; Abate, Z.; Chao, S.; Cantu, D.; Bossolini, E.; Chen, X.; Pumphrey, M.; Dubcovsky, J. A genome-wide association study of resistance to stripe rust (Puccinia striiformis f. sp. tritici) in a worldwide collection of hexaploid spring wheat (Triticum aestivum L.). G3 Genes Genomes Genet. 2015, 5, 449–465. [Google Scholar] [CrossRef]
- Borrelli, G.M.; Mazzucotelli, E.; Marone, D.; Crosatti, C.; Michelotti, V.; Valè, G.; Mastrangelo, A.M. Regulation and evolution of NLR genes: A close interconnection for plant immunity. Int. J. Mol. Sci. 2018, 19, 1662. [Google Scholar] [CrossRef]
- Ling, H.-Q.; Qiu, J.; Singh, R.P.; Keller, B. Identification and genetic characterization of an Aegilops tauschii ortholog of the wheat leaf rust disease resistance gene Lr1. Theor. Appl. Genet. 2004, 109, 1133–1138. [Google Scholar] [CrossRef] [PubMed]
- Bourras, S.; McNally, K.E.; Ben-David, R.; Parlange, F.; Roffler, S.; Praz, C.R.; Oberhaensli, S.; Menardo, F.; Stirnweis, D.; Frenkel, Z.; et al. Multiple avirulence loci and allele-specific effector recognition control the Pm3 race-specific resistance of wheat to powdery mildew. Plant Cell 2015, 27, 2991–3012. [Google Scholar] [CrossRef]
- Desiderio, F.; Guerra, D.; Rubiales, D.; Piarulli, L.; Pasquini, M.; Mastrangelo, A.M.; Simeone, R.; Blanco, A.; Cattivelli, L.; Valè, G. Identification and mapping of quantitative trait loci for leaf rust resistance derived from a tetraploid wheat Triticum dicoccum accession. Mol. Breed. 2014, 34, 1659–1675. [Google Scholar] [CrossRef]
- Brunner, S.; Hurni, S.; Streckeisen, P.; Mayr, G.; Albrecht, M.; Yahiaoui, N.; Keller, B. Intragenic allele pyramiding combines different specificities of wheat Pm3 resistance alleles. Plant J. 2010, 64, 433–445. [Google Scholar] [CrossRef]
- Jones, B.; Sall, J. JMP statistical discovery software. Wiley Interdiscip. Rev. Comput. Stat. 2011, 3, 188–194. [Google Scholar] [CrossRef]
- Nyquist, W.E.; Baker, R.J. Estimation of heritability and prediction of selection response in plant populations. Crit. Rev. Plant Sci. 1991, 10, 235–322. [Google Scholar] [CrossRef]
- De Givry, S.; Bouchez, M.; Milan, D.; Schiex, T.; Chabrier, P. CARHTA GENE: Multipopulation integrated genetic and radiation hybrid mapping. Bioinformatics 2004, 21, 1703–1704. [Google Scholar] [CrossRef]
- Kosambi, D.D. The estimation of map distances from recombination values. Ann. Eugen. 1943, 12, 172–175. [Google Scholar] [CrossRef]
- Broman, K.W.; Wu, H.; Sen, Ś.; Churchill, G.A. R/qtl: QTL mapping in experimental crosses. Bioinformatics 2003, 19, 889–890. [Google Scholar] [CrossRef] [PubMed]
- Lander, E.S.; Botstein, D. Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 1989, 121, 185–199. [Google Scholar] [CrossRef]
- Churchill, G.A.; Doerge, R.W. Empirical threshold values for quantitative trait mapping. Genetics 1994, 138, 963–971. [Google Scholar] [CrossRef] [PubMed]
- Sen, Ś.; Churchill, G.A. A statistical framework for quantitative trait mapping. Genetics 2001, 159, 371–387. [Google Scholar]
- Darvasi, A.; Soller, M. A simple method to calculate resolving power and confidence interval of QTL map location. Behav. Genet. 1997, 27, 125–132. [Google Scholar] [CrossRef]
- The International Wheat Genome Sequencing Consortium (IWGSC); Appels, R.; Eversole, K.; Stein, N.; Feuillet, C.; Keller, B.; Rogers, J.; Pozniak, C.J.; Choulet, F.; Distelfeld, A.; et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191. [Google Scholar] [CrossRef]
- Borrill, P.; Ramirez-Gonzalez, R.; Uauy, C. expVIP: A customizable RNA-seq data analysis and visualization platform. Plant Physiol. 2016, 170, 2172–2186. [Google Scholar] [CrossRef] [PubMed]

| Pt_Jerez05 | Pt_VMC03 | Bgt_96224 | Bgt_94202 | Bgt_JIW2 | |
|---|---|---|---|---|---|
| Victo | 0 | 0 | 4.3 | 8 | 3.1 |
| Spada | 9 | 9 | 7.8 | 9 | 8 |
| Trait/Disease | R:S | Ratio | p1 |
|---|---|---|---|
| IT Leaf Rust | 83:38 | 1:1 | 4.6 × 10−5 |
| IT Powdery Mildew | 59:61 | 1:1 | 0.797894 |
| Chromosome_ Linkage Group | Total Marker | Total Marker (No Co-Segregant) | cM | cM/Marker 1 | cM/Marker 2 |
|---|---|---|---|---|---|
| 1A_1 | 704 | 102 | 159.2 | 0.23 | 1.56 |
| 1B_1 | 801 | 115 | 186.1 | 0.23 | 1.62 |
| 1D_1 | 146 | 44 | 100.3 | 0.69 | 2.28 |
| 2A_1 | 422 | 63 | 167 | 0.40 | 2.65 |
| 2B_1 | 841 | 113 | 199.3 | 0.24 | 1.76 |
| 2D_1 | 21 | 8 | 9.7 | 0.46 | 1.21 |
| 2D_2 | 40 | 19 | 85.8 | 2.15 | 4.52 |
| 2D_all | 61 | 27 | 95.5 | 1.57 | 3.54 |
| 3A_1 | 383 | 79 | 167.6 | 0.44 | 2.12 |
| 3A_2 | 68 | 14 | 38.4 | 0.56 | 2.74 |
| 3A_all | 451 | 93 | 206 | 0.46 | 2.22 |
| 3B_1 | 708 | 116 | 228.7 | 0.32 | 1.97 |
| 3D_1 | 34 | 6 | 19.1 | 0.56 | 3.18 |
| 4A_1 | 171 | 60 | 161.5 | 0.94 | 2.69 |
| 4A_2 | 84 | 16 | 15.7 | 0.19 | 0.98 |
| 4A_all | 255 | 76 | 177.2 | 0.69 | 2.33 |
| 4B_1 | 305 | 62 | 150.7 | 0.49 | 2.43 |
| 4D_1 | 7 | 6 | 12.4 | 1.77 | 2.07 |
| 4D_2 | 6 | 6 | 9.3 | 1.55 | 1.55 |
| 4D_3 | 3 | 3 | 10.1 | 3.37 | 3.37 |
| 4D_all | 16 | 15 | 31.8 | 1.99 | 2.12 |
| 5A_1 | 578 | 146 | 323.2 | 0.56 | 2.21 |
| 5B_1 | 774 | 139 | 233 | 0.30 | 1.68 |
| 5D_1 | 95 | 18 | 104.2 | 1.10 | 5.79 |
| 6A_1 | 644 | 75 | 157.7 | 0.24 | 2.10 |
| 6B_1 | 448 | 78 | 197.1 | 0.44 | 2.53 |
| 6D_1 | 22 | 7 | 22.5 | 1.02 | 3.21 |
| 6D_2 | 136 | 14 | 14.2 | 0.10 | 1.01 |
| 6D_all | 158 | 21 | 36.7 | 0.23 | 1.75 |
| 7A_1 | 611 | 115 | 198 | 0.32 | 1.72 |
| 7A_2 | 99 | 15 | 20 | 0.20 | 1.33 |
| 7A_all | 710 | 130 | 218 | 0.31 | 1.68 |
| 7B_1 | 423 | 84 | 131.3 | 0.31 | 1.56 |
| 7B_2 | 91 | 15 | 49.3 | 0.54 | 3.29 |
| 7B_all | 514 | 99 | 180.6 | 0.35 | 1.82 |
| 7D_1 | 13 | 5 | 12.2 | 0.94 | 2.44 |
| 7D_2 | 48 | 25 | 107.9 | 2.25 | 4.32 |
| 7D_all | 61 | 30 | 120.1 | 1.97 | 4.00 |
| Total | 8726 | 1568 | 3291.5 | 0.38 | 2.11 |
| Genome | |||||
| A | 3764 | 685 | 1408.3 | 0.37 | 2.06 |
| B | 4391 | 722 | 1375.5 | 0.31 | 1.91 |
| D | 571 | 161 | 507.7 | 0.89 | 3.15 |
| Group | |||||
| 1 | 1651 | 261 | 445.6 | 0.27 | 1.71 |
| 2 | 1324 | 203 | 461.8 | 0.35 | 2.27 |
| 3 | 1193 | 215 | 453.8 | 0.38 | 2.11 |
| 4 | 576 | 153 | 359.7 | 0.62 | 2.35 |
| 5 | 1447 | 303 | 660.4 | 0.46 | 2.18 |
| 6 | 1250 | 174 | 391.5 | 0.31 | 2.25 |
| 7 | 1285 | 259 | 518.7 | 0.40 | 2.00 |
| QTL | Trait Disease | Chr | Peak Marker | cM | LOD | R2 (%) | Additive Effect 1 | CI |
|---|---|---|---|---|---|---|---|---|
| QPm.gb-7A | IT powdery mildew | 7A | IWB55071 | 14.8 | 60 | 90 | −3.8 | 14.1–15.5 |
| QLr.gb-2B | IT leaf rust | 2B | IWB32376 | 113.9 | 10.5 | 15.2 | −1.16 | 109–118.1 |
| QLr.gb-5D | IT leaf rust | 5D | IWB11400 | 104.2 | 28.1 | 59.3 | −2.43 | 103.1–105.3 |
| QLr.gb-2B*QLr.gb-5D | IT leaf rust | 4.74 | 6.11 | 0.93 | ||||
| QLr.gb-2B | RDS leaf rust | 2B | IWB32376 | 113.9 | 11.2 | 16.2 | −14.153 | 109.9–117.8 |
| QLr.gb-5D | RDS leaf rust | 5D | IWB11400 | 104.2 | 28.4 | 59.3 | −28.599 | 103.1–105.3 |
| QLr.gb-2B*QLr.gb-5D | RDS leaf rust | 5.1 | 6.51 | 11.335 |
| QTL | Chr | Genetic Interval (cM) | Start (cM) | End (cM) | Mbp | Start (bp) | End (bp) |
|---|---|---|---|---|---|---|---|
| QPm.gb-7A | 7A | 1.41 | 14.1 | 15.5 | 6.35 | 724,084,703 | 730,433,215 |
| QLr.gb-5D | 5D | 2.15 | 103.1 | 105.3 | 2.49 | 560,228,250 | 562,726,656 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Desiderio, F.; Bourras, S.; Mazzucotelli, E.; Rubiales, D.; Keller, B.; Cattivelli, L.; Valè, G. Characterization of the Resistance to Powdery Mildew and Leaf Rust Carried by the Bread Wheat Cultivar Victo. Int. J. Mol. Sci. 2021, 22, 3109. https://doi.org/10.3390/ijms22063109
Desiderio F, Bourras S, Mazzucotelli E, Rubiales D, Keller B, Cattivelli L, Valè G. Characterization of the Resistance to Powdery Mildew and Leaf Rust Carried by the Bread Wheat Cultivar Victo. International Journal of Molecular Sciences. 2021; 22(6):3109. https://doi.org/10.3390/ijms22063109
Chicago/Turabian StyleDesiderio, Francesca, Salim Bourras, Elisabetta Mazzucotelli, Diego Rubiales, Beat Keller, Luigi Cattivelli, and Giampiero Valè. 2021. "Characterization of the Resistance to Powdery Mildew and Leaf Rust Carried by the Bread Wheat Cultivar Victo" International Journal of Molecular Sciences 22, no. 6: 3109. https://doi.org/10.3390/ijms22063109
APA StyleDesiderio, F., Bourras, S., Mazzucotelli, E., Rubiales, D., Keller, B., Cattivelli, L., & Valè, G. (2021). Characterization of the Resistance to Powdery Mildew and Leaf Rust Carried by the Bread Wheat Cultivar Victo. International Journal of Molecular Sciences, 22(6), 3109. https://doi.org/10.3390/ijms22063109









