Long-Distance Movement of Mineral Deficiency-Responsive mRNAs in Nicotiana Benthamiana/Tomato Heterografts
Abstract
1. Introduction
2. Results
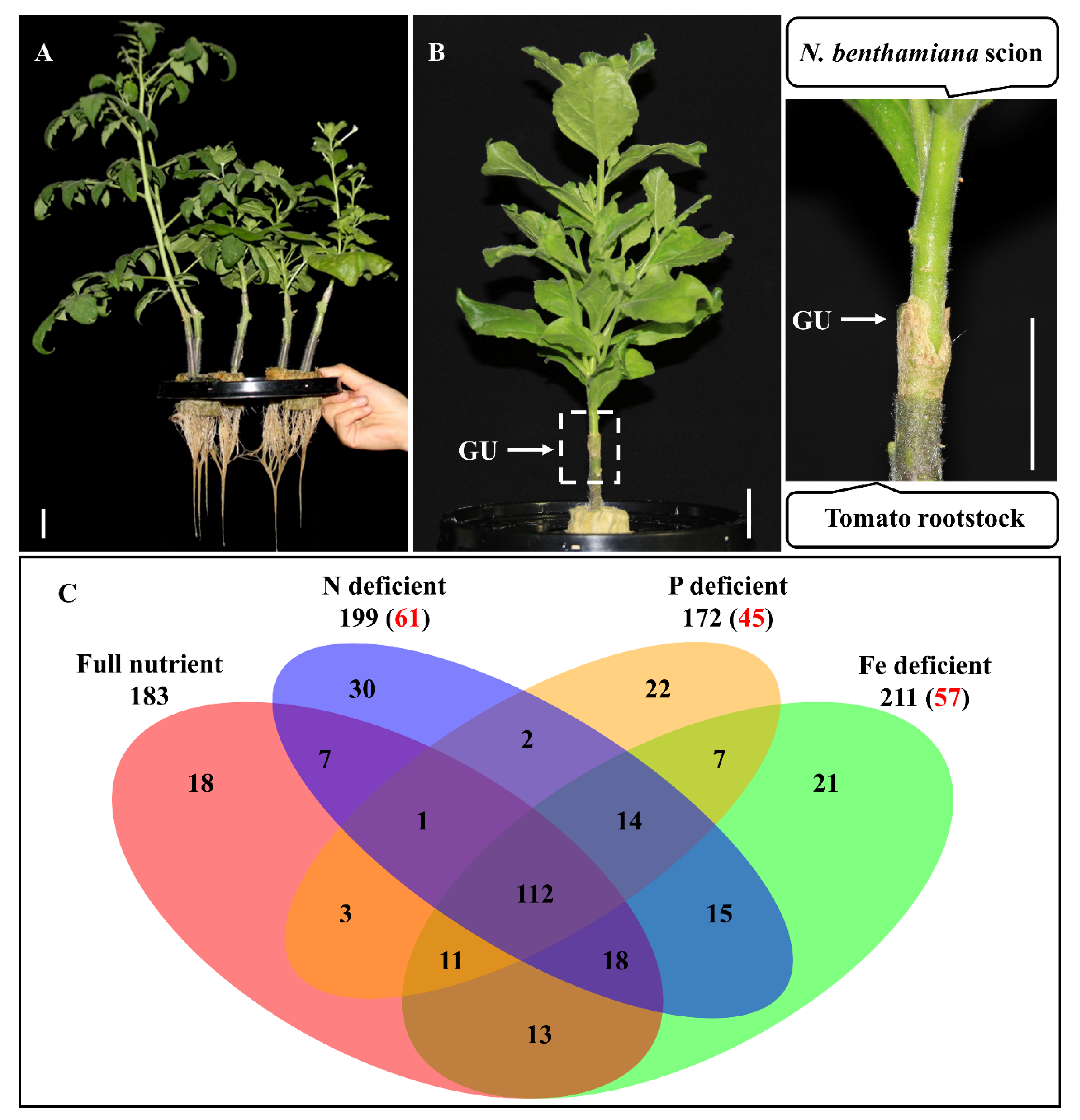
2.1. Shoot-to-Root mRNA Migration under Mineral Deficient Conditions
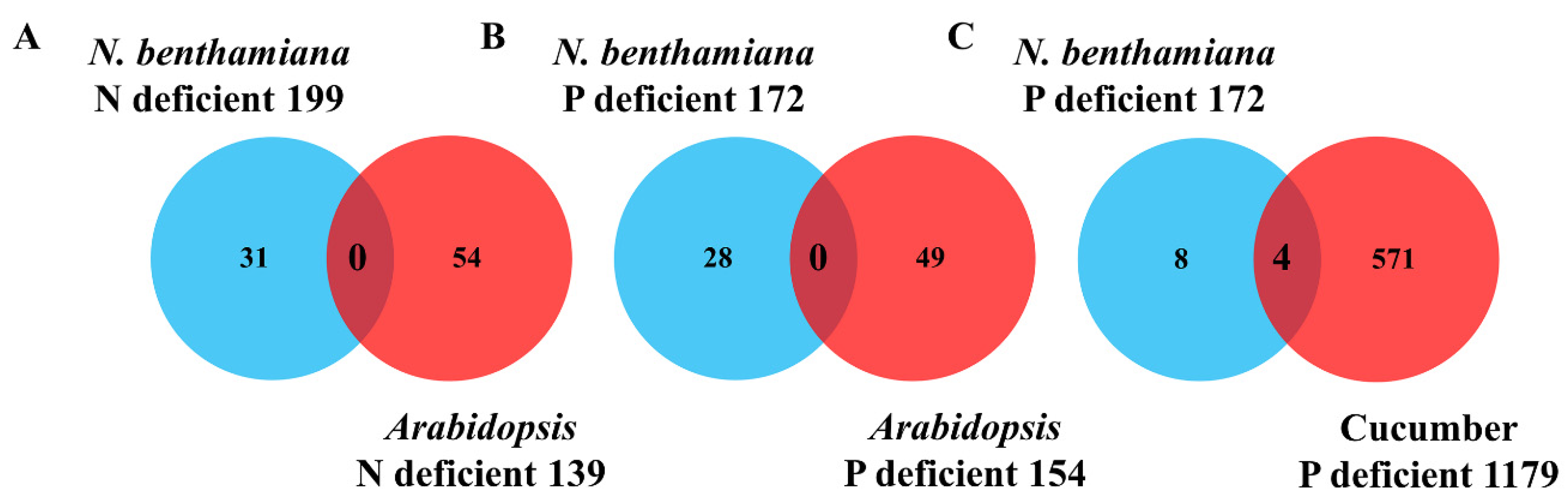
2.2. Identification of the Conserved mRNAs by Comparison with Other Heterografts
2.3. Mobile mRNAs Transported to the Roots are not Enriched in Phloem Transcripts Identified by Other Phloem Sap Collection Methods
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Grafting Procedure
5.2. RNA-Seq and Bioinformatic Analysis
5.3. Orthology Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zhang, Z.; Liao, H.; Lucas, W.J. Molecular mechanisms underlying phosphate sensing, signaling, and adaptation in plants. J. Integr. Plant Biol. 2014, 56, 192–220. [Google Scholar] [CrossRef] [PubMed]
- Jia, Z.; Wirén, N.V. Signaling pathways underlying nitrogen-dependent changes in root system architecture: From model to crop species. J. Exp. Bot. 2020. [Google Scholar] [CrossRef] [PubMed]
- Xuan, W.; Beeckman, T.; Xu, G. Plant nitrogen nutrition: Sensing and signaling. Curr. Opin. Plant Biol. 2017, 39, 57–65. [Google Scholar] [CrossRef]
- Gayomba, S.R.; Zhai, Z.; Jung, H.I.; Vatamaniuk, O.K. Local and systemic signaling of iron status and its interactions with homeostasis of other essential elements. Front. Plant Sci. 2015, 6, 716. [Google Scholar] [CrossRef]
- Burleigh, S.H.; Harrison, M.J. The down-regulation of Mt4-like genes by phosphate fertilization occurs systemically and involves phosphate translocation to the shoots. Plant Physiol. 1999, 119, 241–248. [Google Scholar] [CrossRef]
- Liu, C.; Muchhal, U.S.; Uthappa, M.; Kononowicz, A.K.; Raghothama, K.G. Tomato phosphate transporter genes are differentially regulated in plant tissues by phosphorus. Plant Physiol. 1998, 116, 91–99. [Google Scholar] [CrossRef]
- Thibaud, M.C.; Arrighi, J.F.; Bayle, V.; Chiarenza, S.; Creff, A.; Bustos, R.; Paz-Ares, J.; Poirier, Y.; Nussaume, L. Dissection of local and systemic transcriptional responses to phosphate starvation in Arabidopsis. Plant J. 2010, 64, 775–789. [Google Scholar] [CrossRef] [PubMed]
- Guan, P.; Wang, R.; Nacry, P.; Breton, G.; Kay, S.A.; Pruneda-Paz, J.L.; Davani, A.; Crawford, N.M. Nitrate foraging by Arabidopsis roots is mediated by the transcription factor TCP20 through the systemic signaling pathway. Proc. Natl. Acad. Sci. USA 2014, 111, 15267–15272. [Google Scholar] [CrossRef]
- Ota, R.; Ohkubo, Y.; Yamashita, Y.; Ogawa-Ohnishi, M.; Matsubayashi, Y. Shoot-to-root mobile CEPD-like 2 integrates shoot nitrogen status to systemically regulate nitrate uptake in Arabidopsis. Nat. Commun. 2020, 11, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Ohkubo, Y.; Tanaka, M.; Tabata, R.; Ogawa-Ohnishi, M.; Matsubayashi, Y. Shoot-to-root mobile polypeptides involved in systemic regulation of nitrogen acquisition. Nat. Plants 2017, 3, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Aung, K.; Lin, S.I.; Wu, C.C.; Huang, Y.T.; Su, C.L.; Chiou, T.J. pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiol. 2006, 141, 1000–1011. [Google Scholar] [CrossRef] [PubMed]
- Bari, R.; Pant, B.D.; Stitt, M.; Scheible, W.R. PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol. 2006, 141, 988–999. [Google Scholar] [CrossRef] [PubMed]
- Grillet, L.; Lan, P.; Li, W.; Mokkapati, G.; Schmidt, W. IRON MAN is a ubiquitous family of peptides that control iron transport in plants. Nat. Plants 2018, 4, 953–963. [Google Scholar] [CrossRef] [PubMed]
- Turgeon, R.; Wolf, S. Phloem transport: Cellular pathways and molecular trafficking. Annu. Rev. Plant Biol. 2009, 60, 207–221. [Google Scholar] [CrossRef] [PubMed]
- Xia, C.; Zhang, C. Long-distance movement of mRNAs in plants. Plants 2020, 9, 731. [Google Scholar] [CrossRef]
- Notaguchi, M.; Wolf, S.; Lucas, W.J. Phloem-mobile Aux/IAA transcripts target to the root tip and modify root architecture. J. Integr. Plant Biol. 2012, 54, 760–772. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Perrera, V.; Saplaoura, E.; Apelt, F.; Bahin, M.; Kramdi, A.; Olas, J.; Mueller-Roeber, B.; Sokolowska, E.; Zhang, W.; et al. m(5)C methylation guides systemic transport of messenger RNA over graft junctions in plants. Curr. Biol. 2019, 29, 2465–2476. [Google Scholar] [CrossRef]
- Branco-Price, C.; Kaiser, K.A.; Jang, C.J.; Larive, C.K.; Bailey-Serres, J. Selective mRNA translation coordinates energetic and metabolic adjustments to cellular oxygen deprivation and reoxygenation in Arabidopsis thaliana. Plant J. 2008, 56, 743–755. [Google Scholar] [CrossRef]
- Banerjee, A.K.; Chatterjee, M.; Yu, Y.; Suh, S.G.; Miller, W.A.; Hannapel, D.J. Dynamics of a mobile RNA of potato involved in a long-distance signaling pathway. Plant Cell 2006, 18, 3443–3457. [Google Scholar] [CrossRef]
- Hannapel, D.J.; Banerjee, A.K. Multiple mobile mRNA signals regulate tuber development in potato. Plants 2017, 6, 8. [Google Scholar] [CrossRef]
- Thieme, C.J.; Rojas-Triana, M.; Stecyk, E.; Schudoma, C.; Zhang, W.; Yang, L.; Miñambres, M.; Walther, D.; Schulze, W.X.; Paz-Ares, J.; et al. Endogenous Arabidopsis messenger RNAs transported to distant tissues. Nat. Plants 2015, 1, 15025. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zheng, Y.; Ham, B.K.; Chen, J.; Yoshida, A.; Kochian, L.V.; Fei, Z.; Lucas, W.J. Vascular-mediated signalling involved in early phosphate stress response in plants. Nat. Plants 2016, 2, 16033. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Wu, F.; Shi, J.; Zhu, Y.; Zhu, Z.; Gong, Q.; Hu, J. ROOT HAIR DEFECTIVE3 family of dynamin-like GTPases mediates homotypic endoplasmic reticulum fusion and is essential for Arabidopsis development. Plant Physiol. 2013, 163, 713–720. [Google Scholar] [CrossRef] [PubMed]
- To, J.P.; Deruère, J.; Maxwell, B.B.; Morris, V.F.; Hutchison, C.E.; Ferreira, F.J.; Schaller, G.E.; Kieber, J.J. Cytokinin regulates type-A Arabidopsis Response Regulator activity and protein stability via two-component phosphorelay. Plant Cell 2007, 19, 3901–3914. [Google Scholar] [CrossRef]
- Kinoshita, A.; Colette, A.; Tabata, R.; Yamada, M.; Shimizu, N.; Ishida, T.; Yamaguchi, K.; Shigenobu, S.; Takebayashi, Y.; Iuchi, S.; et al. A plant U-box protein, PUB4, regulates asymmetric cell division and cell proliferation in the root meristem. Development 2015, 142, 444–453. [Google Scholar] [CrossRef]
- Zhao, W.; Yang, X.; Yu, H.; Jiang, W.; Sun, N.; Liu, X.; Liu, X.; Zhang, X.; Wang, Y.; Gu, X. RNA-Seq-based transcriptome profiling of early nitrogen deficiency response in cucumber seedlings provides new insight into the putative nitrogen regulatory network. Plant Cell Physiol. 2015, 56, 455–467. [Google Scholar] [CrossRef]
- Liu, X.; Bartholomew, E.; Cai, Y.; Ren, H. Trichome-related mutants provide a new perspective on multicellular trichome initiation and development in cucumber (Cucumis sativus L). Front. Plant Sci. 2016, 7, 1187. [Google Scholar] [CrossRef]
- Ivashikina, N.; Deeken, R.; Ache, P.; Kranz, E.; Pommerrenig, B.; Sauer, N.; Hedrich, R. Isolation of AtSUC2 promoter-GFP-marked companion cells for patch-clamp studies and expression profiling. Plant J. 2003, 36, 931–945. [Google Scholar] [CrossRef]
- Deeken, R.; Ache, P.; Kajahn, I.; Klinkenberg, J.; Bringmann, G.; Hedrich, R. Identification of Arabidopsis thaliana phloem RNAs provides a search criterion for phloem-based transcripts hidden in complex datasets of microarray experiments. Plant J. 2008, 55, 746–759. [Google Scholar] [CrossRef]
- Doering-Saad, C.; Newbury, H.J.; Couldridge, C.E.; Bale, J.S.; Pritchard, J. A phloem-enriched cDNA library from Ricinus: Insights into phloem function. J. Exp. Bot. 2006, 57, 3183–3193. [Google Scholar] [CrossRef]
- Kanehira, A.; Yamada, K.; Iwaya, T.; Tsuwamoto, R.; Kasai, A.; Nakazono, M.; Harada, T. Apple phloem cells contain some mRNAs transported over long distances. Tree Genet. Genomes 2010, 6, 635–642. [Google Scholar] [CrossRef]
- Guo, S.; Zhang, J.; Sun, H.; Salse, J.; Lucas, W.J.; Zhang, H.; Zheng, Y.; Mao, L.; Ren, Y.; Wang, Z.; et al. The draft genome of watermelon (Citrullus lanatus) and resequencing of 20 diverse accessions. Nat. Genet. 2013, 45, 51–58. [Google Scholar] [CrossRef]
- Omid, A.; Keilin, T.; Glass, A.; Leshkowitz, D.; Wolf, S. Characterization of phloem-sap transcription profile in melon plants. J. Exp. Bot. 2007, 58, 3645–3656. [Google Scholar] [CrossRef] [PubMed]
- Xia, C.; Zheng, Y.; Huang, J.; Zhou, X.; Li, R.; Zha, M.; Wang, S.; Huang, Z.; Lan, H.; Turgeon, R.; et al. Elucidation of the mechanisms of long-distance mRNA movement in a Nicotiana benthamiana/tomato heterograft system. Plant Physiol. 2018, 177, 745–758. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, W.; Schikora, A. Different pathways are involved in phosphate and iron stress-induced alterations of root epidermal cell development. Plant Physiol. 2001, 125, 2078–2084. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gruber, B.D.; Giehl, R.F.; Friedel, S.; von Wirén, N. Plasticity of the Arabidopsis root system under nutrient deficiencies. Plant Physiol. 2013, 163, 161–179. [Google Scholar] [CrossRef] [PubMed]
- Medici, A.; Szponarski, W.; Dangeville, P.; Safi, A.; Dissanayake, I.M.; Saenchai, C.; Emanuel, A.; Rubio, V.; Lacombe, B.; Ruffel, S.; et al. Identification of molecular integrators shows that nitrogen actively controls the phosphate starvation response in plants. Plant Cell 2019, 31, 1171–1184. [Google Scholar] [CrossRef]
- Maeda, Y.; Konishi, M.; Kiba, T.; Sakuraba, Y.; Sawaki, N.; Kurai, T.; Ueda, Y.; Sakakibara, H.; Yanagisawa, S. A NIGT1-centred transcriptional cascade regulates nitrate signalling and incorporates phosphorus starvation signals in Arabidopsis. Nat. Commun. 2018, 9, 1376. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Z.; Gayomba, S.R.; Jung, H.I.; Vimalakumari, N.K.; Piñeros, M.; Craft, E.; Rutzke, M.A.; Danku, J.; Lahner, B.; Punshon, T.; et al. OPT3 is a phloem-specific iron transporter that is essential for systemic iron signaling and redistribution of iron and cadmium in Arabidopsis. Plant Cell 2014, 26, 2249–2264. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Tomato Genome Consortium. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 2012, 485, 635. [Google Scholar] [CrossRef] [PubMed]
- Bombarely, A.; Rosli, H.G.; Vrebalov, J.; Moffett, P.; Mueller, L.A.; Martin, G.B. A draft genome sequence of Nicotiana benthamiana to enhance molecular plant-microbe biology research. Mol. Plant Microbe Interact. 2012, 25, 1523–1530. [Google Scholar] [CrossRef] [PubMed]
| Arabidopsis Thaliana [25] | Arabidopsis Thaliana [26] | Ricinus Communis [27] | Malus Prunifolia [28] | Citrullus Lanatus [29] | Cucumis Sativus [29] | Cucumis Melo [30] | All Sets | |
|---|---|---|---|---|---|---|---|---|
| Number of mRNAs in previous studies | 147 | 950 | 141 | 113 | 701 | 365 | 332 | 2352 |
| Number of mobile mRNAs | 0 | 3 | 2 | 0 | 1 | 0 | 1 | 7 |
| Covered % | 0.00 | 0.32 | 1.42 | 0.00 | 0.14 | 0.00 | 0.30 | 0.30 |
| Over- or under-enrichment | N/A | N/A | N/A | N/A | N/A | N/A | N/A | under- |
| P-Value (Hypergeometric test) | N/A | 0.13 | 0.23 | N/A | 0.06 | N/A | 0.36 | 0.01 |
| Arabidopsis Thaliana [25] | Arabidopsis Thaliana [26] | Ricinus Communis [27] | Malus Prunifolia [28] | Citrullus Lanatus [29] | Cucumis Sativus [29] | Cucumis Melo [30] | All Sets | |
|---|---|---|---|---|---|---|---|---|
| Long-stem: 854 mobile mRNAs | ||||||||
| Number of mRNAs in previous studies | 147 | 950 | 141 | 113 | 701 | 365 | 332 | 2352 |
| Number of mobile mRNAs | 10 | 48 | 11 | 9 | 33 | 13 | 13 | 114 |
| Covered % | 6.80 | 5.89 | 9.22 | 10.62 | 4.85 | 3.56 | 3.92 | 5.44 |
| Over- or under-enrichment | over- | over- | over- | over- | over- | over- | over- | over- |
| P-Value (Hypergeometric test) | 5.00 × 10−4 | 9.59 × 10−10 | 7.95 × 10−5 | 2.99 × 10−4 | 1.98 × 10−6 | 0.02 | 0.01 | 2.76 × 10−20 |
| Root: 242 mobile mRNAs | ||||||||
| Number of mRNAs in previous studies | 147 | 950 | 141 | 113 | 701 | 365 | 332 | 2352 |
| Number of mobile mRNAs | 0 | 7 | 0 | 3 | 1 | 0 | 2 | 13 |
| Covered % | 0.00 | 0.74 | 0.00 | 2.65 | 0.14 | 0.00 | 0.60 | 0.55 |
| Over- or under-enrichment | N/A | N/A | N/A | over- | N/A | N/A | N/A | N/A |
| P-Value (Hypergeometric test) | N/A | 0.25 | N/A | 0.02 | 0.11 | N/A | 0.53 | 0.49 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xia, C.; Huang, J.; Lan, H.; Zhang, C. Long-Distance Movement of Mineral Deficiency-Responsive mRNAs in Nicotiana Benthamiana/Tomato Heterografts. Plants 2020, 9, 876. https://doi.org/10.3390/plants9070876
Xia C, Huang J, Lan H, Zhang C. Long-Distance Movement of Mineral Deficiency-Responsive mRNAs in Nicotiana Benthamiana/Tomato Heterografts. Plants. 2020; 9(7):876. https://doi.org/10.3390/plants9070876
Chicago/Turabian StyleXia, Chao, Jing Huang, Hai Lan, and Cankui Zhang. 2020. "Long-Distance Movement of Mineral Deficiency-Responsive mRNAs in Nicotiana Benthamiana/Tomato Heterografts" Plants 9, no. 7: 876. https://doi.org/10.3390/plants9070876
APA StyleXia, C., Huang, J., Lan, H., & Zhang, C. (2020). Long-Distance Movement of Mineral Deficiency-Responsive mRNAs in Nicotiana Benthamiana/Tomato Heterografts. Plants, 9(7), 876. https://doi.org/10.3390/plants9070876