Transcriptomic Landscape of Circulating Extracellular Vesicles in Heart Transplant Ischemia–Reperfusion
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Sample Collection and Processing
2.3. Transmission Electron Microscopy and Nanoparticle Tracking Analysis
2.4. RNA Sequencing
2.5. Bioinformatics
3. Results
3.1. Recipient Characteristics
3.2. EV Characterization
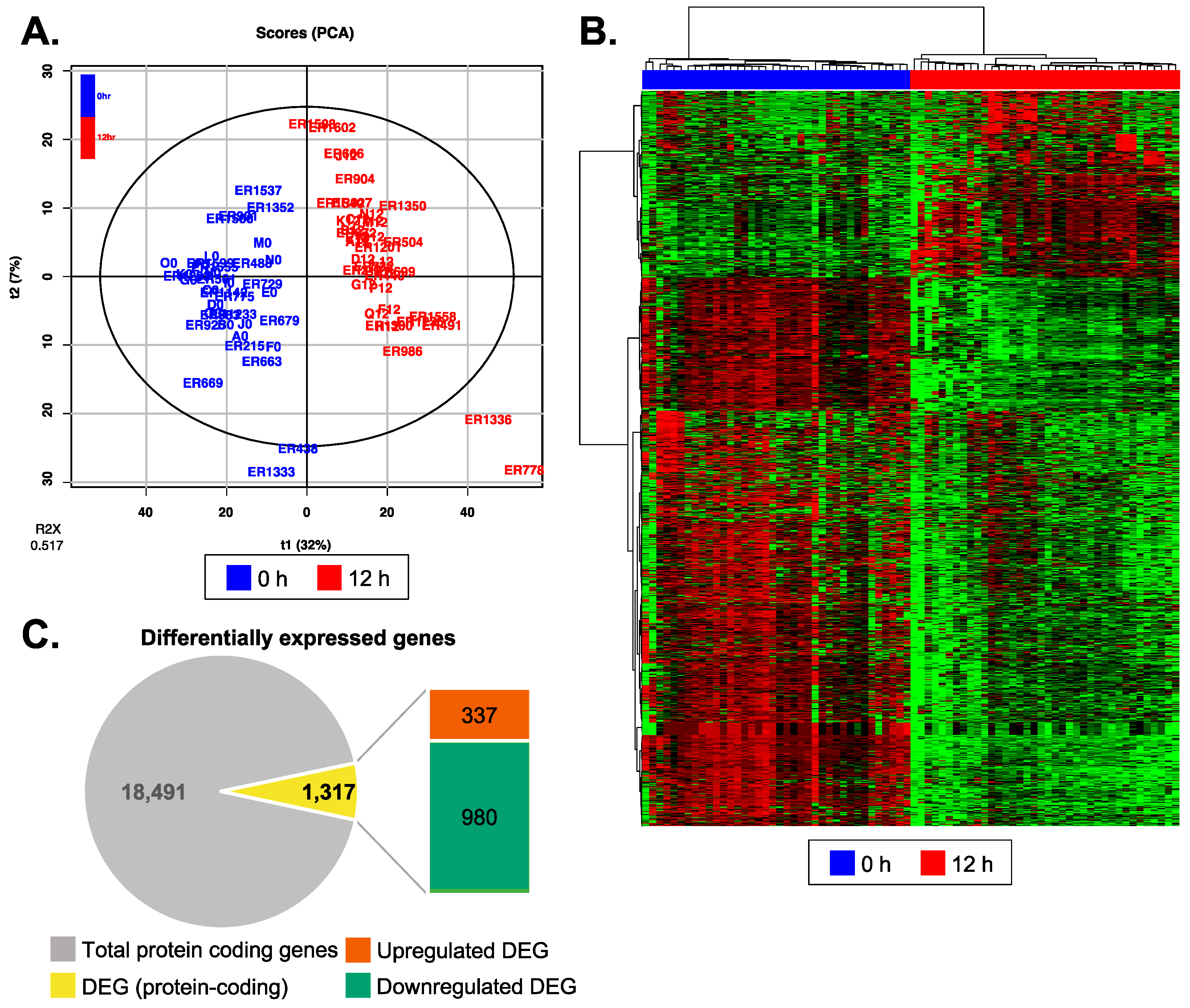
3.3. Reperfusion Altered mRNA Expression Profiles of Recipient Plasma EV
3.4. Biological Functions Related to the Differentially Expressed Genes after Reperfusion
3.5. Post-Reperfusion EVs Carry More Adaptive Immunity-Related Genes but Less Extracellular Matrix Component Protein-Coding Genes
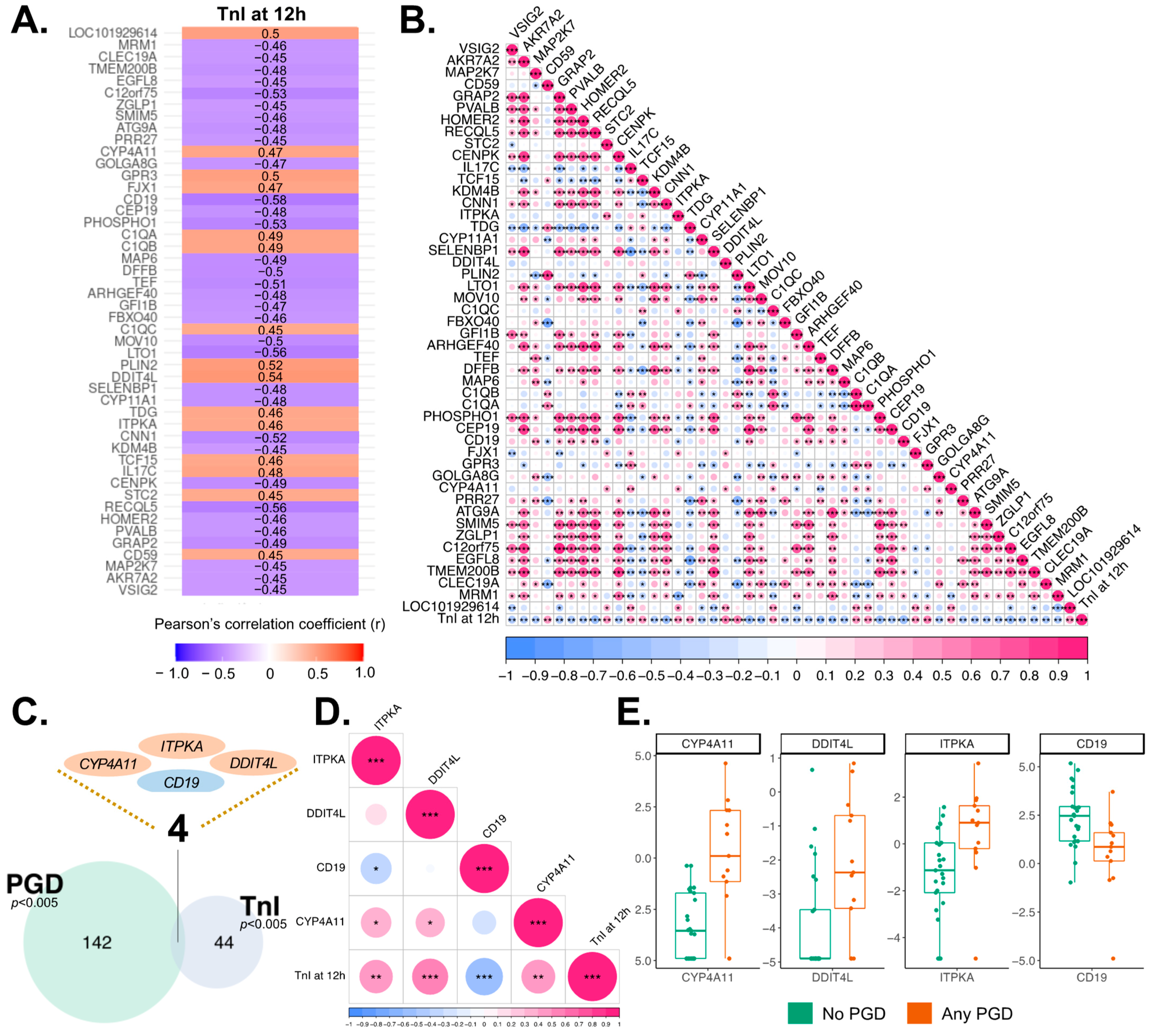
3.6. Correlation of EV Transcriptome with Cardiomyocyte Injury
3.7. Association of EV Transcriptome and Outcomes after Heart Transplantation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tang, Q.; Dong, C.; Sun, Q. Immune Response Associated with Ischemia and Reperfusion Injury during Organ Transplantation. Inflamm. Res. 2022, 71, 1463–1476. [Google Scholar] [CrossRef] [PubMed]
- Fernández, A.R.; Sánchez-Tarjuelo, R.; Cravedi, P.; Ochando, J.; López-Hoyos, M. Review: Ischemia Reperfusion Injury—A Translational Perspective in Organ Transplantation. Int. J. Mol. Sci. 2020, 21, 8549. [Google Scholar] [CrossRef]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-Mediated Transfer of MRNAs and MicroRNAs Is a Novel Mechanism of Genetic Exchange between Cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef]
- Ravichandran, R.; Bansal, S.; Rahman, M.; Sureshbabu, A.; Sankpal, N.; Fleming, T.; Bharat, A.; Mohanakumar, T. Extracellular Vesicles Mediate Immune Responses to Tissue-Associated Self-Antigens: Role in Solid Organ Transplantations. Front. Immunol. 2022, 13, 861583. [Google Scholar] [CrossRef]
- Ge, X.; Meng, Q.; Wei, L.; Liu, J.; Li, M.; Liang, X.; Lin, F.; Zhang, Y.; Li, Y.; Liu, Z.; et al. Myocardial Ischemia-reperfusion Induced Cardiac Extracellular Vesicles Harbour Proinflammatory Features and Aggravate Heart Injury. J. Extracell. Vesicles 2021, 10, e12072. [Google Scholar] [CrossRef] [PubMed]
- Tuuminen, R.; Syrjälä, S.; Krebs, R.; Keränen, M.A.I.; Koli, K.; Abo-Ramadan, U.; Neuvonen, P.J.; Tikkanen, J.M.; Nykänen, A.I.; Lemström, K.B. Donor Simvastatin Treatment Abolishes Rat Cardiac Allograft Ischemia/Reperfusion Injury and Chronic Rejection Through Microvascular Protection. Circulation 2011, 124, 1138–1150. [Google Scholar] [CrossRef] [PubMed]
- Nykänen, A.I.; Holmström, E.J.; Tuuminen, R.; Krebs, R.; Dhaygude, K.; Kankainen, M.; Jokinen, J.J.; Lommi, J.; Helanterä, I.; Räisänen-Sokolowski, A.; et al. Donor Simvastatin Treatment in Heart Transplantation. Circulation 2019, 140, 627–640. [Google Scholar] [CrossRef]
- Kobashigawa, J.; Zuckermann, A.; Macdonald, P.; Leprince, P.; Esmailian, F.; Luu, M.; Mancini, D.; Patel, J.; Razi, R.; Reichenspurner, H.; et al. Report from a Consensus Conference on Primary Graft Dysfunction after Cardiac Transplantation. J. Heart Lung Transplant. 2014, 33, 327–340. [Google Scholar] [CrossRef] [PubMed]
- Puhka, M.; Nordberg, M.-E.; Valkonen, S.; Rannikko, A.; Kallioniemi, O.; Siljander, P.; Af Hällström, T.M. KeepEX, a Simple Dilution Protocol for Improving Extracellular Vesicle Yields from Urine. Eur. J. Pharm. Sci. 2017, 98, 30–39. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. The Subread Aligner: Fast, Accurate and Scalable Read Mapping by Seed-and-Vote. Nucleic Acids Res. 2013, 41, e108. [Google Scholar] [CrossRef]
- Thévenot, E.A.; Roux, A.; Xu, Y.; Ezan, E.; Junot, C. Analysis of the Human Adult Urinary Metabolome Variations with Age, Body Mass Index, and Gender by Implementing a Comprehensive Workflow for Univariate and OPLS Statistical Analyses. J. Proteome Res. 2015, 14, 3322–3335. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Kolde, R. Pheatmap: Pretty Heatmaps. Available online: https://CRAN.R-project.org/package=pheatmap (accessed on 11 August 2022).
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. ClusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Supek, F.; Bošnjak, M.; Škunca, N.; Šmuc, T. REVIGO Summarizes and Visualizes Long Lists of Gene Ontology Terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef] [PubMed]
- Sayols, S. Rrvgo: A Bioconductor Package for Interpreting Lists of Gene Ontology Terms. MicroPubl. Biol. 2023, 2023. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING V11: Protein–Protein Association Networks with Increased Coverage, Supporting Functional Discovery in Genome-Wide Experimental Datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Dai, Y.; Hu, R.; Liu, A.; Cho, K.S.; Manuel, A.M.; Li, X.; Dong, X.; Jia, P.; Zhao, Z. WebCSEA: Web-Based Cell-Type-Specific Enrichment Analysis of Genes. Nucleic Acids Res. 2022, 50, W782–W790. [Google Scholar] [CrossRef] [PubMed]
- Schlicker, A.; Domingues, F.S.; Rahnenführer, J.; Lengauer, T. A New Measure for Functional Similarity of Gene Products Based on Gene Ontology. BMC Bioinform. 2006, 7, 302. [Google Scholar] [CrossRef]
- Jones, R.C.; Karkanias, J.; Krasnow, M.A.; Pisco, A.O.; Quake, S.R.; Salzman, J.; Yosef, N.; Bulthaup, B.; Brown, P.; Harper, W.; et al. The Tabula Sapiens: A Multiple-Organ, Single-Cell Transcriptomic Atlas of Humans. Science 2022, 376, eabl4896. [Google Scholar] [CrossRef]
- Holmström, E.J.; Syrjälä, S.O.; Dhaygude, K.; Tuuminen, R.; Krebs, R.; Nykänen, A.; Lemström, K.B. Severe Primary Graft Dysfunction of the Heart Transplant Is Associated with Increased Plasma and Intragraft Proinflammatory Cytokine Expression. J. Heart Lung Transplant. 2023, 42, 807–818. [Google Scholar] [CrossRef]
- Movahed, M.; Brockie, S.; Hong, J.; Fehlings, M.G. Transcriptomic Hallmarks of Ischemia-Reperfusion Injury. Cells 2021, 10, 1838. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.; Fedak, P.W.M.; Weisel, R.D.; Butany, J.; Rao, V.; Maitland, A.; Li, R.-K.; Dhillon, B.; Yau, T.M. Fundamentals of Reperfusion Injury for the Clinical Cardiologist. Circulation 2002, 105, 2332–2336. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Gao, L.; Thakur, A.; Siu, P.M.; Lai, C.W.K. Role of Free Fatty Acids in Endothelial Dysfunction. J. Biomed. Sci. 2017, 24, 50. [Google Scholar] [CrossRef] [PubMed]
- Kim, F.; Tysseling, K.A.; Rice, J.; Pham, M.; Haji, L.; Gallis, B.M.; Baas, A.S.; Paramsothy, P.; Giachelli, C.M.; Corson, M.A.; et al. Free Fatty Acid Impairment of Nitric Oxide Production in Endothelial Cells Is Mediated by IKKβ. Arterioscler. Thromb. Vasc. Biol. 2005, 25, 989–994. [Google Scholar] [CrossRef]
- Zhang, C.; He, M.; Ni, L.; He, K.; Su, K.; Deng, Y.; Li, Y.; Xia, H. The Role of Arachidonic Acid Metabolism in Myocardial Ischemia–Reperfusion Injury. Cell Biochem. Biophys. 2020, 78, 255–265. [Google Scholar] [CrossRef]
- Chauhan, P.K.; Sowdhamini, R. Transcriptome Data Analysis of Primary Cardiomyopathies Reveals Perturbations in Arachidonic Acid Metabolism. Front. Cardiovasc. Med. 2023, 10, 1111019. [Google Scholar] [CrossRef]
- Zhou, Y.; Khan, H.; Xiao, J.; Cheang, W.S. Effects of Arachidonic Acid Metabolites on Cardiovascular Health and Disease. Int. J. Mol. Sci. 2021, 22, 12029. [Google Scholar] [CrossRef]
- Gao, H.; Cao, Y.; Xia, H.; Zhu, X.; Jin, Y. CYP4A11 Is Involved in the Development of Nonalcoholic Fatty Liver Disease via ROS-induced Lipid Peroxidation and Inflammation. Int. J. Mol. Med. 2020, 45, 1121–1129. [Google Scholar] [CrossRef] [PubMed]
- Elbekai, R.H.; El-Kadi, A.O.S. Cytochrome P450 Enzymes: Central Players in Cardiovascular Health and Disease. Pharmacol. Ther. 2006, 112, 564–587. [Google Scholar] [CrossRef]
- Sirotina, S.; Ponomarenko, I.; Kharchenko, A.; Bykanova, M.; Bocharova, A.; Vagaytseva, K.; Stepanov, V.; Churnosov, M.; Solodilova, M.; Polonikov, A. A Novel Polymorphism in the Promoter of the CYP4A11 Gene Is Associated with Susceptibility to Coronary Artery Disease. Dis. Markers 2018, 2018, 5812802. [Google Scholar] [CrossRef]
- Hernández, G.; Lal, H.; Fidalgo, M.; Guerrero, A.; Zalvide, J.; Force, T.; Pombo, C.M. A Novel Cardioprotective P38-MAPK/MTOR Pathway. Exp. Cell Res. 2011, 317, 2938–2949. [Google Scholar] [CrossRef]
- Cuaz-Pérolin, C.; Furman, C.; Larigauderie, G.; Legedz, L.; Lasselin, C.; Copin, C.; Jaye, M.; Searfoss, G.; Yu, K.T.; Duverger, N.; et al. REDD2 Gene Is Upregulated by Modified LDL or Hypoxia and Mediates Human Macrophage Cell Death. Arterioscler. Thromb. Vasc. Biol. 2004, 24, 1830–1835. [Google Scholar] [CrossRef]
- Simonson, B.; Subramanya, V.; Chan, M.C.; Zhang, A.; Franchino, H.; Ottaviano, F.; Mishra, M.K.; Knight, A.C.; Hunt, D.; Ghiran, I.; et al. DDiT4L Promotes Autophagy and Inhibits Pathological Cardiac Hypertrophy in Response to Stress. Sci. Signal. 2017, 10, eaaf5967. [Google Scholar] [CrossRef] [PubMed]
- Fearnley, C.J.; Roderick, H.L.; Bootman, M.D. Calcium Signaling in Cardiac Myocytes. Cold Spring Harb. Perspect. Biol. 2011, 3, a004242. [Google Scholar] [CrossRef] [PubMed]
- Buhl, A.M.; Pleiman, C.M.; Rickert, R.C.; Cambier, J.C. Qualitative Regulation of B Cell Antigen Receptor Signaling by CD19: Selective Requirement for PI3-Kinase Activation, Inositol-1,4,5-Trisphosphate Production and Ca2+ Mobilization. J. Exp. Med. 1997, 186, 1897–1910. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, B.; Chen, W.-D.; Wang, Y.-D. Role of G-Protein Coupled Receptors in Cardiovascular Diseases. Front. Cardiovasc. Med. 2023, 10, 1130312. [Google Scholar] [CrossRef]
- Xu, Y.; Fairfax, K.; Light, A.; Huntington, N.D.; Tarlinton, D.M. CD19 Differentially Regulates BCR Signalling through the Recruitment of PI3K. Autoimmunity 2014, 47, 430–437. [Google Scholar] [CrossRef] [PubMed]
- Adamo, L.; Rocha-Resende, C.; Lin, C.-Y.; Evans, S.; Williams, J.; Dun, H.; Li, W.; Mpoy, C.; Andhey, P.S.; Rogers, B.E.; et al. Myocardial B Cells Are a Subset of Circulating Lymphocytes with Delayed Transit through the Heart. JCI Insight 2020, 5, e134700. [Google Scholar] [CrossRef]
- Gorski, P.A.; Ceholski, D.K.; Hajjar, R.J. Altered Myocardial Calcium Cycling and Energetics in Heart Failure—A Rational Approach for Disease Treatment. Cell Metab. 2015, 21, 183–194. [Google Scholar] [CrossRef] [PubMed]




| Total (n = 38) | No PGD (n = 25) | PGD (n = 13) | p | |
|---|---|---|---|---|
| Female, n (%) | 9 (23.7%) | 7 (28%) | 2 (15.4%) | 0.46 |
| Sex mismatch, n (%) | 2 (5.3%) | 2 (8%) | 0 (0%) | 0.54 |
| Age, median, y (range) | 59 (27–67) | 60 (27–67) | 58 (46–66) | 0.46 |
| Weight, median, kg (range) | 82 (41–120) | 84 (41–112) | 77 (62–120) | 0.58 |
| BMI, median, kg/m2 (range) | 27.5 (15.6–36.3) | 27.6 (15.6–36.3) | 25.7 (21.5–36.2) | 0.66 |
| Panel reactive antibodies (PRA) | ||||
| PRA I, % (range) | 0 (0–60) | 7 (0–60) | 0 (0–20) | 0.14 |
| PRA II, % (range) | 0 (0) | 0 (0) | 0 (0) | 0.34 |
| Original diagnosis, % of known | 0.16 | |||
| Amyloidosis | 1 (2.6%) | 0 (0%) | 1 (7.7%) | |
| Dilated cardiomyopathy | 19 (50%) | 13 (52%) | 6 (46.2%) | |
| Hypertrophic cardiomyopathy | 0 (0%) | 0 (0%) | 0 (0%) | |
| Ischemic cardiomyopathy | 2 (5.3%) | 0 (0%) | 2 (15.4%) | |
| Congenital | 1 (2.6%) | 1 (4%) | 0 (0%) | |
| End-stage coronary disease | 8 (21.1%) | 7 (28%) | 1 (7.7%) | |
| Myocarditis | 3 (7.9%) | 2 (8%) | 1 (7.7%) | |
| Sarcoidosis | 1 (2.6%) | 1 (4%) | 0 (0%) | |
| Other | 3 (7.9%) | 1 (4%) | 2 (15.4%) | |
| Chronic illnesses, % of known | ||||
| Hypertension | 8 (23.5%) | 6 (26.1%) | 2 (18.2%) | 1.00 |
| Diabetes mellitus (I or II) | 8 (26.7%) | 6 (30%) | 2 (20%) | 0.68 |
| Coronary artery disease | 10 (29.4%) | 9 (39.1%) | 1 (9.1%) | 0.11 |
| Peripheral vascular disease | 0 (0%) | 0 (0%) | 0 (0%) | 1.00 |
| Malignancy | 2 (5.3%) | 2 (8.7%) | 0 (0%) | 0.53 |
| Prior stroke | 7 (20.6%) | 6 (26.1%) | 1 (9.1%) | 0.38 |
| Prior heart surgery | 8 (21.6%) | 5 (20%) | 3 (25%) | 1.00 |
| Prior sternotomy | 10 (29.4%) | 6 (26.1%) | 4 (36.4%) | 0.69 |
| Organ-specific parameters prior to HTx | ||||
| EF, % (range) | 20 (10–50) | 22 (10–50) | 20 (15–50) | 0.89 |
| PVR, Wood (range) | 2 (1–6) | 3 (1–6) | 2 (1–4) | 0.06 |
| TPG, mmHg (range) | 8 (2–17) | 10 (2–17) | 7 (3–11) | 0.12 |
| sPAP, mmHg (range) | 39 (22–62) | 43 (22–62) | 37 (27–62) | 0.39 |
| dPAP, mmHg (range) | 21 (8–39) | 21 (8–39) | 18 (9–33) | 0.45 |
| mPAP, mmHg (range) | 28 (13–43) | 28 (13–43) | 26 (15–41) | 0.78 |
| Absolute FEV1, L/min (range) | 2 (1–3) | 2 (1–3) | 2 (2–3) | 0.30 |
| Relative FEV1, % (range) | 65% (14–96) | 65% (14–96) | 65% (55–85) | 0.56 |
| P-Bilirubin, µmol/L (range) | 12 (5–46) | 11 (5–46) | 13 (6–40) | 0.98 |
| Pre-op GFR, mL/min/1.73 m2 (range) | 51 (26–128) | 48 (26–120) | 51 (34–128) | 0.71 |
| Smoking, n (%) | 0.44 | |||
| Current | 1 (2.6%) | 0 (0%) | 1 (7.7%) | |
| Former | 13 (34.2%) | 8 (34.8%) | 5 (41.7%) | |
| Never | 22 (62.9%) | 15 (65.2%) | 7 (58.3%) | |
| History unknown; only current known | 3 (7.9%) | 2 (8%) | 1 (7.7%) | |
| Preoperative VAD, n (%) | 0.40 | |||
| Continuous flow | 3 (7.9%) | 3 (12%) | 0 (0%) | |
| Pulsatile flow | 3 (7.9%) | 1 (4%) | 2 (15.4%) | |
| Preoperative ECMO, n (%) | 4 (10.5%) | 3 (13.6%) | 1 (8.3%) | 1.00 |
| Time on organ waiting list, median days (range) | 117 (1–1020) | 120 (1–840) | 60 (2–1020) | 0.77 |
| Graft ischemia, median min (range) | ||||
| Cold | 105 (7–165) | 110 (7–165) | 73 (9–149) | 0.54 |
| Warm | 77 (30–120) | 78 (30–117) | 75 (40–120) | 0.65 |
| Total | 160 (58–265) | 170 (58–238) | 156 (80–265) | 0.94 |
| Perfusion support | 80 (40–186) | 80 (40–125) | 80 (45–186) | 0.28 |
| Nitric oxide | 22 (57.9%) | 11 (44%) | 11 (84.6%) | 0.004 |
| RBC transfusions during Tx, units | 4 (0–17) | 3 (0–10) | 4 (1–17) | 0.07 |
| Recipient EBV | 34 (94.4%) | 22 (91.7%) | 12 (100%) | 0.54 |
| Recipient CMV | 31 (83.8%) | 22 (88%) | 9 (75%) | 0.37 |
| CMV prophylaxis | 35 (94.6%) | 22 (91.7%) | 13 (100%) | 0.53 |
| Primary graft dysfunction, n (%) | ||||
| No PGD | 25 (65.8%) | 25 (100%) | 0 (0%) | |
| Any PGD | 13 (34.2%) | 0 (0%) | 13 (100%) | |
| Mild PGD | 2 (5.3%) | 0 (0%) | 2 (15.4%) | |
| Moderate PGD | 6 (15.8%) | 0 (0%) | 6 (46.2%) | |
| Severe PGD | 4 (10.5%) | 0 (0%) | 4 (30.8%) | |
| RV-PGD | 1 (2.6%) | 0 (0%) | 1 (7.7%) | |
| Myocardial injury marker, median (range) | ||||
| TnI at 12 h after reperfusion | 90,434 (22,757–50,000) | 60,640 (22,757–35,405) | 287,445 (28,711–50,000) | 0.008 |
| TnT at 12 h after reperfusion | 6805 (1565–48,650) | 5462 (1565–18,470) | 21,040 (3356–48,650) | 0.007 |
| CPK-MB at 12 h after reperfusion | 228 (51–600) | 156 (51–431) | 390 (95–600) | 0.001 |
| Symbol | Description | Coefficient r | p-Value |
|---|---|---|---|
| DDIT4L | DNA damage inducible transcript 4 like | 0.54 | 4.99 × 10–4 |
| PLIN2 | Perilipin 2 | 0.52 | 8.25 × 10–4 |
| GPR3 | G-protein-coupled receptor 3 | 0.50 | 1.26 × 10–3 |
| LOC101929614 | Proline-rich receptor-like protein kinase PERK10 | 0.50 | 1.43 × 10–3 |
| C1QB | Complement C1q B chain | 0.49 | 1.71 × 10–3 |
| C1QA | Complement C1q A chain | 0.49 | 1.92 × 10–3 |
| IL17C | Interleukin 17C | 0.48 | 2.44 × 10–3 |
| FJX1 | Four-jointed box kinase 1 | 0.47 | 2.93 × 10–3 |
| CYP4A11 | Cytochrome P450 family 4 subfamily A member 11 | 0.47 | 2.94 × 10–3 |
| TDG | Thymine DNA glycosylase | 0.46 | 3.42 × 10–3 |
| ITPKA | Inositol-trisphosphate 3-kinase A | 0.46 | 3.65 × 10–3 |
| TCF15 | Transcription factor 15 | 0.46 | 3.94 × 10–3 |
| CD59 | CD59 molecule (CD59 blood group) | 0.45 | 4.24 × 10–3 |
| STC2 | Stanniocalcin 2 | 0.45 | 4.54 × 10–3 |
| C1QC | Complement C1q C chain | 0.45 | 4.96 × 10–3 |
| VSIG2 | V-set and immunoglobulin domain containing 2 | –0.45 | 4.98 × 10–3 |
| PRR27 | Proline-rich 27 | –0.45 | 4.70 × 10–3 |
| KDM4B | Lysine demethylase 4B | –0.45 | 4.64 × 10–3 |
| EGFL8 | EGF-like domain multiple 8 | –0.45 | 4.56 × 10–3 |
| AKR7A2 | Aldo-keto reductase family 7 member A2 | –0.45 | 4.53 × 10–3 |
| ZGLP1 | Zinc finger GATA-like protein 1 | –0.45 | 4.36 × 10–3 |
| MAP2K7 | Mitogen-activated protein kinase kinase 7 | –0.45 | 4.33 × 10–3 |
| CLEC19A | C-type lectin domain containing 19A | –0.45 | 4.21 × 10–3 |
| HOMER2 | Homer scaffold protein 2 | –0.46 | 4.09 × 10–3 |
| MRM1 | Mitochondrial rRNA methyltransferase 1 | –0.46 | 3.75 × 10–3 |
| PVALB | Parvalbumin | –0.46 | 3.69 × 10–3 |
| FBXO40 | F-box protein 40 | –0.46 | 3.47 × 10–3 |
| SMIM5 | Small integral membrane protein 5 | –0.46 | 3.44 × 10–3 |
| GFI1B | Growth-factor-independent 1B transcriptional repressor | –0.47 | 3.24 × 10–3 |
| GOLGA8G | Golgin A8 family member G | –0.47 | 3.03 × 10–3 |
| CEP19 | Centrosomal protein 19 | –0.48 | 2.47 × 10–3 |
| SELENBP1 | Selenium-binding protein 1 | –0.48 | 2.32 × 10–3 |
| ARHGEF40 | Rho guanine nucleotide exchange factor 40 | –0.48 | 2.28 × 10–3 |
| TMEM200B | Transmembrane protein 200B | –0.48 | 2.25 × 10–3 |
| ATG9A | Autophagy-related 9A | –0.48 | 2.23 × 10–3 |
| CYP11A1 | Cytochrome P450 family 11 subfamily A member 1 | –0.48 | 2.11 × 10–3 |
| GRAP2 | GRB2-related adaptor protein 2 | –0.49 | 1.99 × 10–3 |
| MAP6 | Microtubule-associated protein 6 | –0.49 | 1.85 × 10–3 |
| CENPK | Centromere protein K | –0.49 | 1.60 × 10–3 |
| MOV10 | Mov10 RISC complex RNA helicase | –0.50 | 1.34 × 10–3 |
| DFFB | DNA fragmentation factor subunit β | –0.50 | 1.32 × 10–3 |
| TEF | TEF transcription factor, PAR bZIP family member | –0.51 | 1.13 × 10–3 |
| CNN1 | Calponin 1 | –0.52 | 7.64 × 10–4 |
| C12orf75 | Chromosome 12 open reading frame 75 | –0.53 | 6.80 × 10–4 |
| PHOSPHO1 | Phosphoethanolamine/phosphocholine phosphatase 1 | –0.53 | 6.34 × 10–4 |
| LTO1 | LTO1 maturation factor of ABCE1 | –0.56 | 2.55 × 10–4 |
| RECQL5 | RecQ like helicase 5 | –0.56 | 2.43 × 10–4 |
| CD19 | CD19 molecule | –0.58 | 1.34 × 10–4 |
| Symbol | Description | p-Value |
|---|---|---|
| SULT2A1 | Sulfotransferase family 2A member 1 | 8.05 × 10–5 |
| ZFTA | Zinc finger translocation associated | 1.15 × 10–4 |
| ITIH3 | Inter-α-trypsin inhibitor heavy chain 3 | 1.94 × 10–4 |
| ATAT1 | α tubulin acetyltransferase 1 | 2.82 × 10–4 |
| LOC124900286 | Putative uncharacterized protein DIP2C-AS1 | 4.01 × 10–4 |
| SLC38A3 | Solute carrier family 38 member 3 | 4.02 × 10–4 |
| AHSG | α 2-HS glycoprotein | 4.16 × 10–4 |
| HAO1 | Hydroxyacid oxidase 1 | 4.25 × 10–4 |
| NPBWR1 | Neuropeptides B and W receptor 1 | 4.37 × 10–4 |
| OOEP | Oocyte-expressed protein | 5.00 × 10–4 |
| LOC102724250 | Neuroblastoma breakpoint family member 1-like | 5.06 × 10–4 |
| APOH | Apolipoprotein H | 5.07 × 10–4 |
| KRT17 | Keratin 17 | 5.68 × 10–4 |
| CYP4A11 | Cytochrome P450 family 4 subfamily A member 11 | 6.55 × 10–4 |
| MT1H | Metallothionein 1H | 6.59 × 10–4 |
| FGFR1 | Fibroblast growth factor receptor 1 | 6.66 × 10–4 |
| TPSD1 | Tryptase delta 1 | 6.96 × 10–4 |
| SLC39A14 | Solute carrier family 39 member 14 | 7.12 × 10–4 |
| TAP2 | Transporter 2, ATP-binding cassette subfamily B member | 7.60 × 10–4 |
| CEP295NL | CEP295 N-terminal like | 7.60 × 10–4 |
| CPB2 | Carboxypeptidase B2 | 7.79 × 10–4 |
| CYP3A7 | Cytochrome P450 family 3 subfamily A member 7 | 7.92 × 10–4 |
| PRR15 | Proline-rich 15 | 8.70 × 10–4 |
| UCP1 | Uncoupling protein 1 | 9.73 × 10–4 |
| HOXD3 | Homeobox D3 | 9.81 × 10–4 |
| ITIH1 | Inter-α-trypsin inhibitor heavy chain 1 | 9.86 × 10–4 |
| RIPOR2 | RHO family-interacting cell polarization regulator 2 | 9.86 × 10–4 |
| NBR1 | NBR1 autophagy cargo receptor | 9.86 × 10–4 |
| PTPRU | Protein tyrosine phosphatase receptor type U | 9.93 × 10–4 |
| PAH | Phenylalanine hydroxylase | 9.93 × 10–4 |
| SERPIND1 | Serpin family D member 1 | 1.03 × 10–3 |
| IGFL4 | IGF-like family member 4 | 1.09 × 10–3 |
| TMEM150B | Transmembrane protein 150B | 1.10 × 10–3 |
| TRIM69 | Tripartite motif containing 69 | 1.12 × 10–3 |
| CSK | C-terminal Src kinase | 1.12 × 10–3 |
| YJU2 | YJU2 splicing factor homolog | 1.12 × 10–3 |
| LRCOL1 | Leucine-rich colipase like 1 | 1.15 × 10–3 |
| PRG3 | Proteoglycan 3, pro eosinophil major basic protein 2 | 1.23 × 10–3 |
| C2 | Complement C2 | 1.27 × 10–3 |
| SEPTIN1 | Septin 1 | 1.27 × 10–3 |
| PRSS37 | Serine protease 37 | 1.33 × 10–3 |
| UGT2A3 | UDP glucuronosyltransferase family 2 member A3 | 1.33 × 10–3 |
| BTG2 | BTG anti-proliferation factor 2 | 1.44 × 10–3 |
| HAMP | Hepcidin antimicrobial peptide | 1.51 × 10–3 |
| CYP2A6 | Cytochrome P450 family 2 subfamily A member 6 | 1.52 × 10–3 |
| GPR135 | G-protein-coupled receptor 135 | 1.53 × 10–3 |
| FGF23 | Fibroblast growth factor 23 | 1.61 × 10–3 |
| PRSS22 | Serine protease 22 | 1.62 × 10–3 |
| TXNIP | Thioredoxin-interacting protein | 1.62 × 10–3 |
| TNK2 | Tyrosine kinase non receptor 2 | 1.62 × 10–3 |
| ZNG1C | Zn-regulated GTPase metalloprotein activator 1C | 1.62 × 10–3 |
| DDX43 | DEAD-box helicase 43 | 1.70 × 10–3 |
| LGI2 | Leucine-rich repeat LGI family member 2 | 1.80 × 10–3 |
| TNFRSF21 | TNF receptor superfamily member 21 | 1.83 × 10–3 |
| ADAR | Adenosine deaminase RNA specific | 1.83 × 10–3 |
| MATN1 | Matrilin 1 | 1.83 × 10–3 |
| IKZF3 | IKAROS family zinc finger 3 | 1.83 × 10–3 |
| CSF2RB | Colony-stimulating factor 2 receptor subunit β | 1.83 × 10–3 |
| UGT1A7 | UDP glucuronosyltransferase family 1 member A7 | 1.84 × 10–3 |
| STRC | Stereocilin | 1.88 × 10–3 |
| MT1G | Metallothionein 1G | 1.88 × 10–3 |
| CPA5 | Carboxypeptidase A5 | 1.97 × 10–3 |
| DCSTAMP | Dendrocyte-expressed seven transmembrane protein | 2.03 × 10–3 |
| HJV | Hemojuvelin BMP co-receptor | 2.05 × 10–3 |
| PLCG2 | Phospholipase C γ 2 | 2.06 × 10–3 |
| GUCY2D | Guanylate cyclase 2D, retinal | 2.06 × 10–3 |
| DACT1 | Dishevelled-binding antagonist of β catenin 1 | 2.09 × 10–3 |
| SLC10A1 | Solute carrier family 10 member 1 | 2.16 × 10–3 |
| FGF17 | Fibroblast growth factor 17 | 2.20 × 10–3 |
| SAA4 | Serum amyloid A4, constitutive | 2.22 × 10–3 |
| SLC23A1 | Solute carrier family 23 member 1 | 2.27 × 10–3 |
| HERC4 | HECT and RLD domain containing E3 ubiquitin protein ligase 4 | 2.32 × 10–3 |
| OR8K1 | Olfactory receptor family 8 subfamily K member 1 | 2.39 × 10–3 |
| CHRM4 | Cholinergic receptor muscarinic 4 | 2.47 × 10–3 |
| RAB7B | RAB7B, member RAS oncogene family | 2.54 × 10–3 |
| KCNS2 | Potassium voltage-gated channel modifier subfamily S member 2 | 2.54 × 10–3 |
| CYP3A4 | Cytochrome P450 family 3 subfamily A member 4 | 2.55 × 10–3 |
| BANK1 | B-cell scaffold protein with ankyrin repeats 1 | 2.56 × 10–3 |
| CEL | Carboxyl ester lipase | 2.56 × 10–3 |
| CRYBG2 | Crystallin β-γ domain containing 2 | 2.61 × 10–3 |
| IRF1 | Interferon regulatory factor 1 | 2.61 × 10–3 |
| HP | Haptoglobin | 2.61 × 10–3 |
| TMEM221 | Transmembrane protein 221 | 2.66 × 10–3 |
| EPPIN-WFDC6 | EPPIN-WFDC6 readthrough | 2.67 × 10–3 |
| BAAT | Bile acid-CoA:amino acid N-acyltransferase | 2.76 × 10–3 |
| HS3ST3A1 | Heparan sulfate-glucosamine 3-sulfotransferase 3A1 | 2.79 × 10–3 |
| IDUA | α-L-iduronidase | 2.81 × 10–3 |
| APOA1 | Apolipoprotein A1 | 2.82 × 10–3 |
| KIAA1522 | KIAA1522 | 2.92 × 10–3 |
| DTX3L | Deltex E3 ubiquitin ligase 3L | 2.92 × 10–3 |
| ORC3 | Origin recognition complex subunit 3 | 2.92 × 10–3 |
| STX17 | Syntaxin 17 | 2.92 × 10–3 |
| ANKRD13A | Ankyrin repeat domain 13A | 2.92 × 10–3 |
| ZSWIM4 | Zinc finger SWIM-type containing 4 | 2.92 × 10–3 |
| ITPKA | Inositol-trisphosphate 3-kinase A | 2.97 × 10–3 |
| AKR7A3 | Aldo-keto reductase family 7 member A3 | 2.98 × 10–3 |
| SALL1 | Spalt-like transcription factor 1 | 3.09 × 10–3 |
| PCDHAC1 | Protocadherin α subfamily C, 1 | 3.16 × 10–3 |
| C9 | Complement C9 | 3.16 × 10–3 |
| RGS18 | Regulator of G protein signaling 18 | 3.27 × 10–3 |
| DIAPH1 | Diaphanous-related formin 1 | 3.27 × 10–3 |
| IGBP1 | Immunoglobulin-binding protein 1 | 3.27 × 10–3 |
| TRIM22 | Tripartite motif containing 22 | 3.27 × 10–3 |
| L3MBTL1 | L3MBTL histone methyl-lysine binding protein 1 | 3.27 × 10–3 |
| ALX4 | ALX homeobox 4 | 3.37 × 10–3 |
| CXCL14 | C-X-C motif chemokine ligand 14 | 3.40 × 10–3 |
| IFNL3 | Interferon lambda 3 | 3.42 × 10–3 |
| SH2D1B | SH2 domain containing 1B | 3.46 × 10–3 |
| LRRC56 | Leucine-rich repeat containing 56 | 3.46 × 10–3 |
| EHD1 | EH domain containing 1 | 3.65 × 10–3 |
| SMARCC2 | SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin subfamily c member 2 | 3.65 × 10–3 |
| RDH8 | Retinol dehydrogenase 8 | 3.72 × 10–3 |
| ABCA8 | ATP-binding cassette subfamily A member 8 | 3.79 × 10–3 |
| UNC93A | unc-93 homolog A | 3.82 × 10–3 |
| USP21 | Ubiquitin-specific peptidase 21 | 3.82 × 10–3 |
| PCDH11Y | Protocadherin 11 Y-linked | 3.99 × 10–3 |
| HOXB9 | Homeobox B9 | 4.07 × 10–3 |
| ZMYM6 | Zinc finger MYM-type containing 6 | 4.08 × 10–3 |
| POLR1A | RNA polymerase I subunit A | 4.08 × 10–3 |
| RETREG1 | Reticulophagy regulator 1 | 4.08 × 10–3 |
| AKAP17A | A-kinase-anchoring protein 17A | 4.08 × 10–3 |
| PDCD4 | Programmed cell death 4 | 4.08 × 10–3 |
| DRC3 | Dynein-regulatory complex subunit 3 | 4.08 × 10–3 |
| SPOP | Speckle-type BTB/POZ protein | 4.08 × 10–3 |
| PACSIN2 | Protein kinase C and casein kinase substrate in neurons 2 | 4.08 × 10–3 |
| KCNIP4 | Potassium voltage-gated channel interacting protein 4 | 4.14 × 10–3 |
| TMEM132C | Transmembrane protein 132C | 4.16 × 10–3 |
| C15orf48 | Chromosome 15 open reading frame 48 | 4.17 × 10–3 |
| BTBD19 | BTB domain containing 19 | 4.20 × 10–3 |
| ADAM20 | ADAM metallopeptidase domain 20 | 4.20 × 10–3 |
| C8G | Complement C8 γ chain | 4.25 × 10–3 |
| KRTAP10-8 | Keratin-associated protein 10-8 | 4.27 × 10–3 |
| ADAMTS15 | ADAM metallopeptidase with thrombospondin type 1 motif 15 | 4.35 × 10–3 |
| KRT71 | Keratin 71 | 4.38 × 10–3 |
| TTR | Transthyretin | 4.40 × 10–3 |
| DDX60L | DExD/H-box 60 like | 4.54 × 10–3 |
| CD19 | CD19 molecule | 4.54 × 10–3 |
| DPEP2 | Dipeptidase 2 | 4.54 × 10–3 |
| SOCS7 | Suppressor of cytokine signaling 7 | 4.54 × 10–3 |
| KRT83 | Keratin 83 | 4.59 × 10–3 |
| TGM3 | Transglutaminase 3 | 4.62 × 10–3 |
| ENTPD8 | Ectonucleoside triphosphate diphosphohydrolase 8 | 4.73 × 10–3 |
| TNP2 | Transition protein 2 | 4.84 × 10–3 |
| DDIT4L | DNA damage inducible transcript 4 like | 4.91 × 10–3 |
| F11 | Coagulation factor XI | 4.98 × 10–3 |
| LMX1B | LIM homeobox transcription factor 1 β | 4.98 × 10–3 |
| Symbol | Biological Relevance | TnI R | TnI p-Value | PGD p-Value |
|---|---|---|---|---|
| ITPKA | Calmodulin-binding, calcium signaling | 0.46 | 3.65 × 10–3 | 2.97 × 10–3 |
| Inositol phosphate metabolism | ||||
| Phosphatidylinositol signaling system | ||||
| DDIT4L | Negative regulation of signal transduction | 0.54 | 4.99 × 10–4 | 4.91 × 10–3 |
| Inhibits cell growth via the TOR signaling pathway and downstream of AKT1 | ||||
| CD19 | Normal B-cell differentiation | –0.58 | 1.34 × 10–4 | 4.54 × 10–3 |
| Activation of PI3K and the mobilization of intracellular Ca2+ stores | ||||
| Positive regulation of calcium ion transmembrane transport via BCR signaling pathway | ||||
| CYP4A11 | Metabolism of drugs, fatty acids, arachidonic acids | 0.47 | 2.94 × 10–3 | 6.55 × 10–4 |
| Biosynthesis of cholesterol, steroids, and other lipids PPAR signaling pathway, atherosclerosis |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Joo, S.; Dhaygude, K.; Westerberg, S.; Krebs, R.; Puhka, M.; Holmström, E.; Syrjälä, S.; Nykänen, A.I.; Lemström, K. Transcriptomic Landscape of Circulating Extracellular Vesicles in Heart Transplant Ischemia–Reperfusion. Genes 2023, 14, 2101. https://doi.org/10.3390/genes14112101
Joo S, Dhaygude K, Westerberg S, Krebs R, Puhka M, Holmström E, Syrjälä S, Nykänen AI, Lemström K. Transcriptomic Landscape of Circulating Extracellular Vesicles in Heart Transplant Ischemia–Reperfusion. Genes. 2023; 14(11):2101. https://doi.org/10.3390/genes14112101
Chicago/Turabian StyleJoo, SeoJeong, Kishor Dhaygude, Sofie Westerberg, Rainer Krebs, Maija Puhka, Emil Holmström, Simo Syrjälä, Antti I. Nykänen, and Karl Lemström. 2023. "Transcriptomic Landscape of Circulating Extracellular Vesicles in Heart Transplant Ischemia–Reperfusion" Genes 14, no. 11: 2101. https://doi.org/10.3390/genes14112101
APA StyleJoo, S., Dhaygude, K., Westerberg, S., Krebs, R., Puhka, M., Holmström, E., Syrjälä, S., Nykänen, A. I., & Lemström, K. (2023). Transcriptomic Landscape of Circulating Extracellular Vesicles in Heart Transplant Ischemia–Reperfusion. Genes, 14(11), 2101. https://doi.org/10.3390/genes14112101






