Small Molecule 20S Proteasome Enhancer Regulates MYC Protein Stability and Exhibits Antitumor Activity in Multiple Myeloma
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Primary Cell Culture and Cell Viability
2.3. Proteasome Activity in Purified Protein Assay
2.4. MYC Degradation in MM Cells
2.5. MYC-Luc Reporter Assay
2.6. Gene Expression Profiling of RPMI-8226 Cells
2.7. ROSALIND® NanoString Gene Expression and Pathway Analysis
2.8. Assessment of Apoptosis
2.9. Pharmacokinetic Assessment of TCH-165
2.10. Pharmacokinetic Assessment and Tolerance in Dogs
2.11. Proteasome Activity in Canine PBMC Lysate
2.12. RPMI-8226 Xenograft Tumor Model
3. Results
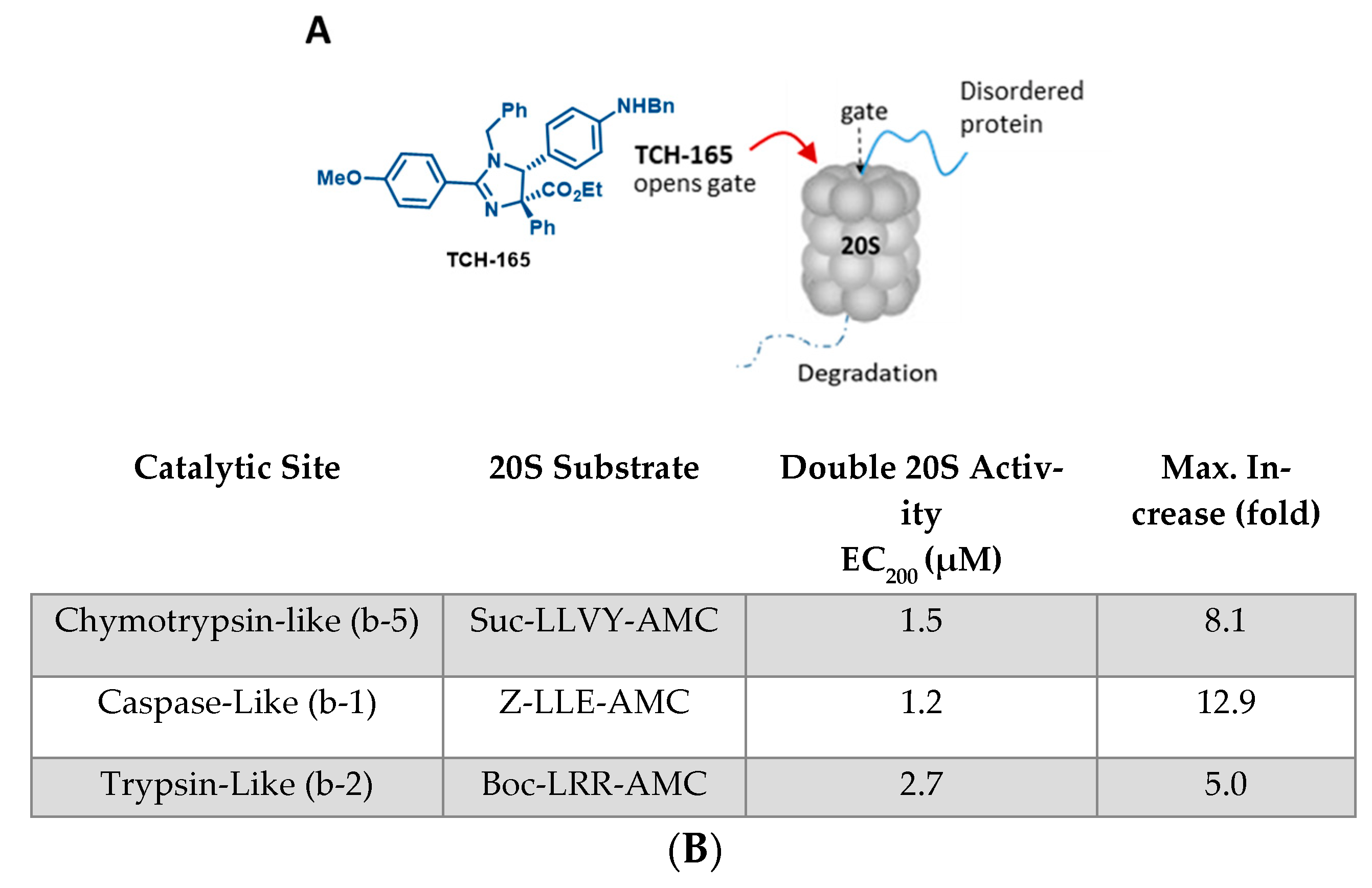
3.1. TCH-165 Enhances the Proteolytic Activity of the 20S Proteasome
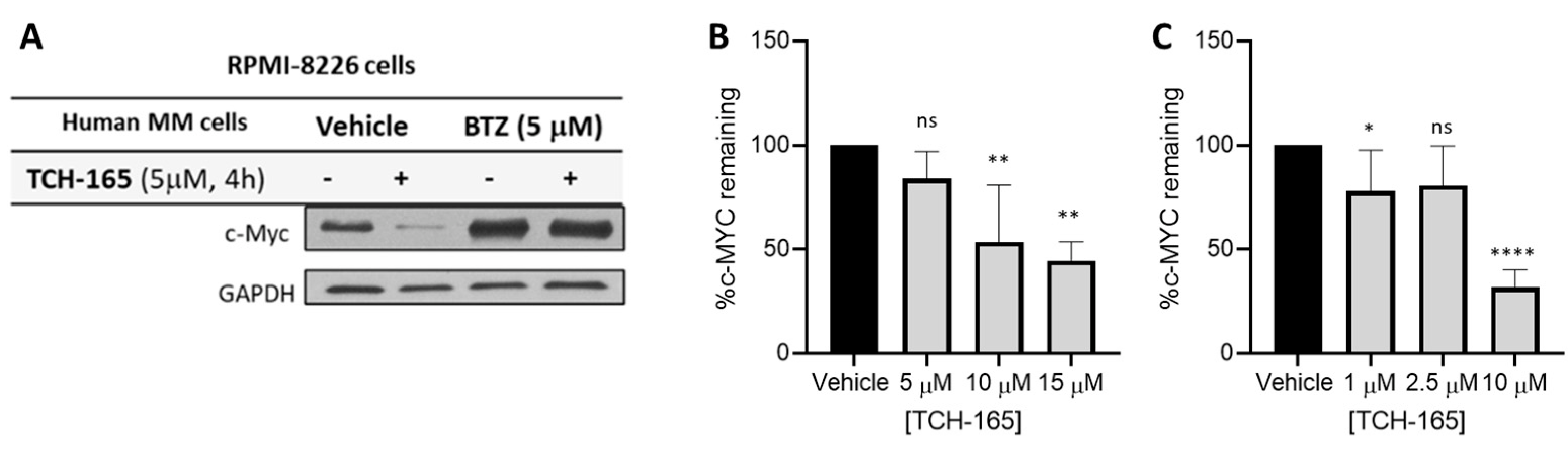
3.2. 20S Proteasome Activation by TCH-165 Regulates MYC Degradation
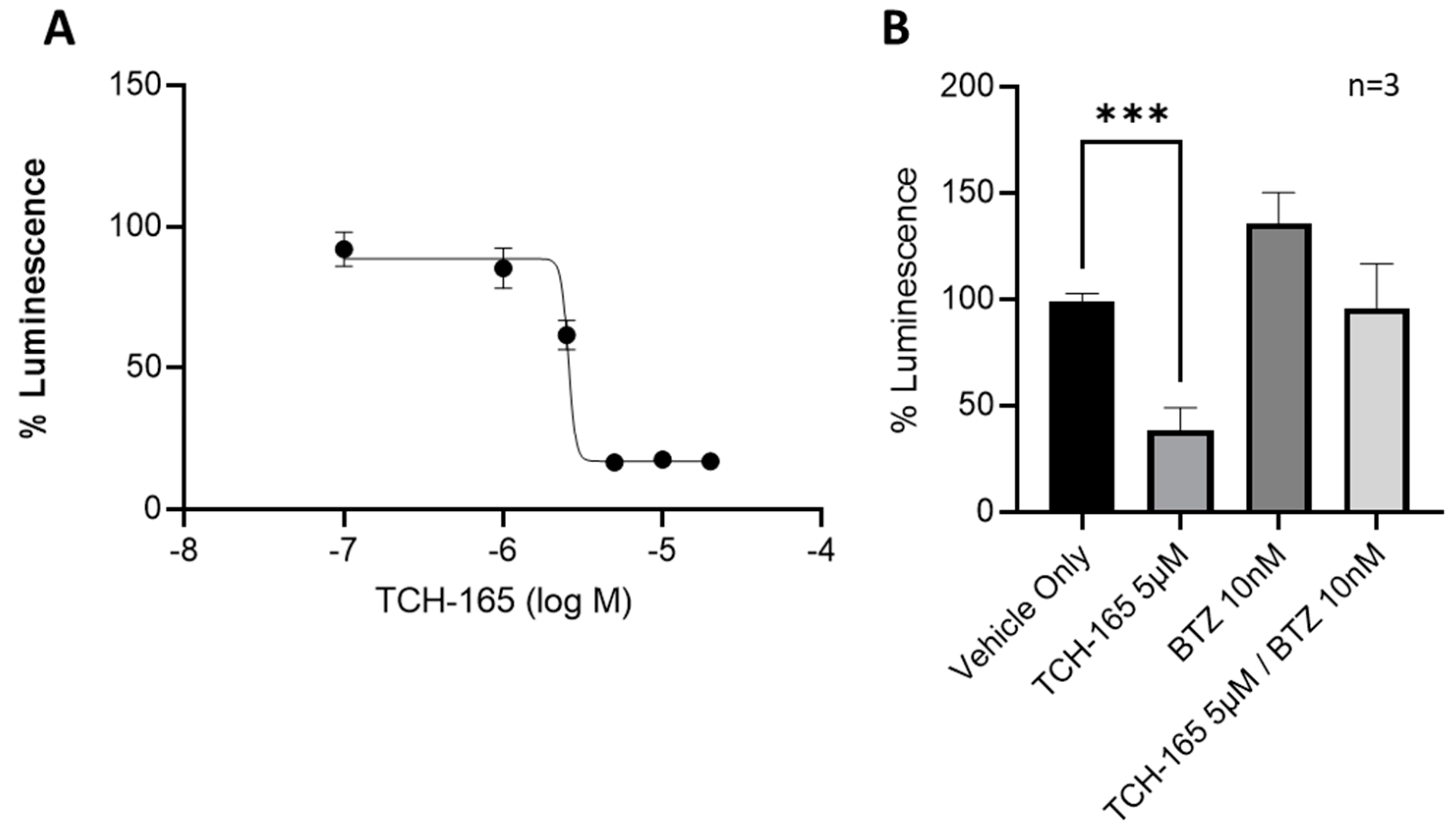
3.3. 20S Proteasome Activation by TCH-165 Regulates MYC-Mediated Gene Transcription
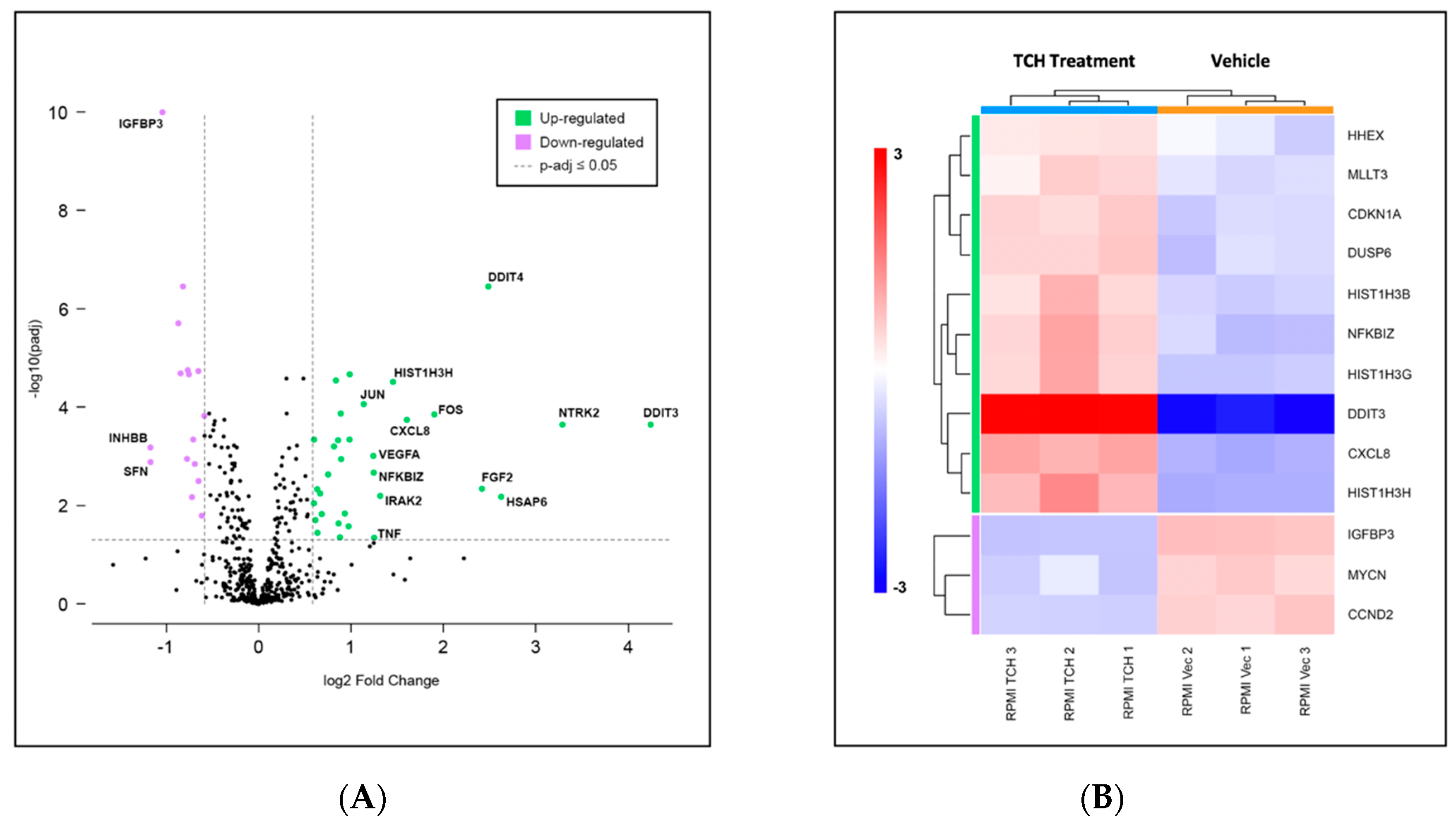
3.4. Gene Expression Profiling Indicates Limited Changes in Gene Expression following TCH-165 Treatment
3.5. Various Multiple Myeloma Cancer Cell Lines, and Patient Derived Primary and Refractory Multiple Myeloma Cells Are Vulnerable to the Proteasome Activator, TCH-165
3.6. Pharmacokinetic Properties and Anti-Tumor Efficacy of 20S Enhancer in In Vivo Xenograft
3.7. In Vivo Tolerance and Target Engagement of the 20S Proteasome Enhancer, TCH-165
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Podar, K.; Leleu, X. Relapsed/Refractory Multiple Myeloma in 2020/2021 and Beyond. Cancers 2021, 13, 5154. [Google Scholar] [CrossRef] [PubMed]
- Schick, M.; Habringer, S.; Nilsson, J.A.; Keller, U. Pathogenesis and therapeutic targeting of aberrant MYC expression in haematological cancers. Br. J. Haematol. 2017, 179, 724–738. [Google Scholar] [CrossRef] [PubMed]
- Dang, C.V. MYC on the path to cancer. Cell 2012, 149, 22–35. [Google Scholar] [CrossRef] [PubMed]
- Farrell, A.S.; Sears, R.C. MYC degradation. Cold Spring Harb. Perspect. Med. 2014, 4, a014365. [Google Scholar] [CrossRef] [PubMed]
- Kalkat, M.; De Melo, J.; Hickman, K.A.; Lourenco, C.; Redel, C.; Resetca, D.; Tamachi, A.; Tu, W.B.; Penn, L.Z. MYC Deregulation in Primary Human Cancers. Genes 2017, 8, 151. [Google Scholar] [CrossRef] [PubMed]
- Dang, C.V.; Reddy, E.P.; Shokat, K.M.; Soucek, L. Drugging the ‘undruggable’ cancer targets. Nat. Rev. Cancer 2017, 17, 502–508. [Google Scholar] [CrossRef]
- Delmore, J.E.; Issa, G.C.; Lemieux, M.E.; Rahl, P.B.; Shi, J.; Jacobs, H.M.; Kastritis, E.; Gilpatrick, T.; Paranal, R.M.; Qi, J.; et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 2011, 146, 904–917. [Google Scholar] [CrossRef]
- Amorim, S.; Stathis, A.; Gleeson, M.; Iyengar, S.; Magarotto, V.; Leleu, X.; Morschhauser, F.; Karlin, L.; Broussais, F.; Rezai, K.; et al. Bromodomain inhibitor OTX015 in patients with lymphoma or multiple myeloma: A dose-escalation, open-label, pharmacokinetic, phase 1 study. Lancet Haematol. 2016, 3, e196–e204. [Google Scholar] [CrossRef]
- Albrecht, B.K.; Gehling, V.S.; Hewitt, M.C.; Vaswani, R.G.; Cote, A.; Leblanc, Y.; Nasveschuk, C.G.; Bellon, S.; Bergeron, L.; Campbell, R.; et al. Identification of a Benzoisoxazoloazepine Inhibitor (CPI-0610) of the Bromodomain and Extra-Terminal (BET) Family as a Candidate for Human Clinical Trials. J. Med. Chem. 2016, 59, 1330–1339. [Google Scholar] [CrossRef]
- Manier, S.; Huynh, D.; Shen, Y.J.; Zhou, J.; Yusufzai, T.; Salem, K.Z.; Ebright, R.Y.; Shi, J.; Park, J.; Glavey, S.V.; et al. Inhibiting the oncogenic translation program is an effective therapeutic strategy in multiple myeloma. Sci. Transl. Med. 2017, 9, eaal2668. [Google Scholar] [CrossRef]
- Thomas, L.R.; Tansey, W.P. Proteolytic control of the oncoprotein transcription factor Myc. Adv. Cancer Res. 2011, 110, 77–106. [Google Scholar] [CrossRef] [PubMed]
- Bard, J.A.M.; Goodall, E.A.; Greene, E.R.; Jonsson, E.; Dong, K.C.; Martin, A. Structure and Function of the 26S Proteasome. Annu. Rev. Biochem. 2018, 87, 697–724. [Google Scholar] [CrossRef] [PubMed]
- Finley, D.; Chen, X.; Walters, K.J. Gates, Channels, and Switches: Elements of the Proteasome Machine. Trends Biochem. Sci. 2016, 41, 77–93. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Jiang, C.; Pan, J.; Wang, X.; Jin, J.; Zhao, L.; Pan, W.; Liao, G.; Cai, X.; Li, X.; et al. Regulation of c-Myc protein stability by proteasome activator REGgamma. Cell Death Differ. 2015, 22, 1000–1011. [Google Scholar] [CrossRef] [PubMed]
- Tsafou, K.; Tiwari, P.B.; Forman-Kay, J.D.; Metallo, S.J.; Toretsky, J.A. Targeting Intrinsically Disordered Transcription Factors: Changing the Paradigm. J. Mol. Biol. 2018, 430, 2321–2341. [Google Scholar] [CrossRef]
- Fiolek, T.J.; Magyar, C.L.; Wall, T.J.; Davies, S.B.; Campbell, M.V.; Savich, C.J.; Tepe, J.J.; Mosey, R.A. Dihydroquinazolines enhance 20S proteasome activity and induce degradation of alpha-synuclein, an intrinsically disordered protein associated with neurodegeneration. Bioorg. Med. Chem. Lett. 2021, 36, 127821. [Google Scholar] [CrossRef]
- Fiolek, T.J.; Keel, K.L.; Tepe, J.J. Fluspirilene Analogs Activate the 20S Proteasome and Overcome Proteasome Impairment by Intrinsically Disordered Protein Oligomers. ACS Chem. Neurosci. 2021, 12, 1438–1448. [Google Scholar] [CrossRef]
- Njomen, E.; Tepe, J.J. Proteasome Activation as a New Therapeutic Approach To Target Proteotoxic Disorders. J. Med. Chem. 2019, 62, 6469–6481. [Google Scholar] [CrossRef]
- Njomen, E.; Tepe, J.J. Regulation of Autophagic Flux by the 20S Proteasome. Cell Chem. Biol. 2019, 26, 1283–1294.e1285. [Google Scholar] [CrossRef]
- Jones, C.L.; Tepe, J.J. Proteasome Activation to Combat Proteotoxicity. Molecules 2019, 24, 2481. [Google Scholar] [CrossRef]
- Njomen, E.; Osmulski, P.A.; Jones, C.L.; Gaczynska, M.; Tepe, J.J. Small Molecule Modulation of Proteasome Assembly. Biochemistry 2018, 57, 4214–4224. [Google Scholar] [CrossRef]
- Jones, C.L.; Njomen, E.; Sjogren, B.; Dexheimer, T.S.; Tepe, J.J. Small Molecule Enhancement of 20S Proteasome Activity Targets Intrinsically Disordered Proteins. ACS Chem. Biol. 2017, 12, 2240–2247. [Google Scholar] [CrossRef] [PubMed]
- Coleman, R.A.; Trader, D.J. Methods to Discover and Evaluate Proteasome Small Molecule Stimulators. Molecules 2019, 24, 2341. [Google Scholar] [CrossRef] [PubMed]
- Coleman, R.A.; Muli, C.S.; Zhao, Y.; Bhardwaj, A.; Newhouse, T.R.; Trader, D.J. Analysis of chain length, substitution patterns, and unsaturation of AM-404 derivatives as 20S proteasome stimulators. Bioorg. Med. Chem. Lett. 2019, 29, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Coleman, R.A.; Trader, D.J. All About the Core: A Therapeutic Strategy to Prevent Protein Accumulation with Proteasome Core Particle Stimulators. ACS Pharmacol. Transl. Sci. 2018, 1, 140–142. [Google Scholar] [CrossRef] [PubMed]
- Trader, D.J.; Simanski, S.; Dickson, P.; Kodadek, T. Establishment of a suite of assays that support the discovery of proteasome stimulators. Biochim. Biophys. Acta 2017, 1861, 892–899. [Google Scholar] [CrossRef]
- Lansdell, T.A.; Hurchla, M.A.; Xiang, J.; Hovde, S.; Weilbaecher, K.N.; Henry, R.W.; Tepe, J.J. Noncompetitive Modulation of the Proteasome by Imidazoline Scaffolds Overcomes Bortezomib Resistance and Delays MM Tumor Growth in Vivo. ACS Chem. Biol. 2013, 8, 578–587. [Google Scholar] [CrossRef]
- Azevedo, L.M.; Lansdell, T.A.; Ludwig, J.R.; Mosey, R.A.; Woloch, D.K.; Cogan, D.P.; Patten, G.P.; Kuszpit, M.R.; Fisk, J.S.; Tepe, J.J. Inhibition of the human proteasome by imidazoline scaffolds. J. Med. Chem. 2013, 56, 5974–5978. [Google Scholar] [CrossRef]
- Kahlon, D.K.; Lansdell, T.A.; Fisk, J.S.; Tepe, J.J. Structural-activity relationship study of highly-functionalized imidazolines as potent inhibitors of nuclear transcription factor-kappaB mediated IL-6 production. Bioorg. Med. Chem. 2009, 17, 3093–3103. [Google Scholar] [CrossRef]
- Kahlon, D.K.; Lansdell, T.A.; Fisk, J.S.; Hupp, C.D.; Friebe, T.L.; Hovde, S.; Jones, A.D.; Dyer, R.D.; Henry, R.W.; Tepe, J.J. Nuclear Factor-kappaB Mediated Inhibition of Cytokine Production by Imidazoline Scaffolds. J. Med. Chem. 2009, 52, 1302–1309. [Google Scholar] [CrossRef]
- Sharma, V.; Hupp, C.D.; Tepe, J.J. Enhancement of chemotherapeutic efficacy by small molecule inhibition of NF-kappaB and checkpoint kinases. Curr. Med. Chem. 2007, 14, 1061–1074. [Google Scholar] [CrossRef]
- Sharma, V.; Peddibhotla, S.; Tepe, J.J. Sensitization of cancer cells to DNA damaging agents by imidazolines. J. Am. Chem. Soc. 2006, 128, 9137–9143. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.; Lansdell, T.A.; Peddibhotla, S.; Tepe, J.J. Sensitization of tumor cells towards chemotherapy: Enhancing the efficacy of camptothecin by novel imidazolines. Chem. Biol. 2004, 11, 1689–1699. [Google Scholar] [CrossRef] [PubMed]
- Council, N.R. Guide for the Care and Use of Laboratory Animals: Eighth Edition, 8th ed.; The National Academies Press: Washington, DC, USA, 2011. [Google Scholar]
- Percie du Sert, N.; Hurst, V.; Ahluwalia, A.; Alam, S.; Avey, M.T.; Baker, M.; Browne, W.J.; Clark, A.; Cuthill, I.C.; Dirnagl, U.; et al. The ARRIVE guidelines 2.0: Updated guidelines for reporting animal research. PLoS Biol. 2020, 18, e3000410. [Google Scholar] [CrossRef]
- Available online: https://rosalind.bio/ (accessed on 22 October 2021).
- Perkins, J.R.; Dawes, J.M.; McMahon, S.B.; Bennett, D.L.; Orengo, C.; Kohl, M. ReadqPCR and NormqPCR: R packages for the reading, quality checking and normalisation of RT-qPCR quantification cycle (Cq) data. BMC Genomics 2012, 13, 296. [Google Scholar] [CrossRef] [PubMed]
- Hennig C: Cran-package fpc. Available online: https://cran.r-project.org/web/packages/fpc/index.html (accessed on 11 October 2021).
- Alexa, A.J.R. topGO: Enrichment Analysis for Gene Ontology. In R package version 1.38.1; R Core Team: Vienna, Austria, 2019. [Google Scholar]
- Mitchell, A.L.; Attwood, T.K.; Babbitt, P.C.; Blum, M.; Bork, P.; Bridge, A.; Brown, S.D.; Chang, H.Y.; El-Gebali, S.; Fraser, M.I.; et al. InterPro in 2019: Improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 2019, 47, D351–D360. [Google Scholar] [CrossRef]
- Geer, L.Y.; Marchler-Bauer, A.; Geer, R.C.; Han, L.; He, J.; He, S.; Liu, C.; Shi, W.; Bryant, S.H. The NCBI BioSystems database. Nucleic Acids Res. 2010, 38, D492–D496. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Liberzon, A.; Subramanian, A.; Pinchback, R.; Thorvaldsdottir, H.; Tamayo, P.; Mesirov, J.P. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011, 27, 1739–1740. [Google Scholar] [CrossRef]
- Fabregat, A.; Jupe, S.; Matthews, L.; Sidiropoulos, K.; Gillespie, M.; Garapati, P.; Haw, R.; Jassal, B.; Korninger, F.; May, B.; et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 2018, 46, D649–D655. [Google Scholar] [CrossRef]
- Slenter, D.N.; Kutmon, M.; Hanspers, K.; Riutta, A.; Windsor, J.; Nunes, N.; Melius, J.; Cirillo, E.; Coort, S.L.; Digles, D.; et al. WikiPathways: A multifaceted pathway database bridging metabolomics to other omics research. Nucleic Acids Res. 2018, 46, D661–D667. [Google Scholar] [CrossRef] [PubMed]
- Groll, M.; Heinemeyer, W.; Jager, S.; Ullrich, T.; Bochtler, M.; Wolf, D.H.; Huber, R. The catalytic sites of 20S proteasomes and their role in subunit maturation: A mutational and crystallographic study. Proc. Natl. Acad. Sci. USA 1999, 96, 10976–10983. [Google Scholar] [CrossRef]
- Ubeda, M.; Vallejo, M.; Habener, J.F. CHOP enhancement of gene transcription by interactions with Jun/Fos AP-1 complex proteins. Mol. Cell Biol. 1999, 19, 7589–7599. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Richardson, P.G.; Anderson, K.C. Bortezomib: A novel therapy approved for multiple myeloma. Clin. Adv. Hematol. Oncol. 2003, 1, 596–600. [Google Scholar]
- Andreu-Vieyra, C.; Berenson, J.R. Carfilzomib in multiple myeloma. Exp. Opin. Biol. Ther. 2014, 14, 1685–1699. [Google Scholar] [CrossRef] [PubMed]
- Shirley, M. Ixazomib: First Global Approval. Drugs 2016, 76, 405–411. [Google Scholar] [CrossRef] [PubMed]
- Arnall, J.R.; Maples, K.T.; Harvey, R.D.; Moore, D.C. Daratumumab for the Treatment of Multiple Myeloma: A Review of Clinical Applicability and Operational Considerations. Ann. Pharmacother. 2021, 10600280211058754. [Google Scholar] [CrossRef]
- Munshi, N.C.; Anderson, L.D., Jr.; Shah, N.; Madduri, D.; Berdeja, J.; Lonial, S.; Raje, N.; Lin, Y.; Siegel, D.; Oriol, A.; et al. Idecabtagene Vicleucel in Relapsed and Refractory Multiple Myeloma. N. Engl. J. Med. 2021, 384, 705–716. [Google Scholar] [CrossRef]
- Li, J.; Smith, A.; Crouch, S.; Oliver, S.; Roman, E. Estimating the prevalence of hematological malignancies and precursor conditions using data from Haematological Malignancy Research Network (HMRN). Cancer Causes Control 2016, 27, 1019–1026. [Google Scholar] [CrossRef][Green Version]
- FDA Approves First Cell-Based Gene Therapy for Adult Patients with Multiple Myeloma. Available online: https://www.fda.gov/news-events/press-announcements/fda-approves-first-cell-based-gene-therapy-adult-patients-multiple-myeloma (accessed on 13 January 2022).
- Anderson, L.D., Jr. Idecabtagene vicleucel (ide-cel) CAR T-cell therapy for relapsed and refractory multiple myeloma. Future Oncol. 2022, 18, 277–289. [Google Scholar] [CrossRef]
- Adhikary, S.; Eilers, M. Transcriptional regulation and transformation by Myc proteins. Nat. Rev. Mol. Cell. Biol. 2005, 6, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Bhowmick, P.; Pancsa, R.; Guharoy, M.; Tompa, P. Functional diversity and structural disorder in the human ubiquitination pathway. PLoS ONE 2013, 8, e65443. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.X.; Li, Y.; Sears, R.C.; Dai, M.S. Targeting the MYC Ubiquitination-Proteasome Degradation Pathway for Cancer Therapy. Front. Oncol. 2021, 11, 679445. [Google Scholar] [CrossRef] [PubMed]
- Llombart, V.; Mansour, M.R. Therapeutic targeting of “undruggable” MYC. EBioMedicine 2021, 75, 103756. [Google Scholar] [CrossRef]
- Rishi, G.; Wallace, D.F.; Subramaniam, V.N. Hepcidin: Regulation of the master iron regulator. Biosci. Rep. 2015, 35, e00192. [Google Scholar] [CrossRef]
- Vogt, A.S.; Arsiwala, T.; Mohsen, M.; Vogel, M.; Manolova, V.; Bachmann, M.F. On Iron Metabolism and Its Regulation. Int. J. Mol. Sci. 2021, 22, 4591. [Google Scholar] [CrossRef]
- Bayele, H.K.; McArdle, H.; Srai, S.K. Cis and trans regulation of hepcidin expression by upstream stimulatory factor. Blood 2006, 108, 4237–4245. [Google Scholar] [CrossRef][Green Version]
- Cazzola, M.; Huebers, H.A.; Sayers, M.H.; MacPhail, A.P.; Eng, M.; Finch, C.A. Transferrin saturation, plasma iron turnover, and transferrin uptake in normal humans. Blood 1985, 66, 935–939. [Google Scholar] [CrossRef]
- Wu, K.J.; Polack, A.; Dalla-Favera, R. Coordinated regulation of iron-controlling genes, H-ferritin and IRP2, by c-MYC. Science 1999, 283, 676–679. [Google Scholar] [CrossRef]
- Rossi, M.; Botta, C.; Arbitrio, M.; Grembiale, R.D.; Tagliaferri, P.; Tassone, P. Mouse models of multiple myeloma: Technologic platforms and perspectives. Oncotarget 2018, 9, 20119–20133. [Google Scholar] [CrossRef]

| MM Cells Treated with TCH-165 | CC50 (μM) & 95% Confidence Interval (CI) |
| RPMI-8226 | 0.9 (95% CI 0.8–1.2 μM) |
| L363 | 5.0 (95% CI 4.1 -5.1 μM) |
| NCI-H929 | 4.3 (95% CI 2.2–6.6 μM *) |
| Cells Treated with TCH-165 | CC50 (μM) |
| Primary MM patient cells | 1.0 (95% CI 0.6–1.5 μM) |
| Refractory MM patient cells | 8.1 (95% CI 7.1–9.0 μM) |
| Cells Treated with BTZ | CC50 (nM) |
| Primary MM patient cells | 4.0 (95% CI 2.3–7.1 nM) |
| Refractory MM patient cells | >1000 (95% CI N/A) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Njomen, E.; Vanecek, A.; Lansdell, T.A.; Yang, Y.-T.; Schall, P.Z.; Harris, C.M.; Bernard, M.P.; Isaac, D.; Alkharabsheh, O.; Al-Janadi, A.; et al. Small Molecule 20S Proteasome Enhancer Regulates MYC Protein Stability and Exhibits Antitumor Activity in Multiple Myeloma. Biomedicines 2022, 10, 938. https://doi.org/10.3390/biomedicines10050938
Njomen E, Vanecek A, Lansdell TA, Yang Y-T, Schall PZ, Harris CM, Bernard MP, Isaac D, Alkharabsheh O, Al-Janadi A, et al. Small Molecule 20S Proteasome Enhancer Regulates MYC Protein Stability and Exhibits Antitumor Activity in Multiple Myeloma. Biomedicines. 2022; 10(5):938. https://doi.org/10.3390/biomedicines10050938
Chicago/Turabian StyleNjomen, Evert, Allison Vanecek, Theresa A. Lansdell, Ya-Ting Yang, Peter Z. Schall, Christi M. Harris, Matthew P. Bernard, Daniel Isaac, Omar Alkharabsheh, Anas Al-Janadi, and et al. 2022. "Small Molecule 20S Proteasome Enhancer Regulates MYC Protein Stability and Exhibits Antitumor Activity in Multiple Myeloma" Biomedicines 10, no. 5: 938. https://doi.org/10.3390/biomedicines10050938
APA StyleNjomen, E., Vanecek, A., Lansdell, T. A., Yang, Y.-T., Schall, P. Z., Harris, C. M., Bernard, M. P., Isaac, D., Alkharabsheh, O., Al-Janadi, A., Giletto, M. B., Ellsworth, E., Taylor, C., Tang, T., Lau, S., Bailie, M., Bernard, J. J., Yuzbasiyan-Gurkan, V., & Tepe, J. J. (2022). Small Molecule 20S Proteasome Enhancer Regulates MYC Protein Stability and Exhibits Antitumor Activity in Multiple Myeloma. Biomedicines, 10(5), 938. https://doi.org/10.3390/biomedicines10050938






