Molecular Diagnostic Tools Applied for Assessing Microbial Water Quality
Abstract
:1. Introduction
2. Presence/Absence Examination by End-Point PCR
3. DNA Microarray
4. Quantitative Real-Time PCR (qPCR)
5. Multiplex qPCR (mqPCR)
6. Loop-Mediated Isothermal Amplification (LAMP)
7. Droplet Digital PCR (ddPCR)
8. Next-Generation Sequencing (NGS)-Based Method
9. The Surveillance of SARS-CoV-2 in Wastewater Using Different Molecular Methods—A Showcase
10. Conclusions and Future Perspectives
- End-point PCR can facilitate the “presence/absence” diagnostics of target species of interests and is superior to traditional culture-based characterization considering time and labor aspects.
- DNA microarray enables the simultaneous detection of multiple targets in a single run, offering a high degree of assay parallelism.
- Both qPCR and mqPCR can provide quantitative and more rapid and sensitive assays than conventional PCR, and are among the most applied methods for water pathogen detection.
- LAMP offers a unique isothermal amplification-based detection approach and has shown great potential for the development of novel affordable and portable biosensors for on-site water quality biomonitoring.
- The emergence of ddPCR symbolized a significant concept revolution and technology leap in molecular diagnostics. It holds great promise in realizing the precise quantification of microbial pathogens sparsely present in water, with ultra-high sensitivity, precision, and reproducibility.
- With the use of NGS-based high-throughput approaches, e.g., amplicon deep sequencing, metagenomics, meta-transcriptomics, and WGS, multiple environmental pathogens can be simultaneously characterized in depth (at the genus or species level) in various waters and wastewater. Moreover, the structure and functioning of a microbial community can be deciphered using HT-NGS, facilitating the assessment of microbial water quality and the identification of the major anthropogenic or environmental drivers responsible for ecosystem fluctuations.
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization (WHO). Drinking-Water. Available online: https://www.who.int/news-room/fact-sheets/detail/drinking-water (accessed on 11 February 2022).
- Alhamlan, F.S.; Al-Qahtani, A.A.; Al-Ahdal, M.N. Recommended advanced techniques for waterborne pathogen detection in developing countries. J. Infect. Dev. Ctries. 2015, 9, 128–135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, Q.; Huang, Y.; Wang, H.; Fang, T. Diversity and abundance of bacterial pathogens in urban rivers impacted by domestic sewage. Environ. Pollut. 2019, 249, 24–35. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wang, C.; Wang, P.; Chen, J.; Wang, X.; Yuan, Q. Variation of bacterioplankton community along an urban river impacted by touristic city: With a focus on pathogen. Ecotoxicol. Environ. Saf. 2018, 165, 573–581. [Google Scholar] [CrossRef] [PubMed]
- D’Ugo, E.; Sdanganelli, M.; Grasso, C.; Magurano, F.; Marcheggiani, S.; Boots, B.; Baggieri, M.; Mancini, L. Detection of Coxiella burnetii in Urban River Water. Vector Borne Zoonotic Dis. 2017, 17, 514–516. [Google Scholar] [CrossRef]
- Baker-Austin, C.; Oliver, J.D.; Alam, M.; Ali, A.; Waldor, M.K.; Qadri, F.; Martinez-Urtaza, J. Vibrio spp. infections. Nat. Rev. Dis. Primers 2018, 4, 1–19. [Google Scholar] [CrossRef]
- Baker-Austin, C.; Oliver, J.D. Vibrio vulnificus: New insights into a deadly opportunistic pathogen. Environ. Microbiol. 2018, 20, 423–430. [Google Scholar] [CrossRef] [Green Version]
- Simmons, K.J.; Eason, T.N.; Curioso, C.L.; Griffin, S.M.; Ramudit, M.K.D.; Oshima, K.H.; Sams, E.A.; Wade, T.J.; Grimm, A.; Dufour, A.; et al. Visitors to a Tropical Marine Beach Show Evidence of Immunoconversions to Multiple Waterborne Pathogens. Front. Public Health 2019, 7, 231. [Google Scholar] [CrossRef] [Green Version]
- Kauppinen, A.; Pitkänen, T.; Al-Hello, H.; Maunula, L.; Hokajärvi, A.M.; Rimhanen-Finne, R.; Miettinen, I.T. Two Drinking Water Outbreaks Caused by Wastewater Intrusion Including Sapovirus in Finland. Int. J. Environ. Res. Public Health 2019, 16, 4376. [Google Scholar] [CrossRef] [Green Version]
- Moreira, N.A.; Bondelind, M. Safe drinking water and waterborne outbreaks. J. Water Health 2017, 15, 83–96. [Google Scholar] [CrossRef]
- Cui, B.J.; Gao, F.; Hu, C.; Li, Z.Y.; Fan, X.Y.; Cui, E.P. Effect of Different Reclaimed Water Irrigation Methods on Bacterial Community Diversity and Pathogen Abundance in the Soil-Pepper Ecosystem. Huan Jing Ke Xue 2019, 40, 5151–5163. [Google Scholar]
- Fernandez-Cassi, X.; Silvera, C.; Cervero-Aragó, S.; Rusiñol, M.; Latif-Eugeni, F.; Bruguera-Casamada, C.; Civit, S.; Araujo, R.M.; Figueras, M.J.; Girones, R.; et al. Evaluation of the microbiological quality of reclaimed water produced from a lagooning system. Environ. Sci. Pollut. Res. Int. 2016, 23, 16816–16833. [Google Scholar] [CrossRef] [PubMed]
- Chahal, C.; van den Akker, B.; Young, F.; Franco, C.; Blackbeard, J.; Monis, P. Pathogen and Particle Associations in Wastewater: Significance and Implications for Treatment and Disinfection Processes. Adv. Appl. Microbiol. 2016, 97, 63–119. [Google Scholar] [PubMed]
- Jahne, M.A.; Brinkman, N.E.; Keely, S.P.; Zimmerman, B.D.; Wheaton, E.A.; Garland, J.L. Droplet digital PCR quantification of norovirus and adenovirus in decentralized wastewater and graywater collections: Implications for onsite reuse. Water Res. 2020, 169, 115213. [Google Scholar] [CrossRef] [PubMed]
- Paruch, L.; Paruch, A.M.; Sørheim, R. DNA-based faecal source tracking of contaminated drinking water causing a large Campylobacter outbreak in Norway 2019. Int. J. Hyg. Environ. Health 2020, 224, 113420. [Google Scholar] [CrossRef] [PubMed]
- Garvey, P.; Carroll, A.; McNamara, E.; McKeown, P.J. Verotoxigenic Escherichia coli transmission in Ireland: A review of notified outbreaks, 2004–2012. Epidemiol. Infect. 2016, 144, 917–926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guix, S.; Fuentes, C.; Pintó, R.M.; Blanco, A.; Sabrià, A.; Anfruns-Estrada, E.; Garrido, V.R.; Alonso, M.; Bartolomé, R.; Cornejo, T.; et al. Infectivity of Norovirus GI and GII from Bottled Mineral Water during a Waterborne Outbreak, Spain. Emerg. Infect. Dis. 2020, 26, 134–137. [Google Scholar] [CrossRef]
- Joshi, M.S.; Lole, K.S.; Barve, U.S.; Salve, D.S.; Ganorkar, N.N.; Chavan, N.A.; Shinde, M.S.; Gopalkrishna, V. Investigation of a large waterborne acute gastroenteritis outbreak caused by group B rotavirus in Maharashtra state, India. J. Med. Virol. 2019, 91, 1877–1881. [Google Scholar] [CrossRef] [Green Version]
- Bonadonna, L.; La Rosa, G. A Review and Update on Waterborne Viral Diseases Associated with Swimming Pools. Int. J. Environ. Res. Public Health 2019, 16, 166. [Google Scholar] [CrossRef] [Green Version]
- Ćirković, V.; Klun, I.; Utaaker, K.S.; Uzelac, A.; Tysnes, K.R.; Robertson, L.J.; Djurković-Djaković, O. Surface waters as a potential source of Giardia and Cryptosporidium in Serbia. Exp. Parasitol. 2019, 209, 107824. [Google Scholar] [CrossRef]
- Chalmers, R.M.; Robinson, G.; Elwin, K.; Elson, R. Analysis of the Cryptosporidium spp. and gp60 subtypes linked to human outbreaks of cryptosporidiosis in England and Wales, 2009 to 2017. Parasit. Vectors 2019, 12, 95. [Google Scholar] [CrossRef]
- Xiao, S.; Yin, P.; Zhang, Y.; Zhao, X.; Sun, L.; Yuan, H.; Lu, J.; Hu, S. Occurrence, genotyping, and health risk of Cryptosporidium and Giardia in recreational lakes in Tianjin, China. Water Res. 2018, 141, 46–56. [Google Scholar] [CrossRef] [PubMed]
- Almakki, A.; Jumas-Bilak, E.; Marchandin, H.; Licznar-Fajardo, P. Antibiotic resistance in urban runoff. Sci. Total Environ. 2019, 667, 64–76. [Google Scholar] [CrossRef] [PubMed]
- Szekeres, E.; Chiriac, C.M.; Baricz, A.; Szőke-Nagy, T.; Lung, I.; Soran, M.L.; Rudi, K.; Dragos, N.; Coman, C. Investigating antibiotics, antibiotic resistance genes, and microbial contaminants in groundwater in relation to the proximity of urban areas. Environ. Pollut. 2018, 236, 734–744. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Y.; Kuang, Z.; Xu, J.; Li, C.; Li, Y.; Jiang, Y.; Xie, J. Comparison of Microbiomes and Resistomes in Two Karst Groundwater Sites in Chongqing, China. Ground Water 2019, 57, 807–818. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Wan, K.; Zeng, J.; Lin, W.; Ye, C.; Yu, X. Co-selection and stability of bacterial antibiotic resistance by arsenic pollution accidents in source water. Environ. Int. 2020, 135, 105351. [Google Scholar] [CrossRef]
- Harrison, K.R.; Kappell, A.D.; McNamara, P.J. Benzalkonium chloride alters phenotypic and genotypic antibiotic resistance profiles in a source water used for drinking water treatment. Environ. Pollut. 2019, 26, 113472. [Google Scholar] [CrossRef] [PubMed]
- Proia, L.; Anzil, A.; Subirats, J.; Borrego, C.; Farrè, M.; Llorca, M.; Balcázar, J.L.; Servais, P. Antibiotic resistance along an urban river impacted by treated wastewaters. Sci. Total Environ. 2018, 628–629, 453–466. [Google Scholar] [CrossRef] [PubMed]
- Su, H.C.; Liu, Y.S.; Pan, C.G.; Chen, J.; He, L.Y.; Ying, G.G. Persistence of antibiotic resistance genes and bacterial community changes in drinking water treatment system: From drinking water source to tap water. Sci. Total Environ. 2018, 616–617, 453–461. [Google Scholar] [CrossRef]
- Xu, L.; Ouyang, W.; Qian, Y.; Su, C.; Su, J.; Chen, H. High-throughput profiling of antibiotic resistance genes in drinking water treatment plants and distribution systems. Environ. Pollut. 2016, 213, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Sanganyado, E.; Gwenzi, W. Antibiotic resistance in drinking water systems: Occurrence, removal, and human health risks. Sci. Total Environ. 2019, 669, 785–797. [Google Scholar] [CrossRef]
- Tsen, H.Y.; Lin, C.K.; Chi, W.R. Development and use of 16S rRNA gene targeted PCR primers for the identification of Escherichia coli cells in water. J. Appl. Microbiol. 1998, 85, 554–560. [Google Scholar] [CrossRef]
- Saxena, T.; Kaushik, P.; Krishna Mohan, M. Prevalence of E. coli O157:H7 in water sources: An overview on associated diseases, outbreaks and detection methods. Diagn. Microbiol. Infect. Dis. 2015, 82, 249–264. [Google Scholar] [CrossRef] [PubMed]
- Maheux, A.F.; Picard, F.J.; Boissinot, M.; Bissonnette, L.; Paradis, S.; Bergeron, M.G. Analytical comparison of nine PCR primer sets designed to detect the presence of Escherichia coli/Shigella in water samples. Water Res. 2009, 43, 3019–3028. [Google Scholar] [CrossRef] [PubMed]
- Mueller-Spitz, S.R.; Stewart, L.B.; Klump, J.V.; McLellan, S.L. Freshwater suspended sediments and sewage are reservoirs for enterotoxin-positive Clostridium perfringens. Appl. Environ. Microbiol. 2010, 76, 5556–5562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, M.S.; Ahn, T.Y.; Joh, K.; Lee, E.S.; Park, D.S. Improved PCR assay for the species-specific identification and quantitation of Legionella pneumophila in water. Appl. Microbiol. Biotechnol. 2015, 99, 9227–9236. [Google Scholar] [CrossRef] [PubMed]
- Waage, A.S.; Vardund, T.; Lund, V.; Kapperud, G. Detection of small numbers of Campylobacter jejuni and Campylobacter coli cells in environmental water, sewage, and food samples by a seminested PCR assay. Appl. Environ. Microbiol. 1999, 65, 1636–1643. [Google Scholar] [CrossRef] [Green Version]
- Pollard, D.R.; Johnson, W.M.; Lior, H.; Tyler, S.D.; Rozee, K.R. Rapid and specific detection of verotoxin genes in Escherichia coli by the polymerase chain reaction. J. Clin. Microbiol. 1990, 28, 540–545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ram, S.; Vajpayee, P.; Shanker, R. Contamination of potable water distribution systems by multiantimicrobial-resistant enterohemorrhagic Escherichia coli. Environ. Health Perspect. 2008, 116, 448–452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamner, S.; Broadaway, S.C.; Mishra, V.B.; Tripathi, A.; Mishra, R.K.; Pulcini, E.; Pyle, B.H.; Ford, T.E. Isolation of potentially pathogenic Escherichia coli O157:H7 from the Ganges River. Appl. Environ. Microbiol. 2007, 73, 2369–2372. [Google Scholar] [CrossRef] [Green Version]
- Abong’o, B.O.; Momba, M.N. Prevalence and potential link between E. coli O157:H7 isolated from drinking water, meat and vegetables and stools of diarrhoeic confirmed and non-confirmed HIV/AIDS patients in the Amathole District—South Africa. J. Appl. Microbiol. 2008, 105, 424–431. [Google Scholar] [CrossRef]
- Momba, M.N.; Malakate, V.K.; Theron, J. Abundance of pathogenic Escherichia coli, Salmonella typhimurium and Vibrio cholerae in Nkonkobe drinking water sources. J. Water Health 2006, 4, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Xie, G.; Yu, S.; Li, W.; Mu, D.; Aguilar, Z.P.; Xu, H. Simultaneous detection of Salmonella spp., Pseudomonas aeruginosa, Bacillus cereus, and Escherichia coli O157:H7 in environmental water using PMA combined with mPCR. J. Microbiol. 2020, 58, 668–674. [Google Scholar] [CrossRef]
- Gomi, R.; Matsuda, T.; Fujimori, Y.; Harada, H.; Matsui, Y.; Yoneda, M. Characterization of Pathogenic Escherichia coli in River Water by Simultaneous Detection and Sequencing of 14 Virulence Genes. Environ. Sci. Technol. 2015, 49, 6800–6807. [Google Scholar] [CrossRef]
- Triviño-Valencia, J.; Lora, F.; Zuluaga, J.D.; Gomez-Marin, J.E. Detection by PCR of pathogenic protozoa in raw and drinkable water samples in Colombia. Parasitol. Res. 2016, 115, 1789–1797. [Google Scholar] [CrossRef] [PubMed]
- Bae, K.S.; Lee, S.; Lee, J.Y.; Kim, J.H.; Joo, Y.L.; Lee, S.H.; Chung, H.M.; You, K.A. Development of diagnostic systems for wide range and highly sensitive detection of two waterborne hepatitis viruses from groundwater using the conventional reverse transcription nested PCR assay. J. Virol. Methods 2022, 299, 114344. [Google Scholar] [CrossRef] [PubMed]
- Kluge, M.; Fleck, J.D.; Soliman, M.C.; Luz, R.B.; Fabres, R.B.; Comerlato, J.; Silva, J.V.; Staggemeier, R.; Vecchia, A.D.; Capalonga, R.; et al. Human adenovirus (HAdV), human enterovirus (hEV), and genogroup A rotavirus (GARV) in tap water in southern Brazil. J. Water Health 2014, 12, 526–532. [Google Scholar] [CrossRef] [Green Version]
- Lin, M.; Wu, X.; Yan, Q.; Ma, Y.; Huang, L.; Qin, Y.; Xu, X. Incidence of antimicrobial-resistance genes and integrons in antibiotic-resistant bacteria isolated from eels and aquaculture ponds. Dis. Aquat. Organ. 2016, 120, 115–123. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Li, X.; Wu, J.; Coin, L.; O’Brien, J.; Hai, F.; Jiang, G. Molecular Methods for Pathogenic Bacteria Detection and Recent Advances in Wastewater Analysis. Water 2021, 13, 3551. [Google Scholar] [CrossRef]
- Inoue, D.; Hinoura, T.; Suzuki, N.; Pang, J.; Malla, R.; Shrestha, S.; Chapagain, S.K.; Matsuzawa, H.; Nakamura, T.; Tanaka, Y.; et al. High-throughput DNA microarray detection of pathogenic bacteria in shallow well groundwater in the Kathmandu Valley, Nepal. Curr. Microbiol. 2015, 70, 43–50. [Google Scholar] [CrossRef]
- Weidhaas, J.; Anderson, A.; Jamal, R. Elucidating Waterborne Pathogen Presence and Aiding Source Apportionment in an Impaired Stream. Appl. Environ. Microbiol. 2018, 84, e02510-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomes, M.; Vieira, H.; Vale, F.F. Characterization, validation and application of a DNA microarray for the detection of mandatory and other waterborne pathogens. J. Biochem. 2015, 158, 393–401. [Google Scholar] [CrossRef]
- Lee, D.Y.; Seto, P.; Korczak, R. DNA microarray-based detection and identification of waterborne protozoan pathogens. J. Microbiol. Methods 2010, 80, 129–133. [Google Scholar] [CrossRef]
- Brinkman, N.E.; Francisco, R.; Nichols, T.L.; Robinson, D.; Schaefer, F.W.; Schaudies, R.P.; Villegas, E.N. Detection of multiple waterborne pathogens using microsequencing arrays. J. Appl. Microbiol. 2013, 114, 564–573. [Google Scholar] [CrossRef]
- Wong, M.V.; Hashsham, S.A.; Gulari, E.; Rouillard, J.M.; Aw, T.G.; Rose, J.B. Detection and characterization of human pathogenic viruses circulating in community wastewater using multi target microarrays and polymerase chain reaction. J. Water Health 2013, 11, 659–670. [Google Scholar] [CrossRef] [PubMed]
- Unc, A.; Zurek, L.; Peterson, G.; Narayanan, S.; Springthorpe, S.V.; Sattar, S.A. Microarray assessment of virulence, antibiotic, and heavy metal resistance in an agricultural watershed creek. J. Environ. Qual. 2012, 41, 534–543. [Google Scholar] [CrossRef] [PubMed]
- Bartley, P.S.; Domitrovic, T.N.; Moretto, V.T.; Santos, C.S.; Ponce-Terashima, R.; Reis, M.G.; Barbosa, L.M.; Blanton, R.E.; Bonomo, R.A.; Perez, F. Antibiotic Resistance in Enterobacteriaceae from Surface Waters in Urban Brazil Highlights the Risks of Poor Sanitation. Am. J. Trop. Med. Hyg. 2019, 100, 1369–1377. [Google Scholar] [PubMed]
- Lemuth, K.; Rupp, S. Microarrays as Research Tools and Diagnostic Devices. In RNA and DNA Diagnostics; Erdmann, V., Jurga, S., Barciszewski, J., Eds.; Springer: Cham, Switzerland, 2015; pp. 259–280. [Google Scholar]
- Jaksik, R.; Iwanaszko, M.; Rzeszowska-Wolny, J.; Kimmel, M. Microarray experiments and factors which affect their reliability. Biol. Direct. 2015, 10, 46. [Google Scholar] [CrossRef]
- Palka-Santini, M.; Cleven, B.E.; Eichinger, L.; Krönke, M.; Krut, O. Large scale multiplex PCR improves pathogen detection by DNA microarrays. BMC Microbiol. 2009, 9, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kostić, T.; Sessitsch, A. Microbial Diagnostic Microarrays for the Detection and Typing of Food- and Water-Borne (Bacterial) Pathogens. Microarrays 2012, 1, 3–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Lin, H.R.; Zhang, S.T.; Yu, X. Real-time PCR detection and quantification of emerging waterborne pathogens (EWPs) and antibiotic resistance genes (ARGs) in the downstream area of Jiulong River. Huan Jing Ke Xue 2012, 33, 2685–2690. [Google Scholar] [PubMed]
- Bonetta, S.; Borelli, E.; Bonetta, S.; Conio, O.; Palumbo, F.; Carraro, E. Development of a PCR protocol for the detection of Escherichia coli O157:H7 and Salmonella spp. in surface water. Environ. Monit. Assess. 2011, 177, 493–503. [Google Scholar] [CrossRef] [PubMed]
- Patel, C.B.; Shanker, R.; Gupta, V.K.; Upadhyay, R.S. Q-PCR Based Culture-Independent Enumeration and Detection of Enterobacter: An Emerging Environmental Human Pathogen in Riverine Systems and Potable Water. Front. Microbiol. 2016, 7, 172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, N.; Neumann, N.F.; Ruecker, N.; Alderisio, K.A.; Sturbaum, G.D.; Villegas, E.N.; Chalmers, R.; Monis, P.; Feng, Y.; Xiao, L. Development and Evaluation of Three Real-Time PCR Assays for Genotyping and Source Tracking Cryptosporidium spp. in Water. Appl. Environ. Microbiol. 2015, 81, 5845–5854. [Google Scholar] [CrossRef] [Green Version]
- Iaconelli, M.; Muscillo, M.; Della Libera, S.; Fratini, M.; Meucci, L.; De Ceglia, M.; Giacosa, D.; La Rosa, G. One-year Surveillance of Human Enteric Viruses in Raw and Treated Wastewaters, Downstream River Waters, and Drinking Waters. Food Environ. Virol. 2017, 9, 79–88. [Google Scholar] [CrossRef]
- Jia, J.; Guan, Y.; Cheng, M.; Chen, H.; He, J.; Wang, S.; Wang, Z. Occurrence and distribution of antibiotics and antibiotic resistance genes in Ba River, China. Sci. Total Environ. 2018, 642, 1136–1144. [Google Scholar] [CrossRef]
- Ahmed, W.; Gyawali, P.; Hamilton, K.A.; Joshi, S.; Aster, D.; Donner, E.; Simpson, S.L.; Symonds, E.M. Antibiotic Resistance and Sewage-Associated Marker Genes in Untreated Sewage and a River Characterized During Baseflow and Stormflow. Front. Microbiol. 2021, 12, 632850. [Google Scholar] [CrossRef] [PubMed]
- Paruch, L.; Paruch, A.M.; Blankenberg, A.-G.B.; Bechmann, M.; Mæhlum, T. Application of host-specific genetic markers for microbial source tracking of faecal water contamination in an agricultural catchment. Acta Agric. Scand. 2015, 65, 164–172. [Google Scholar] [CrossRef]
- Staley, Z.R.; Boyd, R.J.; Shum, P.; Edge, T.A. Microbial Source Tracking Using Quantitative and Digital PCR To Identify Sources of Fecal Contamination in Stormwater, River Water, and Beach Water in a Great Lakes Area of Concern. Appl. Environ. Microbiol. 2018, 84, e01634-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vadde, K.K.; McCarthy, A.J.; Rong, R.; Sekar, R. Quantification of Microbial Source Tracking and Pathogenic Bacterial Markers in Water and Sediments of Tiaoxi River (Taihu Watershed). Front. Microbiol. 2019, 10, 699. [Google Scholar] [CrossRef]
- Sinigalliano, C.; Kim, K.; Gidley, M.; Yuknavage, K.; Knee, K.; Palacios, D.; Bautista, C.; Bonacolta, A.; Lee, H.W.; Maurin, L. Microbial Source Tracking of Fecal Indicating Bacteria in Coral Reef Waters, Recreational Waters, and Groundwater of Saipan by Real-Time Quantitative PCR. Front. Microbiol. 2021, 11, 596650. [Google Scholar] [CrossRef]
- Deshmukh, R.A.; Joshi, K.; Bhand, S.; Roy, U. Recent developments in detection and enumeration of waterborne bacteria: A retrospective minireview. MicrobiologyOpen 2016, 5, 901–922. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inoue, D.; Tsutsui, H.; Yamazaki, Y.; Sei, K.; Soda, S.; Fujita, M.; Ike, M. Application of real-time polymerase chain reaction (PCR) coupled with ethidium monoazide treatment for selective quantification of viable bacteria in aquatic environment. Water Sci. Technol. 2008, 58, 1107–1112. [Google Scholar] [CrossRef] [PubMed]
- Banihashemi, A.; van Dyke, M.I.; Huck, P.M. Detection of viable bacterial pathogens in a drinking water source using propidium monoazide-quantitative PCR. J. Water Supply 2015, 64, 139–148. [Google Scholar] [CrossRef]
- Alygizakis, N.; Markou, A.N.; Rousis, N.I.; Galani, A.; Avgeris, M.; Adamopoulos, P.G.; Scorilas, A.; Lianidou, E.S.; Paraskevis, D.; Tsiodras, S.; et al. Analytical methodologies for the detection of SARS-CoV-2 in wastewater: Protocols and future perspectives. Trends Analyt. Chem. 2021, 134, 116125. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Qu, W.; Wang, W.; Lu, Y.; Wu, Y.; Li, Z.; Hang, X.; Wang, X.; Zhao, D.; Zhang, C. MPprimer: A program for reliable multiplex PCR primer design. BMC Bioinform. 2010, 11, 143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, J.; Yi, J.; Zhan, M.; Xie, Q.; Zhen, T.T.; Zhou, J.; Li, Z.; Li, Z. The web-based multiplex PCR primer design software Ultiplex and the associated experimental workflow: Up to 100-plex multiplicity. BMC Genom. 2021, 22, 835. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.C.; Jinneman, K.C.; Neal-McKinney, J.; Wu, W.H.; Rice, D.H. Simultaneous Identification of Campylobacter jejuni, Campylobacter coli, and Campylobacter lari with Smart Cycler-Based Multiplex Quantitative Polymerase Chain Reaction. Foodborne Pathog. Dis. 2017, 14, 371–378. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Xin, H.; Li, S.F. Multiplex PMA-qPCR Assay with Internal Amplification Control for Simultaneous Detection of Viable Legionella pneumophila, Salmonella typhimurium, and Staphylococcus aureus in Environmental Waters. Environ. Sci. Technol. 2015, 49, 14249–14256. [Google Scholar] [CrossRef] [PubMed]
- Guy, R.A.; Payment, P.; Krull, U.J.; Horgen, P.A. Real-time PCR for quantification of Giardia and Cryptosporidium in environmental water samples and sewage. Appl. Environ. Microbiol. 2003, 69, 5178–5185. [Google Scholar] [CrossRef] [Green Version]
- Kang, L.H.; Oh, S.H.; Park, J.W.; Won, Y.J.; Ryu, S.; Paik, S.Y. Simultaneous detection of waterborne viruses by multiplex real-time PCR. J. Microbiol. 2013, 51, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Navarro, A.; Gómez, L.; Sanseverino, I.; Niegowska, M.; Roka, E.; Pedraccini, R.; Vargha, M.; Lettieri, T. SARS-CoV-2 detection in wastewater using multiplex quantitative PCR. Sci. Total Environ. 2021, 797, 148890. [Google Scholar] [CrossRef] [PubMed]
- Mondal, S.; Feirer, N.; Brockman, M.; Preston, M.A.; Teter, S.J.; Ma, D.; Goueli, S.A.; Moorji, S.; Saul, B.; Cali, J.J. A direct capture method for purification and detection of viral nucleic acid enables epidemiological surveillance of SARS-CoV-2. Sci. Total Environ. 2021, 795, 148834. [Google Scholar] [CrossRef] [PubMed]
- Paulus, G.K.; Hornstra, L.M.; Alygizakis, N.; Slobodnik, J.; Thomaidis, N.; Medema, G. The impact of on-site hospital wastewater treatment on the downstream communal wastewater system in terms of antibiotics and antibiotic resistance genes. Int. J. Hyg. Environ. Health 2019, 222, 635–644. [Google Scholar] [CrossRef] [PubMed]
- Paulus, G.K.; Hornstra, L.M.; Medema, G. International tempo-spatial study of antibiotic resistance genes across the Rhine river using newly developed multiplex qPCR assays. Sci. Total Environ. 2020, 706, 135733. [Google Scholar] [CrossRef]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 128, E63. [Google Scholar] [CrossRef] [Green Version]
- Hardinge, P.; Murray, J.A.H. Reduced False Positives and Improved Reporting of Loop-Mediated Isothermal Amplification using Quenched Fluorescent Primers. Sci. Rep. 2019, 9, 7400. [Google Scholar] [CrossRef]
- Kim, D.W.; Kilgore, P.E.; Kim, E.J.; Kim, S.A.; Anh, D.D.; Seki, M. Loop-mediated isothermal amplification assay for detection of Haemophilus influenzae type b in cerebrospinal fluid. J. Clin. Microbiol. 2011, 49, 3621–3626. [Google Scholar] [CrossRef] [Green Version]
- Bentaleb, E.M.; Abid, M.; El Messaoudi, M.D.; Lakssir, B.; Ressami, E.M.; Amzazi, S.; Sefrioui, H.; Ait Benhassou, H. Development and evaluation of an in-house single step loop-mediated isothermal amplification (SS-LAMP) assay for the detection of Mycobacterium tuberculosis complex in sputum samples from Moroccan patients. BMC Infect. Dis. 2016, 16, 517. [Google Scholar] [CrossRef] [Green Version]
- Seki, M.; Kilgore, P.E.; Kim, E.J.; Ohnishi, M.; Hayakawa, S.; Kim, D.W. Loop-Mediated Isothermal Amplification Methods for Diagnosis of Bacterial Meningitis. Front. Pediatr. 2018, 6, 57. [Google Scholar] [CrossRef] [Green Version]
- Foo, P.C.; Nurul Najian, A.B.; Muhamad, N.A.; Ahamad, M.; Mohamed, M.; Yean Yean, C.; Lim, B.H. Loop-mediated isothermal amplification (LAMP) reaction as viable PCR substitute for diagnostic applications: A comparative analysis study of LAMP, conventional PCR, nested PCR (nPCR) and real-time PCR (qPCR) based on Entamoeba histolytica DNA derived from faecal sample. BMC Biotechnol. 2020, 20, 34. [Google Scholar]
- Huang, X.; Tang, G.; Ismail, N.; Wang, X. Developing RT-LAMP Assays for Detection of SARS-CoV-2 in Saliva. medRxiv 2021. [Google Scholar] [CrossRef]
- Lalli, M.A.; Langmade, J.S.; Chen, X.; Fronick, C.C.; Sawyer, C.S.; Burcea, L.C.; Wilkinson, M.N.; Fulton, R.S.; Heinz, M.; Buchser, W.J.; et al. Rapid and Extraction-Free Detection of SARS-CoV-2 from Saliva by Colorimetric Reverse-Transcription Loop-Mediated Isothermal Amplification. Clin. Chem. 2021, 67, 415–424. [Google Scholar] [CrossRef] [PubMed]
- Sherrill-Mix, S.; Hwang, Y.; Roche, A.M.; Glascock, A.; Weiss, S.R.; Li, Y.; Haddad, L.; Deraska, P.; Monahan, C.; Kromer, A.; et al. Detection of SARS-CoV-2 RNA using RT-LAMP and molecular beacons. Genome Biol. 2021, 22, 169. [Google Scholar] [CrossRef]
- Kreitlow, A.; Becker, A.; Schotte, U.; Malorny, B.; Plötz, M.; Abdulmawjood, A. Evaluation of different target genes for the detection of Salmonella sp. by loop-mediated isothermal amplification. Lett. Appl. Microbiol. 2021, 72, 420–426. [Google Scholar] [CrossRef] [PubMed]
- Quyen, T.L.; Nordentoft, S.; Vinayaka, A.C.; Ngo, T.A.; Engelsmenn, P.; Sun, Y.; Madsen, M.; Bang, D.D.; Wolff, A. A Sensitive, Specific and Simple Loop Mediated Isothermal Amplification Method for Rapid Detection of Campylobacter spp. in Broiler Production. Front. Microbiol. 2019, 10, 2443. [Google Scholar] [CrossRef]
- Sridapan, T.; Tangkawsakul, W.; Janvilisri, T.; Kiatpathomchai, W.; Dangtip, S.; Ngamwongsatit, N.; Nacapricha, D.; Ounjai, P.; Chankhamhaengdecha, S. Rapid detection of Clostridium perfringens in food by loop-mediated isothermal amplification combined with a lateral flow biosensor. PLoS ONE 2021, 16, e0245144. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Fu, H.; Chen, D.; Shao, Z.; Zhu, J.; Alali, W.Q.; Chen, L. Simple Visualized Detection Method of Virulence-Associated Genes of Vibrio cholerae by Loop-Mediated Isothermal Amplification. Front. Microbiol. 2019, 10, 2899. [Google Scholar] [CrossRef]
- Chamanrokh, P.; Colwell, R.R.; Huq, A. Loop-mediated isothermal amplification (LAMP) assay for rapid detection of viable but non-culturable Vibrio cholerae O1. Can. J. Microbiol. 2022, 68, 103–110. [Google Scholar] [CrossRef]
- Amoah, I.D.; Mthethwa, N.P.; Pillay, L.; Deepnarain, N.; Pillay, K.; Awolusi, O.O.; Kumari, S.; Bux, F. RT-LAMP: A Cheaper, Simpler and Faster Alternative for the Detection of SARS-CoV-2 in Wastewater. Food Environ. Virol. 2021, 13, 447–456. [Google Scholar] [CrossRef]
- Ramírez-Chavarría, R.G.; Castillo-Villanueva, E.; Alvarez-Serna, B.E.; Carrillo-Reyes, J.; Ramírez-Zamora, R.M.; Buitrón, G.; Alvarez-Icaza, L. Loop-mediated isothermal amplification-based electrochemical sensor for detecting SARS-CoV-2 in wastewater samples. J. Environ. Chem. Eng. 2022, 10, 107488. [Google Scholar] [CrossRef]
- Ongerth, J.E.; Danielson, R.E. RT qLAMP-Direct Detection of SARS-CoV-2 in Raw Sewage. J. Biomol. Tech. 2021, 32, 206–213. [Google Scholar] [CrossRef] [PubMed]
- Haque, M.F.U.; Bukhari, S.S.; Ejaz, R.; Zaman, F.U.; Sreejith, K.R.; Rashid, N.; Umer, M.; Shahzad, N. A novel RdRp-based colorimetric RT-LAMP assay for rapid and sensitive detection of SARS-CoV-2 in clinical and sewage samples from Pakistan. Virus Res. 2021, 302, 198484. [Google Scholar] [CrossRef] [PubMed]
- Reuter, C.; Slesiona, N.; Hentschel, S.; Aehlig, O.; Breitenstein, A.; Csáki, A.; Henkel, T.; Fritzsche, W. Loop-mediated amplification as promising on-site detection approach for Legionella pneumophila and Legionella spp. Appl. Microbiol. Biotechnol. 2020, 104, 405–415. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, F.; Stedtfeld, R.D.; Waseem, H.; Williams, M.R.; Cupples, A.M.; Tiedje, J.M.; Hashsham, S.A. Most probable number—loop mediated isothermal amplification (MPN-LAMP) for quantifying waterborne pathogens in <25 min. J. Microbiol. Methods 2017, 132, 27–33. [Google Scholar] [CrossRef] [Green Version]
- Wong, Y.P.; Othman, S.; Lau, Y.L.; Radu, S.; Chee, H.Y. Loop-mediated isothermal amplification (LAMP): A versatile technique for detection of micro-organisms. J. Appl. Microbiol. 2018, 124, 626–643. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, F.; Seyrig, G.; Tourlousse, D.M.; Stedtfeld, R.D.; Tiedje, J.M.; Hashsham, S.A. A CCD-based fluorescence imaging system for real-time loop-mediated isothermal amplification-based rapid and sensitive detection of waterborne pathogens on microchips. Biomed. Microdevices 2011, 13, 929–937. [Google Scholar] [CrossRef]
- Jin, J.; Duan, L.; Fu, J.; Chai, F.; Zhou, Q.; Wang, Y.; Shao, X.; Wang, L.; Yan, M.; Su, X.; et al. A real-time LAMP-based dual-sample microfluidic chip for rapid and simultaneous detection of multiple waterborne pathogenic bacteria from coastal waters. Anal. Methods 2021, 13, 2710–2721. [Google Scholar] [CrossRef]
- Moehling, T.J.; Lee, D.H.; Henderson, M.E.; McDonald, M.K.; Tsang, P.H.; Kaakeh, S.; Kim, E.S.; Wereley, S.T.; Kinzer-Ursem, T.L.; Clayton, K.N.; et al. A smartphone-based particle diffusometry platform for sub-attomolar detection of Vibrio cholerae in environmental water. Biosens. Bioelectron. 2020, 167, 112497. [Google Scholar] [CrossRef]
- Meagher, R.J.; Priye, A.; Light, Y.K.; Huang, C.; Wang, E. Impact of primer dimers and self-amplifying hairpins on reverse transcription loop-mediated isothermal amplification detection of viral RNA. Analyst 2018, 143, 1924–1933. [Google Scholar] [CrossRef]
- Gadkar, V.J.; Goldfarb, D.M.; Gantt, S.; Tilley, P.A.G. Real-time Detection and Monitoring of Loop Mediated Amplification (LAMP) Reaction Using Self-quenching and De-quenching Fluorogenic Probes. Sci. Rep. 2018, 8, 5548. [Google Scholar] [CrossRef] [Green Version]
- Taylor, S.C.; Laperriere, G.; Germain, H. Droplet Digital PCR versus qPCR for gene expression analysis with low abundant targets: From variable nonsense to publication quality data. Sci. Rep. 2017, 7, 2409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heijnen, L.; Elsinga, G.; de Graaf, M.; Molenkamp, R.; Koopmans, M.P.G.; Medema, G. Droplet digital RT-PCR to detect SARS-CoV-2 signature mutations of variants of concern in wastewater. Sci. Total Environ. 2021, 799, 149456. [Google Scholar] [CrossRef] [PubMed]
- Flood, M.T.; D’Souza, N.; Rose, J.B.; Aw, T.G. Methods Evaluation for Rapid Concentration and Quantification of SARS-CoV-2 in Raw Wastewater Using Droplet Digital and Quantitative RT-PCR. Food Environ. Virol. 2021, 13, 303–315. [Google Scholar] [PubMed]
- Cao, Y.; Raith, M.R.; Griffith, J.F. Droplet digital PCR for simultaneous quantification of general and human-associated fecal indicators for water quality assessment. Water Res. 2015, 70, 337–349. [Google Scholar] [CrossRef]
- Kanagal-Shamanna, R. Digital PCR: Principles and Applications. Methods Mol. Biol. 2016, 1392, 43–50. [Google Scholar]
- Mauvisseau, Q.; Davy-Bowker, J.; Bulling, M.; Brys, R.; Neyrinck, S.; Troth, C.; Sweet, M. Combining ddPCR and environmental DNA to improve detection capabilities of a critically endangered freshwater invertebrate. Sci. Rep. 2019, 9, 14064. [Google Scholar] [CrossRef] [Green Version]
- Rothrock, M.J.; Hiett, K.L.; Kiepper, B.H.; Ingram, K.; Hinton, A. Quantification of Zoonotic Bacterial Pathogens within Commercial Poultry Processing Water Samples Using Droplet Digital PCR. Adv. Microbiol. 2013, 3, 403–411. [Google Scholar] [CrossRef] [Green Version]
- Singh, G.; Sithebe, A.; Enitan, A.M.; Kumari, S.; Bux, F.; Stenström, T.A. Comparison of droplet digital PCR and quantitative PCR for the detection of Salmonella and its application for river sediments. J. Water Health 2017, 15, 505–508. [Google Scholar] [CrossRef]
- Lewin, A.S.; Haugen, T.; Netzer, R.; Tøndervik, A.; Dahle, S.W.; Hageskal, G. Multiplex droplet digital PCR assay for detection of Flavobacterium psychrophilum and Yersinia ruckeri in Norwegian aquaculture. J. Microbiol. Methods 2020, 177, 106044. [Google Scholar] [CrossRef] [PubMed]
- Falzone, L.; Gattuso, G.; Lombardo, C.; Lupo, G.; Grillo, C.M.; Spandidos, D.A.; Libra, M.; Salmeri, M. Droplet digital PCR for the detection and monitoring of Legionella pneumophila. Int. J. Mol. Med. 2020, 46, 1777–1782. [Google Scholar] [CrossRef]
- Rački, N.; Morisset, D.; Gutierrez-Aguirre, I.; Ravnikar, M. One-step RT-droplet digital PCR: A breakthrough in the quantification of waterborne RNA viruses. Anal. Bioanal. Chem. 2014, 406, 661–667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ho, J.; Stange, C.; Suhrborg, R.; Wurzbacher, C.; Drewes, J.E.; Tiehm, A. SARS-CoV-2 wastewater surveillance in Germany: Long-term RT-digital droplet PCR monitoring, suitability of primer/probe combinations and biomarker stability. Water Res. 2022, 210, 117977. [Google Scholar] [CrossRef] [PubMed]
- Falzone, L.; Musso, N.; Gattuso, G.; Bongiorno, D.; Palermo, C.I.; Scalia, G.; Libra, M.; Stefani, S. Sensitivity assessment of droplet digital PCR for SARS-CoV-2 detection. Int. J. Mol. Med. 2020, 46, 957–964. [Google Scholar] [CrossRef]
- Wang, X.; Gu, J.; Gao, H.; Qian, X.; Li, H. Abundances of Clinically Relevant Antibiotic Resistance Genes and Bacterial Community Diversity in the Weihe River, China. Int. J. Environ. Res. Public Health 2018, 15, 708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dungan, R.S.; Bjorneberg, D.L. Antibiotic resistance genes, class 1 integrons, and IncP-1/IncQ-1 plasmids in irrigation return flows. Environ. Pollut. 2020, 257, 113568. [Google Scholar] [CrossRef] [PubMed]
- McCombie, W.R.; McPherson, J.D.; Mardis, E.R. Next-Generation Sequencing Technologies. Cold Spring Harb. Perspect. Med. 2019, 9, a036798. [Google Scholar] [CrossRef]
- Greay, T.L.; Gofton, A.W.; Zahedi, A.; Paparini, A.; Linge, K.L.; Joll, C.A.; Ryan, U.M. Evaluation of 16S next-generation sequencing of hypervariable region 4 in wastewater samples: An unsuitable approach for bacterial enteric pathogen identification. Sci. Total Environ. 2019, 670, 1111–1124. [Google Scholar] [CrossRef]
- Zahedi, A.; Greay, T.L.; Paparini, A.; Linge, K.L.; Joll, C.A.; Ryan, U.M. Identification of eukaryotic microorganisms with 18S rRNA next-generation sequencing in wastewater treatment plants, with a more targeted NGS approach required for Cryptosporidium detection. Water Res. 2019, 158, 301–312. [Google Scholar] [CrossRef]
- Giwa, A.S.; Ali, N.; Athar, M.A.; Wang, K. Dissecting microbial community structure in sewage treatment plant for pathogens’ detection using metagenomic sequencing technology. Arch. Microbiol. 2020, 202, 825–833. [Google Scholar] [CrossRef]
- DeMone, C.; Hwang, M.H.; Feng, Z.; McClure, J.T.; Greenwood, S.J.; Fung, R.; Kim, M.; Weese, J.S.; Shapiro, K. Application of next generation sequencing for detection of protozoan pathogens in shellfish. Food Waterborne Parasitol. 2020, 21, e00096. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.C.; Han, M.; Chandrasekaran, S.; Fang, Y.; Kellogg, C.A. Assessing the water quality impacts of two Category-5 hurricanes on St. Thomas, Virgin Islands. Water Res. 2020, 171, 115440. [Google Scholar] [CrossRef]
- Doulgeraki, A.I.; Bikouli, V.; Argyri, A.A.; Chorianopoulos, N.; Mitre, E.; Charvourou, G.; Sourri, P.; Tassou, C.C.; Oikonomou, A. Exploring the Bacterial Communities of the Kaiafas Thermal Spring Anigrides Nymphes in Greece Prior to Rehabilitation Actions. Int. J. Environ. Res. Public Health 2020, 17, 9133. [Google Scholar] [CrossRef]
- Pereira, R.P.A.; Peplies, J.; Höfle, M.G.; Brettar, I. Bacterial community dynamics in a cooling tower with emphasis on pathogenic bacteria and Legionella species using universal and genus-specific deep sequencing. Water Res. 2017, 122, 363–376. [Google Scholar] [CrossRef] [PubMed]
- Vega, L.; Jaimes, J.; Morales, D.; Martínez, D.; Cruz-Saavedra, L.; Muñoz, M.; Ramírez, J.D. Microbial Communities’ Characterization in Urban Recreational Surface Waters Using Next Generation Sequencing. Microb. Ecol. 2021, 81, 847–863. [Google Scholar] [CrossRef] [PubMed]
- Cui, Q.; Fang, T.; Huang, Y.; Dong, P.; Wang, H. Evaluation of bacterial pathogen diversity, abundance and health risks in urban recreational water by amplicon next-generation sequencing and quantitative PCR. J. Environ. Sci. 2017, 57, 137–149. [Google Scholar] [CrossRef] [PubMed]
- Fang, T.; Cui, Q.; Huang, Y.; Dong, P.; Wang, H.; Liu, W.T.; Ye, Q. Distribution comparison and risk assessment of free-floating and particle-attached bacterial pathogens in urban recreational water: Implications for water quality management. Sci. Total Environ. 2018, 613–614, 428–438. [Google Scholar] [CrossRef]
- Pereira, R.P.A.; Peplies, J.; Brettar, I.; Höfle, M.G. Development of a genus-specific next generation sequencing approach for sensitive and quantitative determination of the Legionella microbiome in freshwater systems. BMC Microbiol. 2017, 17, 79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rusiñol, M.; Martínez-Puchol, S.; Timoneda, N.; Fernández-Cassi, X.; Pérez-Cataluña, A.; Fernández-Bravo, A.; Moreno-Mesonero, L.; Moreno, Y.; Alonso, J.L.; Figueras, M.J.; et al. Metagenomic analysis of viruses, bacteria and protozoa in irrigation water. Int. J. Hyg. Environ. Health 2020, 224, 113440. [Google Scholar] [CrossRef] [PubMed]
- Jongman, M.; Chidamba, L.; Korsten, L. Bacterial biomes and potential human pathogens in irrigation water and leafy greens from different production systems described using pyrosequencing. J. Appl. Microbiol. 2017, 123, 1043–1053. [Google Scholar] [CrossRef]
- Li, B.; Saingam, P.; Ishii, S.; Yan, T. Multiplex PCR coupled with direct amplicon sequencing for simultaneous detection of numerous waterborne pathogens. Appl. Microbiol. Biotechnol. 2019, 103, 953–961. [Google Scholar] [CrossRef]
- Crits-Christoph, A.; Kantor, R.S.; Olm, M.R.; Whitney, O.N.; Al-Shayeb, B.; Lou, Y.C.; Flamholz, A.; Kennedy, L.C.; Greenwald, H.; Hinkle, A.; et al. Genome Sequencing of Sewage Detects Regionally Prevalent SARS-CoV-2 Variants. mBio 2021, 12, e02703-20. [Google Scholar] [CrossRef] [PubMed]
- Gregory, D.A.; Wieberg, C.G.; Wenzel, J.; Lin, C.-H.; Johnson, M.C. Monitoring SARS-CoV-2 Populations in Wastewater by Amplicon Sequencing and Using the Novel Program SAM Refiner. Viruses 2021, 13, 1647. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Cataluña, A.; Chiner-Oms, Á.; Cuevas-Ferrando, E.; Díaz-Reolid, A.; Falcó, I.; Randazzo, W.; Girón-Guzmán, I.; Allende, A.; Bracho, M.A.; Comas, I.; et al. Spatial and temporal distribution of SARS-CoV-2 diversity circulating in wastewater. Water Res. 2022, 211, 118007. [Google Scholar] [CrossRef]
- Fongaro, G.; Stoco, P.H.; Souza, D.S.M.; Grisard, E.C.; Magri, M.E.; Rogovski, P.; Schörner, M.A.; Barazzetti, F.H.; Christoff, A.P.; de Oliveira, L.F.V.; et al. The presence of SARS-CoV-2 RNA in human sewage in Santa Catarina, Brazil, November 2019. Sci. Total Environ. 2021, 778, 146198. [Google Scholar] [CrossRef] [PubMed]
- Dharmadhikari, T.; Rajput, V.; Yadav, R.; Boargaonkar, R.; Patil, D.; Kale, S.; Kamble, S.P.; Dastager, S.G.; Dharne, M.S. High throughput sequencing based direct detection of SARS-CoV-2 fragments in wastewater of Pune, West India. Sci. Total Environ. 2022, 807, 151038. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Mao, G.; Yin, X.; Ma, L.; Liu, L.; Bai, Y.; Zhang, T.; Qu, J. Identification and quantification of bacterial genomes carrying antibiotic resistance genes and virulence factor genes for aquatic microbiological risk assessment. Water Res. 2020, 168, 115160. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.K.; Shin, H.; Han, D.; Hur, H.G.; Unno, T. Metagenomic analysis reveals the prevalence and persistence of antibiotic- and heavy metal-resistance genes in wastewater treatment plant. J. Microbiol. 2018, 56, 408–415. [Google Scholar] [CrossRef]
- Che, Y.; Xia, Y.; Liu, L.; Li, A.D.; Yang, Y.; Zhang, T. Mobile antibiotic resistome in wastewater treatment plants revealed by Nanopore metagenomic sequencing. Microbiome 2019, 7, 44. [Google Scholar] [CrossRef] [Green Version]
- Paruch, L.; Paruch, A.M.; Eiken, H.G.; Sørheim, R. Faecal pollution affects abundance and diversity of aquatic microbial community in anthropo-zoogenically influenced lotic ecosystems. Sci. Rep. 2019, 9, 19469. [Google Scholar] [CrossRef] [PubMed]
- Valverde, A.; Cason, E.D.; Gómez-Arias, A.; Bozkale, D.; Govender, D.; Riddell, E.; Cowan, D. Pollution shapes the microbial communities in river water and sediments from the Olifants River catchment, South Africa. Arch. Microbiol. 2021, 203, 295–303. [Google Scholar] [CrossRef]
- Pascual-Benito, M.; Ballesté, E.; Monleón-Getino, T.; Urmeneta, J.; Blanch, A.R.; García-Aljaro, C.; Lucena, F. Impact of treated sewage effluent on the bacterial community composition in an intermittent Mediterranean stream. Environ. Pollut. 2020, 266, 115254. [Google Scholar] [CrossRef]
- Mahmoud, M.A.A.; Magdy, M. Metabarcoding profiling of microbial diversity associated with trout fish farming. Sci. Rep. 2021, 11, 421. [Google Scholar] [CrossRef] [PubMed]
- Picazo, A.; Rochera, C.; Villaescusa, J.A.; Miralles-Lorenzo, J.; Velázquez, D.; Quesada, A.; Camacho, A. Bacterioplankton Community Composition Along Environmental Gradients in Lakes from Byers Peninsula (Maritime Antarctica) as Determined by Next-Generation Sequencing. Front. Microbiol. 2019, 10, 908. [Google Scholar] [CrossRef] [PubMed]
- Betiku, O.C.; Sarjeant, K.C.; Ngatia, L.W.; Aghimien, M.O.; Odewumi, C.O.; Latinwo, L.M. Evaluation of microbial diversity of three recreational water bodies using 16S rRNA metagenomic approach. Sci. Total Environ. 2021, 771, 144773. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Staley, C.; Hamilton, K.A.; Beale, D.J.; Sadowsky, M.J.; Toze, S.; Haas, C.N. Amplicon-based taxonomic characterization of bacteria in urban and peri-urban roof-harvested rainwater stored in tanks. Sci. Total Environ. 2017, 576, 326–334. [Google Scholar] [CrossRef] [PubMed]
- Sutcliffe, B.; Hose, G.C.; Harford, A.J.; Midgley, D.J.; Greenfield, P.; Paulsen, I.T.; Chariton, A.A. Microbial communities are sensitive indicators for freshwater sediment copper contamination. Environ. Pollut. 2019, 247, 1028–1038. [Google Scholar] [CrossRef] [PubMed]
- Aguinaga, O.E.; McMahon, A.; White, K.N.; Dean, A.P.; Pittman, J.K. Microbial Community Shifts in Response to Acid Mine Drainage Pollution within a Natural Wetland Ecosystem. Front. Microbiol. 2018, 9, 1445. [Google Scholar] [CrossRef]
- Buccheri, M.A.; Salvo, E.; Coci, M.; Quero, G.M.; Zoccarato, L.; Privitera, V.; Rappazzo, G. Investigating microbial indicators of anthropogenic marine pollution by 16S and 18S High-Throughput Sequencing (HTS) library analysis. FEMS Microbiol. Lett. 2019, 366, fnz179. [Google Scholar] [CrossRef] [PubMed]
- Yanez-Montalvo, A.; Gómez-Acata, S.; Águila, B.; Hernández-Arana, H.; Falcón, L.I. The microbiome of modern microbialites in Bacalar Lagoon, Mexico. PLoS ONE 2020, 15, e0230071. [Google Scholar]
- Kaestli, M.; Skillington, A.; Kennedy, K.; Majid, M.; Williams, D.; McGuinness, K.; Munksgaard, N.; Gibb, K. Spatial and Temporal Microbial Patterns in a Tropical Macrotidal Estuary Subject to Urbanization. Front. Microbiol. 2017, 8, 1313. [Google Scholar] [CrossRef] [Green Version]
- Iliev, I.; Yahubyan, G.; Marhova, M.; Apostolova, E.; Gozmanova, M.; Gecheva, G.; Kostadinova, S.; Ivanova, A.; Baev, V. Metagenomic profiling of the microbial freshwater communities in two Bulgarian reservoirs. J. Basic Microbiol. 2017, 57, 669–679. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Liu, Y.; Zhao, X.; Zhao, Z.; Zhang, H.; Huang, X.; Xu, W.; Shen, Y.; Lan, W. High-throughput sequencing reveals the spatial distribution variability of microbial community in coastal waters in Shenzhen. Ecotoxicology 2021, 30, 1429–1436. [Google Scholar] [PubMed]
- Sadeepa, D.; Sirisena, K.; Manage, P.M. Diversity of microbial communities in hot springs of Sri Lanka as revealed by 16S rRNA gene high-throughput sequencing analysis. Gene 2022, 812, 146103. [Google Scholar] [CrossRef] [PubMed]
- Paruch, L.; Paruch, A.M.; Eiken, H.G.; Skogen, M.; Sørheim, R. Seasonal dynamics of lotic bacterial communities assessed by 16S rRNA gene amplicon deep sequencing. Sci. Rep. 2020, 10, 16399. [Google Scholar] [CrossRef]
- Eraqi, W.A.; ElRakaiby, M.T.; Megahed, S.A.; Yousef, N.H.; Elshahed, M.S.; Yassin, A.S. The Nile River Microbiome Reveals a Remarkably Stable Community Between Wet and Dry Seasons, and Sampling Sites, in a Large Urban Metropolis (Cairo, Egypt). OMICS 2018, 22, 553–564. [Google Scholar] [CrossRef]
- Roeselers, G.; Coolen, J.; van der Wielen, P.W.; Jaspers, M.C.; Atsma, A.; de Graaf, B.; Schuren, F. Microbial biogeography of drinking water: Patterns in phylogenetic diversity across space and time. Environ. Microbiol. 2015, 17, 2505–2514. [Google Scholar] [CrossRef]
- Boers, S.A.; Prest, E.I.; Taučer-Kapteijn, M.; Knezev, A.; Schaap, P.G.; Hays, J.P.; Jansen, R. Monitoring of microbial dynamics in a drinking water distribution system using the culture-free, user-friendly, MYcrobiota platform. Sci. Rep. 2018, 8, 14727. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.; Giesy, J.P.; Guo, M.; Ung, W.K.; Kong, Y.; Mok, K.M.; Lee, S.M. Temporal Patterns of Bacterial and Viral Communities during Algae Blooms of a Reservoir in Macau. Toxins 2021, 13, 894. [Google Scholar] [CrossRef] [PubMed]
- Aburto-Medina, A.; Shahsavari, E.; Salzman, S.A.; Kramer, A.; Ball, A.S.; Allinson, G. Elucidation of the microbial diversity in rivers in south-west Victoria, Australia impacted by rural agricultural contamination (dairy farming). Ecotoxicol. Environ. Saf. 2019, 172, 356–363. [Google Scholar] [CrossRef]
- Li, C.; Pulin, Z.; Wei, L.; Wei, D.; Jia, O.; Chang, C.C. Molecular biological methods in environmental engineering. Water Environ. Res. 2018, 90, 1371–1391. [Google Scholar]
- Lear, G.; Dickie, I.; Banks, J.; Boyer, S.; Buckley, H.L.; Buckley, T.R.; Holdaway, R. Methods for the extraction, storage, amplification and sequencing of DNA from environmental samples. N. Z. J. Ecol. 2018, 42, 10–50A. [Google Scholar] [CrossRef] [Green Version]
- Dairawan, M.; Shetty, P.J. The Evolution of DNA Extraction Methods. Am. J. Biomed. Sci. Res. 2020, 8, 001234. [Google Scholar]
- Hu, B.; Guo, H.; Zhou, P.; Shi, Z.L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021, 19, 141–154. [Google Scholar] [PubMed]
- Kweinor Tetteh, E.; Opoku Amankwa, M.; Armah, E.K.; Rathilal, S. Fate of COVID-19 occurrences in wastewater systems: Emerging detection and treatment technologies—A review. Water 2020, 12, 2680. [Google Scholar] [CrossRef]
- Foladori, P.; Cutrupi, F.; Segata, N.; Manara, S.; Pinto, F.; Malpei, F.; Bruni, L.; La Rosa, G. SARS-CoV-2 from faeces to wastewater treatment: What do we know? A review. Sci. Total Environ. 2020, 743, 140444. [Google Scholar] [CrossRef] [PubMed]
- Medema, G.; Heijnen, L.; Elsinga, G.; Italiaander, R.; Brouwer, A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in The Netherlands. Environ. Sci. Technol. Lett. 2020, 7, 511–516. [Google Scholar] [CrossRef]
- Wu, F.; Zhang, J.; Xiao, A.; Gu, X.; Lee, W.L.; Armas, F.; Kauffman, K.; Hanage, W.; Matus, M.; Ghaeli, N.; et al. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. mSystems 2020, 5, e00614-20. [Google Scholar] [CrossRef] [PubMed]
- Randazzo, W.; Truchado, P.; Cuevas-Ferrando, E.; Simón, P.; Allende, A.; Sánchez, G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020, 181, 115942. [Google Scholar] [CrossRef]
- La Rosa, G.; Iaconelli, M.; Mancini, P.; Bonanno Ferraro, G.; Veneri, C.; Bonadonna, L.; Lucentini, L.; Suffredini, E. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 2020, 736, 139652. [Google Scholar] [CrossRef] [PubMed]
- Rimoldi, S.G.; Stefani, F.; Gigantiello, A.; Polesello, S.; Comandatore, F.; Mileto, D.; Maresca, M.; Longobardi, C.; Mancon, A.; Romeri, F.; et al. Presence and infectivity of SARS-CoV-2 virus in wastewaters and rivers. Sci. Total Environ. 2020, 744, 140911. [Google Scholar] [CrossRef]
- Kumar, M.; Patel, A.K.; Shah, A.V.; Raval, J.; Rajpara, N.; Joshi, M.; Joshi, C.G. First proof of the capability of wastewater surveillance for COVID-19 in India through detection of genetic material of SARS-CoV-2. Sci. Total Environ. 2020, 746, 141326. [Google Scholar] [CrossRef] [PubMed]
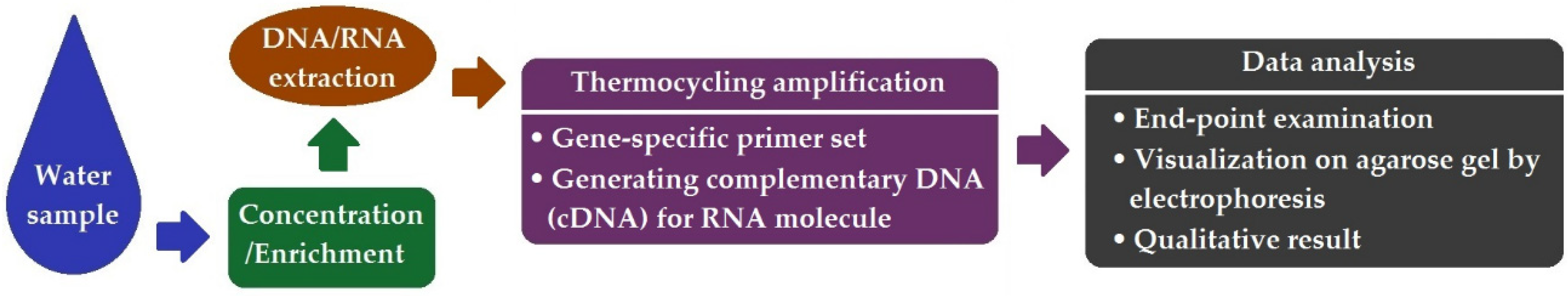



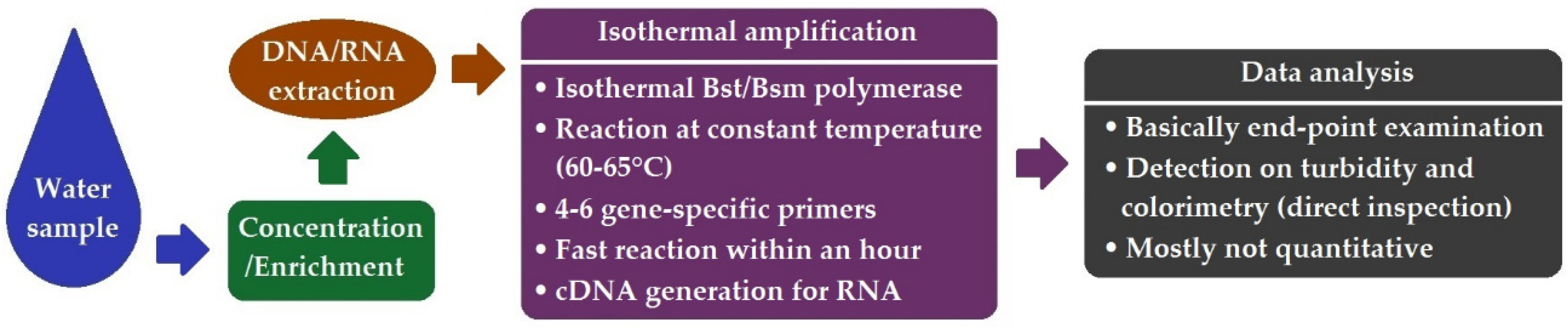

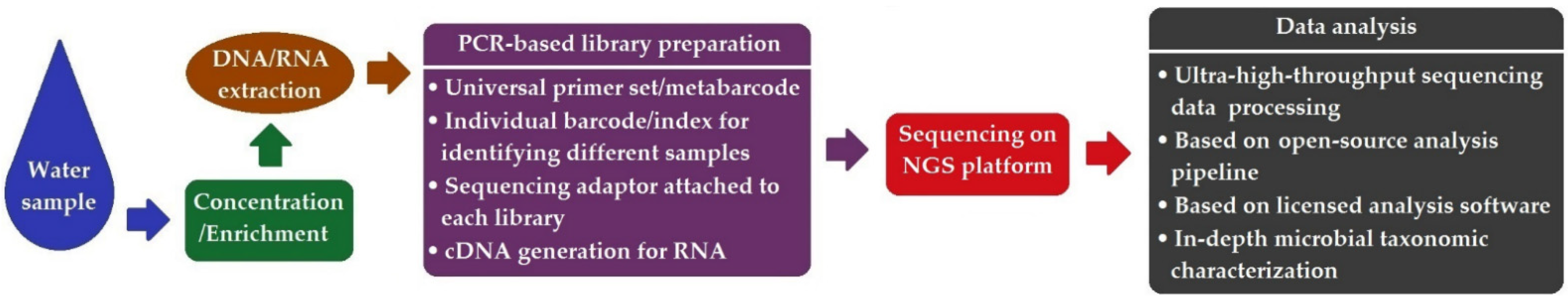
| Method | Pros | Cons |
|---|---|---|
| End-point PCR |
|
|
| DNA microarray |
|
|
| Quantitative real-time PCR (qPCR) |
|
|
| Multiplex qPCR (mqPCR) |
|
|
| Loop-mediated isothermal amplification (LAMP) |
|
|
| Droplet digital PCR (ddPCR) |
|
|
| Next-generation sequencing (NGS)-based |
|
|
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paruch, L. Molecular Diagnostic Tools Applied for Assessing Microbial Water Quality. Int. J. Environ. Res. Public Health 2022, 19, 5128. https://doi.org/10.3390/ijerph19095128
Paruch L. Molecular Diagnostic Tools Applied for Assessing Microbial Water Quality. International Journal of Environmental Research and Public Health. 2022; 19(9):5128. https://doi.org/10.3390/ijerph19095128
Chicago/Turabian StyleParuch, Lisa. 2022. "Molecular Diagnostic Tools Applied for Assessing Microbial Water Quality" International Journal of Environmental Research and Public Health 19, no. 9: 5128. https://doi.org/10.3390/ijerph19095128
APA StyleParuch, L. (2022). Molecular Diagnostic Tools Applied for Assessing Microbial Water Quality. International Journal of Environmental Research and Public Health, 19(9), 5128. https://doi.org/10.3390/ijerph19095128






