Embeddings for Efficient Literature Screening: A Primer for Life Science Investigators
Abstract
1. Introduction
2. A Semantic Space Odyssey
“She took a xylinth, and applying a very light pressure, she was able to remove most of the calculus.”
- (a)
- Red blood cells transport oxygen.
- (b)
- Inflamed tissues are red.
- (a)
- Red blood cells transport oxygen
- (a′)
- Oxygen transports red blood cells
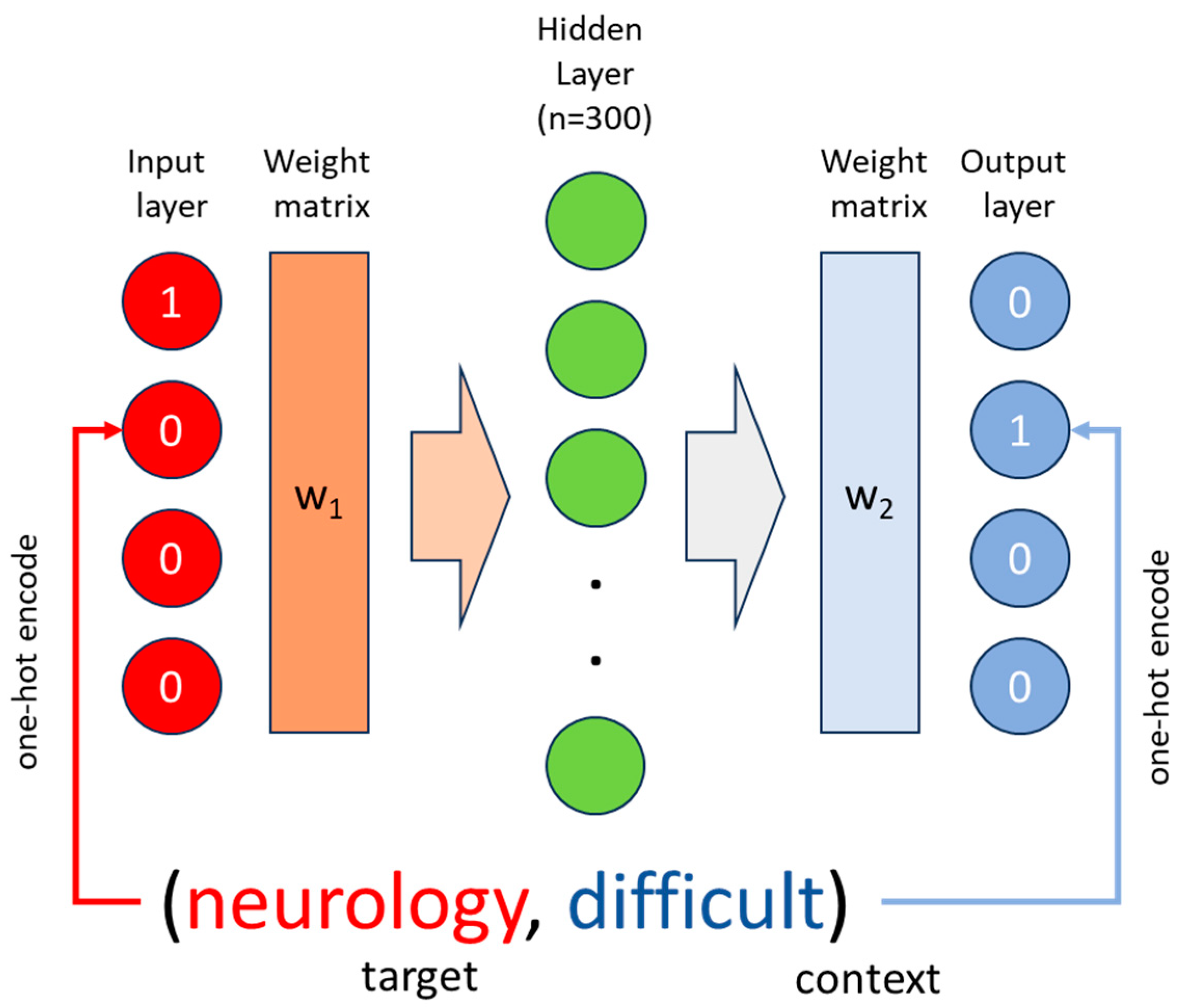
- (a)
- Neurology is a difficult but interesting topic
- (b)
- Neurology difficult interesting topic
| Neurology | = | [1,0,0,0] |
| Difficult | = | [0,1,0,0] |
| Interesting | = | [0,0,1,0] |
| Topic | = | [0,0,0,1] |
3. The Good, the Bad, the Ugly
4. Transformers, More than Meets the Eye
5. Far Away, So Close
- Porous titanium granules in the treatment of peri-implant osseous defects: a 7-year follow-up study, reconstruction of peri-implant osseous defects: a multicenter randomized trial [62],
- Porous titanium granules in the surgical treatment of peri-implant osseous defects: a randomized clinical trial [63],
- D-plex500: a local biodegradable prolonged-release doxycycline-formulated bone graft for the treatment of peri-implantitis. A randomized controlled clinical study [64],
- Surgical treatment of peri-implantitis with or without a deproteinized bovine bone mineral and a native bilayer collagen membrane: a randomized clinical trial [65],
- Effectiveness of the enamel matrix derivative on the clinical and microbiological outcomes following surgical regenerative treatment of peri-implantitis. A randomized controlled trial [66],
- Surgical treatment of peri-implantitis using enamel matrix derivative, an rct: 3- and 5-year follow-up [67],
- Surgical treatment of peri-implantitis lesions with or without the use of a bone substitute—a randomized clinical trial [68],
- Peri-implantitis—reconstructive surgical therapy [69].
| −2.29407530e−02, 1.49187818e−03, 9.16108266e−02, 1.75204929e−02, −8.36422145e−02, −6.10548146e−02, 8.30101445e−02, 3.96910682e−02, 1.58667186e−04, −2.62408387e−02, −7.69069120e−02, 4.60811984e−03, 8.64421800e−02, 7.87990764e−02, −4.33325134e−02, 2.49587372e−02, 2.24952400e−02, −2.90464610e−02, 3.59166898e−02, 4.27976809e−02, 7.94209242e−02, −5.87367006e−02, −6.49892315e−02, −8.70294198e−02, −5.51731326e−02, 4.95349243e−03, −3.01233679e−02, −3.23325321e−02, −1.54273247e−03, 5.24741262e−02, −7.11492598e−02, 5.16711324e−02, −4.42666225e−02, −6.38814121e−02, 6.46011531e−02, −4.63259555e−02, −9.23364013e−02, −3.56980823e−02, −9.30937752e−02, 1.27522862e−02, 5.05894162e−02, 5.07237464e−02, −9.00633708e−02, 6.91129547e−03, 4.79323231e−02, −6.69493945e−03, 1.27279535e−01, −6.33438602e−02, 2.78936550e−02, −3.34392674e−02, −6.21677283e−03, −4.32619415e−02, 5.89787960e−02, −9.10086110e−02, −2.79910862e−02, −5.80033176e−02, −5.82423434e−02, −6.41746866e−03, 4.17577056e−03, −1.90278993e−03, 6.72984421e−02, −4.39932309e−02, … 1.52898552e−02, 9.40597132e−02, −3.60338315e−02 |
6. Everything Everywhere All at Once
7. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Hanson, M.A.; Barreiro, P.G.; Crosetto, P.; Brockington, D. The Strain on Scientific Publishing. Quant. Sci. Stud. 2024, 1–29. [Google Scholar] [CrossRef]
- Landhuis, E. Scientific Literature: Information Overload. Nature 2016, 535, 457–458. [Google Scholar] [CrossRef] [PubMed]
- Dickersin, K.; Scherer, R.; Lefebvre, C. Systematic Reviews: Identifying Relevant Studies for Systematic Reviews. BMJ 1994, 309, 1286–1291. [Google Scholar] [CrossRef]
- Bramer, W.M.; Rethlefsen, M.L.; Kleijnen, J.; Franco, O.H. Optimal Database Combinations for Literature Searches in Systematic Reviews: A Prospective Exploratory Study. Syst. Rev. 2017, 6, 245. [Google Scholar] [CrossRef]
- Lu, Z. PubMed and beyond: A Survey of Web Tools for Searching Biomedical Literature. Database 2011, 2011, baq036. [Google Scholar] [CrossRef] [PubMed]
- Jin, Q.; Leaman, R.; Lu, Z. PubMed and beyond: Biomedical Literature Search in the Age of Artificial Intelligence. EBioMedicine 2024, 100, 104988. [Google Scholar] [CrossRef]
- Galli, C.; Cusano, C.; Meleti, M.; Donos, N. Topic Modeling for Faster Literature Screening Using Transformer-Based Embeddings. 2024. Available online: https://www.preprints.org/manuscript/202407.2198/v1 (accessed on 10 September 2024).
- Grivell, L. Mining the Bibliome: Searching for a Needle in a Haystack? EMBO Rep. 2002, 3, 200–203. [Google Scholar] [CrossRef]
- Wilczynski, N.L.; Haynes, R.B.; Team, H. Developing Optimal Search Strategies for Detecting Clinically Sound Prognostic Studies in MEDLINE: An Analytic Survey. BMC Med. 2004, 2, 23. [Google Scholar] [CrossRef]
- Zhang, L.; Ajiferuke, I.; Sampson, M. Optimizing Search Strategies to Identify Randomized Controlled Trials in MEDLINE. BMC Med. Res. Methodol. 2006, 6, 23. [Google Scholar] [CrossRef]
- Heintz, M.; Hval, G.; Tornes, R.A.; Byelyey, N.; Hafstad, E.; Næss, G.E.; Bakkeli, M. Optimizing the Literature Search: Coverage of Included References in Systematic Reviews in Medline and Embase. J. Med. Libr. Assoc. 2023, 111, 599–605. [Google Scholar] [CrossRef]
- Khalil, H.; Ameen, D.; Zarnegar, A. Tools to Support the Automation of Systematic Reviews: A Scoping Review. J. Clin. Epidemiol. 2022, 144, 22–42. [Google Scholar] [CrossRef] [PubMed]
- Samsir, S.; Saragih, R.S.; Subagio, S.; Aditiya, R.; Watrianthos, R. BERTopic Modeling of Natural Language Processing Abstracts: Thematic Structure and Trajectory. J. Media Inform. Budidarma 2023, 7, 1514. [Google Scholar] [CrossRef]
- Karabacak, M.; Margetis, K. Natural Language Processing Reveals Research Trends and Topics in The Spine Journal over Two Decades: A Topic Modeling Study. Spine J. 2024, 24, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Raman, R.; Pattnaik, D.; Hughes, L.; Nedungadi, P. Unveiling the Dynamics of AI Applications: A Review of Reviews Using Scientometrics and BERTopic Modeling. J. Innov. Knowl. 2024, 9, 100517. [Google Scholar] [CrossRef]
- Jurafsky, D.; Martin, J.H. Vector Semantics and Embeddings. In Speech and Language Processing: An Introduction to Natural Language Processing, Computational Linguistics, and Speech Recognition with Language Models, 3rd ed.; Online manuscript released 20 August; 2024; Available online: https://web.stanford.edu/~jurafsky/slp3 (accessed on 25 September 2024).
- Turney, P.D.; Pantel, P. From Frequency to Meaning: Vector Space Models of Semantics. J. Artif. Intell. Res. 2010, 37, 141–188. [Google Scholar] [CrossRef]
- Harris, Z.S. Distributional Structure. Word 1954, 10, 146–162. [Google Scholar] [CrossRef]
- Erk, K. Vector Space Models of Word Meaning and Phrase Meaning: A Survey. Lang. Linguist. Compass 2012, 6, 635–653. [Google Scholar] [CrossRef]
- Saif, H.; Fernandez, M.; He, Y.; Alani, H. On Stopwords, Filtering and Data Sparsity for Sentiment Analysis of Twitter. In Proceedings of the Ninth International Conference on Language Resources and Evaluation (LREC 2014), Reykjavik, Iceland, 26–31 May 2014. [Google Scholar]
- Zhang, Y.; Jin, R.; Zhou, Z.-H. Understanding Bag-of-Words Model: A Statistical Framework. Int. J. Mach. Learn. Cybern. 2010, 1, 43–52. [Google Scholar] [CrossRef]
- Jing, L.-P.; Huang, H.-K.; Shi, H.-B. Improved Feature Selection Approach TFIDF in Text Mining. In Proceedings of the International Conference on Machine Learning and Cybernetics, Beijing, China, 4–5 November 2002; IEEE: New York, NY, USA, 2002; Volume 2, pp. 944–946. [Google Scholar]
- Wang, S.; Zhou, W.; Jiang, C. A Survey of Word Embeddings Based on Deep Learning. Computing 2020, 102, 717–740. [Google Scholar] [CrossRef]
- Mikolov, T.; Chen, K.; Corrado, G.; Dean, J. Efficient Estimation of Word Representations in Vector Space. arXiv 2013, arXiv:1301.3781. [Google Scholar]
- Di Gennaro, G.; Buonanno, A.; Palmieri, F.A.N. Considerations about Learning Word2Vec. J. Supercomput. 2021, 77, 12320–12335. [Google Scholar] [CrossRef]
- Al-Saqqa, S.; Awajan, A. The Use of Word2vec Model in Sentiment Analysis: A Survey. In Proceedings of the 2019 international Conference on Artificial Intelligence, Robotics and Control, Cairo, Egypt, 14–16 December 2019; pp. 39–43. [Google Scholar]
- Haider, M.M.; Hossin, M.A.; Mahi, H.R.; Arif, H. Automatic Text Summarization Using Gensim Word2vec and K-Means Clustering Algorithm. In Proceedings of the 2020 IEEE Region 10 Symposium (TENSYMP), Dhaka, Bangladesh, 5–7 June 2020; IEEE: New York, NY, USA, 2020; pp. 283–286. [Google Scholar]
- Ibrohim, M.O.; Setiadi, M.A.; Budi, I. Identification of Hate Speech and Abusive Language on Indonesian Twitter Using the Word2vec, Part of Speech and Emoji Features. In Proceedings of the 1st International Conference on Advanced Information Science and System, Singapore, 15–17 November 2019; pp. 1–5. [Google Scholar]
- Jatnika, D.; Bijaksana, M.A.; Suryani, A.A. Word2vec Model Analysis for Semantic Similarities in English Words. Procedia Comput. Sci. 2019, 157, 160–167. [Google Scholar] [CrossRef]
- Hebart, M.N.; Zheng, C.Y.; Pereira, F.; Baker, C.I. Revealing the Multidimensional Mental Representations of Natural Objects Underlying Human Similarity Judgements. Nat. Hum. Behav. 2020, 4, 1173–1185. [Google Scholar] [CrossRef]
- Wang, B.; Sun, Y.; Chu, Y.; Yang, Z.; Lin, H. Global-Locality Preserving Projection for Word Embedding. Int. J. Mach. Learn. Cybern. 2022, 13, 2943–2956. [Google Scholar] [CrossRef]
- Liu, Q.; Kusner, M.J.; Blunsom, P. A Survey on Contextual Embeddings. arXiv 2020, arXiv:2003.07278. [Google Scholar]
- Kruse, R.; Mostaghim, S.; Borgelt, C.; Braune, C.; Steinbrecher, M. Multi-Layer Perceptrons. In Computational Intelligence: A Methodological Introduction; Springer: Cham, Switzerland, 2022; pp. 53–124. [Google Scholar]
- Salehinejad, H.; Sankar, S.; Barfett, J.; Colak, E.; Valaee, S. Recent Advances in Recurrent Neural Networks. arXiv 2017, arXiv:1801.01078. [Google Scholar]
- Hochreiter, S.; Schmidhuber, J. Long Short-Term Memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef]
- Cho, K.; Van Merriënboer, B.; Gulcehre, C.; Bahdanau, D.; Bougares, F.; Schwenk, H.; Bengio, Y. Learning Phrase Representations Using RNN Encoder-Decoder for Statistical Machine Translation. arXiv 2014, arXiv:1406.1078. [Google Scholar]
- Yin, W.; Kann, K.; Yu, M.; Schütze, H. Comparative Study of CNN and RNN for Natural Language Processing. arXiv 2017, arXiv:1702.01923. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention Is All You Need. Adv. Neural Inf. Process Syst. 2017, 30, 6000–6010. [Google Scholar]
- Chernyavskiy, A.; Ilvovsky, D.; Nakov, P. Transformers:“The End of History” for Natural Language Processing? In Machine Learning and Knowledge Discovery in Databases. Research Track: European Conference, ECML PKDD 2021, Bilbao, Spain, 13–17 September 2021; Proceedings, Part III 21; Springer: Cham, Switzerland, 2021; pp. 677–693. [Google Scholar]
- Patwardhan, N.; Marrone, S.; Sansone, C. Transformers in the Real World: A Survey on NLP Applications. Information 2023, 14, 242. [Google Scholar] [CrossRef]
- Goldstein, A.; Grinstein-Dabush, A.; Schain, M.; Wang, H.; Hong, Z.; Aubrey, B.; Schain, M.; Nastase, S.A.; Zada, Z.; Ham, E. Alignment of Brain Embeddings and Artificial Contextual Embeddings in Natural Language Points to Common Geometric Patterns. Nat. Commun. 2024, 15, 2768. [Google Scholar] [CrossRef] [PubMed]
- Wolf, T.; Debut, L.; Sanh, V.; Chaumond, J.; Delangue, C.; Moi, A.; Cistac, P.; Rault, T.; Louf, R.; Funtowicz, M.; et al. Transformers: State-of-the-Art Natural Language Processing. In Proceedings of the 2020 Conference on Empirical Methods in Natural Language Processing: System Demonstrations, Online, 16–20 November 2020; Association for Computational Linguistics: Stroudsburg, PA, USA, 2020; pp. 38–45. [Google Scholar]
- Galli, C.; Donos, N.; Calciolari, E. Performance of 4 Pre-Trained Sentence Transformer Models in the Semantic Query of a Systematic Review Dataset on Peri-Implantitis. Information 2024, 15, 68. [Google Scholar] [CrossRef]
- Guizzardi, S.; Colangelo, M.T.; Mirandola, P.; Galli, C. Modeling New Trends in Bone Regeneration, Using the BERTopic Approach. Regen. Med. 2023, 18, 719–734. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Chen, J.; Chen, J.; Chen, H. Identifying Interdisciplinary Topics and Their Evolution Based on BERTopic. Scientometrics 2023, 1–26. [Google Scholar] [CrossRef]
- Qian, J.; Kang, Y.; He, Y.; Hu, H. Topic Modeling Analysis of Chinese Medicine Literature on Gastroesophageal Reflux Disease: Insights into Potential Treatment. Chin. J. Integr. Med. 2024, 1–9. [Google Scholar] [CrossRef]
- Jeon, E.; Yoon, N.; Sohn, S.Y. Exploring New Digital Therapeutics Technologies for Psychiatric Disorders Using BERTopic and PatentSBERTa. Technol. Forecast. Soc. Chang. 2023, 186, 122130. [Google Scholar] [CrossRef]
- Lindelöf, G.; Aledavood, T.; Keller, B. Dynamics of the Negative Discourse toward COVID-19 Vaccines: Topic Modeling Study and an Annotated Data Set of Twitter Posts. J. Med. Internet Res. 2023, 25, e41319. [Google Scholar] [CrossRef]
- Li, H.; Lu, X.; Wu, Y.; Luo, J. Research on a Data Mining Algorithm Based on BERTopic for Medication Rules in Traditional Chinese Medicine Prescriptions. Med. Adv. 2023, 1, 353–360. [Google Scholar] [CrossRef]
- Karabacak, M.; Jagtiani, P.; Carrasquilla, A.; Jain, A.; Germano, I.M.; Margetis, K. Simplifying Synthesis of the Expanding Glioblastoma Literature: A Topic Modeling Approach. J. Neuro-Oncol. 2024, 169, 601–611. [Google Scholar] [CrossRef]
- Bramer, W.M.; De Jonge, G.B.; Rethlefsen, M.L.; Mast, F.; Kleijnen, J. A systematic approach to searching: An efficient and complete method to develop literature searches. J. Med. Libr. Assoc. JMLA. 2018, 106, 531–541. [Google Scholar] [CrossRef] [PubMed]
- Patrick, L.J.; Munro, S. The Literature Review: Demystifying the Literature Search. Diabetes Educ. 2004, 30, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Farouk, M. Measuring Text Similarity Based on Structure and Word Embedding. Cogn. Syst. Res. 2020, 63, 1–10. [Google Scholar] [CrossRef]
- Li, B.; Han, L. Distance Weighted Cosine Similarity Measure for Text Classification. In Intelligent Data Engineering and Automated Learning–IDEAL 2013, Proceedings of the 14th International Conference, IDEAL 2013, Hefei, China, 20–23 October 2013; Springer: Berlin/Heidelberg, Germany, 2013; pp. 611–618. [Google Scholar]
- Ivchenko, G.I.; Honov, S.A. On the Jaccard Similarity Test. J. Math. Sci. 1998, 88, 789–794. [Google Scholar] [CrossRef]
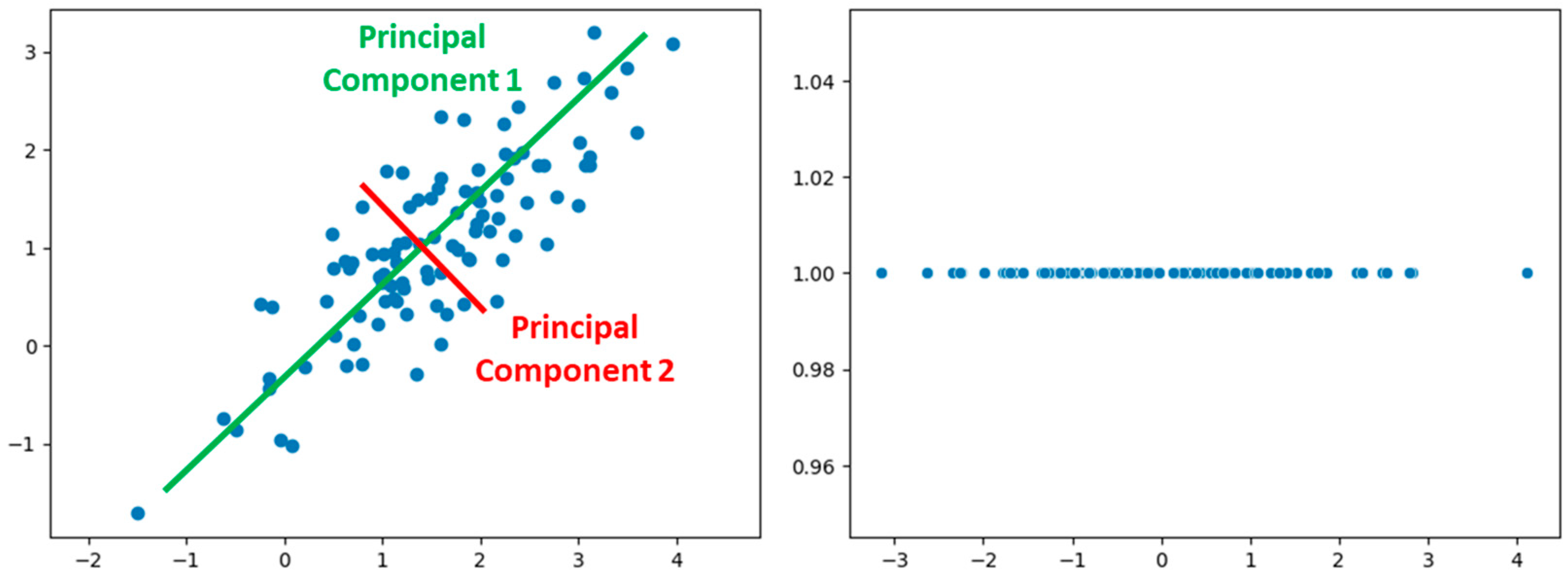
- Pearson, K. LIII. On Lines and Planes of Closest Fit to Systems of Points in Space. Lond. Edinb. Dublin Philos. Mag. J. Sci. 1901, 2, 559–572. [Google Scholar] [CrossRef]
- Labrín, C.; Urdinez, F. Principal Component Analysis. In R for Political Data Science; Chapman and Hall/CRC: Boca Raton, FL, USA, 2020; pp. 375–393. [Google Scholar]
- Van der Maaten, L.; Hinton, G. Visualizing Data Using T-SNE. J. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]
- Xanthopoulos, P.; Pardalos, P.M.; Trafalis, T.B.; Xanthopoulos, P.; Pardalos, P.M.; Trafalis, T.B. Linear Discriminant Analysis. In Robust Data Mining; Springer: New York, NY, USA, 2013; pp. 27–33. [Google Scholar]
- Friedrich, T. Nonlinear Dimensionality Reduction with Locally Linear Embedding and Isomap; University of Sheffield: Sheffield, UK, 2002. [Google Scholar]
- McInnes, L.; Healy, J.; Melville, J. Umap: Uniform Manifold Approximation and Projection for Dimension Reduction. arXiv 2018, arXiv:1802.03426. [Google Scholar]
- Andersen, H.; Aass, A.M.; Wohlfahrt, J.C. Porous Titanium Granules in the Treatment of Peri-Implant Osseous Defects—A 7-Year Follow-up Study. Int. J. Implant. Dent. 2017, 3, 50. [Google Scholar] [CrossRef]
- Wohlfahrt, J.C.; Lyngstadaas, S.P.; Rønold, H.J.; Saxegaard, E.; Ellingsen, J.E.; Karlsson, S.; Aass, A.M. Porous Titanium Granules in the Surgical Treatment of Peri-Implant Osseous Defects: A Randomized Clinical Trial. Int. J. Oral Maxillofac. Implant. 2012, 27, 401–410. [Google Scholar]
- Emanuel, N.; Machtei, E.E.; Reichart, M.; Shapira, L. D-PLEX. Quintessence Int. 2020, 51, 546–553. [Google Scholar]
- Renvert, S.; Giovannoli, J.; Roos-Jansåker, A.; Rinke, S. Surgical Treatment of Peri-implantitis with or without a Deproteinized Bovine Bone Mineral and a Native Bilayer Collagen Membrane: A Randomized Clinical Trial. J. Clin. Periodontol. 2021, 48, 1312–1321. [Google Scholar] [CrossRef] [PubMed]
- Isehed, C.; Holmlund, A.; Renvert, S.; Svenson, B.; Johansson, I.; Lundberg, P. Effectiveness of Enamel Matrix Derivative on the Clinical and Microbiological Outcomes Following Surgical Regenerative Treatment of Peri-implantitis. A randomized controlled trial. J. Clin. Periodontol. 2016, 43, 863–873. [Google Scholar] [PubMed]
- Isehed, C.; Svenson, B.; Lundberg, P.; Holmlund, A. Surgical Treatment of Peri-implantitis Using Enamel Matrix Derivative, an RCT: 3-and 5-year Follow-up. J. Clin. Periodontol. 2018, 45, 744–753. [Google Scholar] [CrossRef]
- Renvert, S.; Roos-Jansåker, A.; Persson, G.R. Surgical Treatment of Peri-implantitis Lesions with or without the Use of a Bone Substitute—A Randomized Clinical Trial. J. Clin. Periodontol. 2018, 45, 1266–1274. [Google Scholar] [CrossRef] [PubMed]
- Nct Peri-Implantitis—Reconstructive Surgical Therapy. 2017. Available online: https://clinicaltrials.gov/show/NCT03077061 (accessed on 10 April 2022).
- Ahmad, N.; Nassif, A.B. Dimensionality Reduction: Challenges and Solutions. In ITM Web of Conferences; EDP Sciences: Les Ulis, France, 2022; Volume 43, p. 01017. [Google Scholar]
- Sumithra, V.; Surendran, S. A Review of Various Linear and Non Linear Dimensionality Reduction Techniques. Int. J. Comput. Sci. Inf. Technol. 2015, 6, 2354–2360. [Google Scholar]
- Zebari, R.; Abdulazeez, A.; Zeebaree, D.; Zebari, D.; Saeed, J. A Comprehensive Review of Dimensionality Reduction Techniques for Feature Selection and Feature Extraction. J. Appl. Sci. Technol. Trends 2020, 1, 56–70. [Google Scholar] [CrossRef]
- Liu, X.; Zheng, Y.; Du, Z.; Ding, M.; Qian, Y.; Yang, Z.; Tang, J. GPT Understands, Too; AI Open: San Francisco, CA, USA, 2023. [Google Scholar]
- Thirunavukarasu, A.J.; Ting, D.S.J.; Elangovan, K.; Gutierrez, L.; Tan, T.F.; Ting, D.S.W. Large Language Models in Medicine. Nat. Med. 2023, 29, 1930–1940. [Google Scholar] [CrossRef]
- Kaddour, J.; Harris, J.; Mozes, M.; Bradley, H.; Raileanu, R.; McHardy, R. Challenges and Applications of Large Language Models. arXiv 2023, arXiv:2307.10169. [Google Scholar]
- Wei, J.; Tay, Y.; Bommasani, R.; Raffel, C.; Zoph, B.; Borgeaud, S.; Yogatama, D.; Bosma, M.; Zhou, D.; Metzler, D. Emergent Abilities of Large Language Models. arXiv 2022, arXiv:2206.07682. [Google Scholar]
- Hersh, W.R. Search Still Matters: Information Retrieval in the Era of Generative AI. arXiv 2023, arXiv:2311.18550. [Google Scholar] [CrossRef]
- Zhu, Y.; Yuan, H.; Wang, S.; Liu, J.; Liu, W.; Deng, C.; Dou, Z.; Wen, J.-R. Large Language Models for Information Retrieval: A Survey. arXiv 2023, arXiv:2308.07107. [Google Scholar]
- Hadi, M.U.; Qureshi, R.; Shah, A.; Irfan, M.; Zafar, A.; Shaikh, M.B.; Akhtar, N.; Wu, J.; Mirjalili, S. A Survey on Large Language Models: Applications, Challenges, Limitations, and Practical Usage. Authorea Preprints 2023. Available online: https://www.techrxiv.org/doi/full/10.36227/techrxiv.23589741.v1 (accessed on 10 September 2024).
- Lozano, A.; Fleming, S.L.; Chiang, C.-C.; Shah, N. Clinfo. Ai: An Open-Source Retrieval-Augmented Large Language Model System for Answering Medical Questions Using Scientific Literature. In Proceedings of the Pacific Symposium on Biocomputing 2024, Waimea, HI, USA, 3–7 January 2024; World Scientific: Singapore, 2023; pp. 8–23. [Google Scholar]
- Agarwal, S.; Laradji, I.H.; Charlin, L.; Pal, C. LitLLM: A Toolkit for Scientific Literature Review. arXiv 2024, arXiv:2402.01788. [Google Scholar]


| W | Red | Blood | Cell | To Transport | Oxygen | Inflamed | Tissue | To Be |
|---|---|---|---|---|---|---|---|---|
| Sent. (A) | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 |
| Sent. (B) | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Galli, C.; Cusano, C.; Guizzardi, S.; Donos, N.; Calciolari, E. Embeddings for Efficient Literature Screening: A Primer for Life Science Investigators. Metrics 2024, 1, 1. https://doi.org/10.3390/metrics1010001
Galli C, Cusano C, Guizzardi S, Donos N, Calciolari E. Embeddings for Efficient Literature Screening: A Primer for Life Science Investigators. Metrics. 2024; 1(1):1. https://doi.org/10.3390/metrics1010001
Chicago/Turabian StyleGalli, Carlo, Claudio Cusano, Stefano Guizzardi, Nikolaos Donos, and Elena Calciolari. 2024. "Embeddings for Efficient Literature Screening: A Primer for Life Science Investigators" Metrics 1, no. 1: 1. https://doi.org/10.3390/metrics1010001
APA StyleGalli, C., Cusano, C., Guizzardi, S., Donos, N., & Calciolari, E. (2024). Embeddings for Efficient Literature Screening: A Primer for Life Science Investigators. Metrics, 1(1), 1. https://doi.org/10.3390/metrics1010001







