Topological Data Analysis (TDA) provides a mathematical framework for uncovering the intrinsic geometric and topological structures underlying data. A central tool in TDA is persistent homology, which tracks the evolution of topological features (e.g., connected components, loops, and voids) across multiple scales. The resulting output is often summarized in the form of a persistence diagram (PD), a multiset of points (A multiset of points means that different topological features can have the same birth and death times in the persistence diagram.) , where each point corresponds to a topological feature that appears (is “born”) at scale b and disappears (or “dies”) at scale d (see Figure 5). These diagrams serve as topological signatures of the data, capturing information such as connected components, loops, and voids across multiple scale values.
To compute a persistence diagram, we must first equip the data (often given as a point cloud, image, or graph) with a topological structure through the construction of a simplicial complex. A simplicial complex provides a combinatorial framework that captures relationships between data points. This is accomplished through various constructions, such as the Vietoris–Rips, Čech, or lower star filtration, which approximate the underlying topological structure of the space from which the data were sampled. This process allows for the systematic study of topological features using tools from algebraic topology, such as homology and persistent homology.
An
(abstract) simplicial complex K is a collection of finite subsets of a vertex set
V, satisfying the following two properties. First, every vertex
must be included as a singleton set
. Second, the complex must be closed under taking subsets, i.e., if
, then any subset
is also in
K. In this context, any subset
containing
vertices is called a
p-simplex and has dimension
p, denoted
. A subset
is called a
face of
, written
. The set of all simplices in
K of dimension
d is denoted by
, and the overall dimension of
K is the highest dimension among its simplices. There is a geometric realization of simplicial complexes in a suitable Euclidean space, in which a vertex corresponds to a 0-simplex, an edge to a 1-simplex, a filled triangle to a 2-simplex, and a tetrahedron to a 3-simplex, and so on (see
Figure 1); in this realization, different simplices intersect only along their common faces. Example 1 illustrates both the concept of a simplicial complex and the process by which it is constructed.
A key step in TDA involves constructing a filtration—a nested sequence of simplicial complexes that progressively captures the evolving topological features of the data at multiple scales or thresholds. One common approach to building filtrations is through the sublevel sets of a monotone function defined on the simplices of a simplicial complex. Specifically, let
K be a simplicial complex, and let
be a real-valued function defined on its simplices. The function
g is said to be monotone if, for every pair of simplices
, it satisfies
. This condition ensures that for each threshold
, the sublevel set
forms a valid simplicial subcomplex of
K, because if a simplex is in
, its faces are also guaranteed to be in
by the monotonicity property. Let
denote the ordered set of distinct function values taken by
g. The sublevel set filtration induced by
g is then given by the nested sequence:
where each
. The number of filtration steps
N corresponds to the number of unique function values attained by
g over the simplices in
K.
2.1.1. Homology and Persistent Homology
To understand how topological features such as connected components, loops, and voids evolve across a filtration, we need algebraic tools that can precisely capture these structures. This is exactly the role of homology, which formalizes these features using algebraic objects called homology groups.
Given a sequence of nested simplicial complexes—called a filtration—we can apply homology to each complex to systematically identify and track topological features across multiple scales. The technique of
persistent homology tracks how these features, such as connected components (
), loops (
), and voids (
), evolve and persist over the course of the filtration. At each stage, a simplicial complex
K is constructed from a set of vertices, edges, triangles, and possibly higher-dimensional simplices. To uncover the topological information contained in these complexes, we associate to them algebraic structures called
chain groups. For each dimension
d, the chain group
is defined as the vector space (or free module) generated by the
d-simplices in
K, with coefficients chosen in a fixed field (
). An element of
is called a
d-chain. It is a linear combination of
d-simplices, written as
where the
are
d-simplices in
K and the
are coefficients from the chosen field. Next, we connect different dimensions using
boundary operators . These operators map each
d-simplex to a sum of its
-dimensional faces. For example, for a simplex
,
where
means that the vertex
is omitted. When working over
, we can ignore the signs and simply track whether faces are present or absent.
A key property of the boundary operator is that applying it twice yields zero, i.e., We define a d-cycle as a d-chain whose boundary is zero, i.e., it is in the kernel of the boundary operator . This space is denoted by . Similarly, a d-boundary is a d-chain that is the boundary of some -chain. These form the image of the boundary operator , denoted by . Because applying the boundary operator twice yields zero (), every boundary is automatically a cycle. This means we have the following subgroup relationship:
The
d-th homology group is then defined as the quotient:
which captures cycles that are not boundaries. The dimension of
, called the
d-th Betti number,
, counts the number of independent
d-dimensional holes. It is calculated as
. For
,
is interpreted as counting the number of independent loops (
) and higher-dimensional voids (
for
). In ordinary homology,
counts the number of connected components. However, in some contexts, particularly for single-point spaces, reduced homology is used, where the zeroth reduced Betti number is one less than the number of connected components [
23].
Persistent homology extends homology to filtrations, allowing us to track how topological features appear and disappear across multiple scales. As we build the simplicial complex in the filtration, topological features are both born and destroyed. For example, new connected components emerge (increasing the Betti number
) and can later merge with other components (decreasing
). Similarly, new loops can be created by the addition of edges (increasing
) and are destroyed when a triangle fills them in (decreasing
). This dynamic process is captured in a persistence diagram, which records the birth and death of each topological feature, providing a concise summary of its lifespan across the filtration. Given a filtration
the inclusion maps
induce homomorphisms between homology groups:
Each homology class can be associated with a birth time,
, indicating when it first appears in the filtration, and a death time,
, indicating when it either becomes trivial or merges into an older feature. Collecting these persistence intervals,
, across all dimensions yields the persistence diagram for a given homological degree
d, which provides a concise summary of the multiscale topological features present in the data.
Although persistent homology is a highly general framework that can be applied to filtrations of any topological space, in this work, we focus on filtrations constructed from scalar functions defined on the nodes of a graph. A simple and widely adopted method for building such filtrations is the lower-star filtration, which constructs nested subcomplexes based on real-valued functions assigned to the vertices. This approach is especially advantageous when distance metrics are not available or do not make sense, or when meaningful scalar attributes—such as structural properties or centrality scores—can be derived from the graph and assigned to its nodes.
Let be an undirected graph, where V is the set of nodes, is the set of edges, and assigns real-valued attributes to the nodes. This graph can be naturally interpreted as a one-dimensional simplicial complex; the nodes are 0-simplices and edges are 1-simplices. To capture more complex topological structures beyond pairwise connections, we extend this to higher-dimensional simplices. For instance, a triangle formed by three mutually connected nodes corresponds to a 2-simplex, and a tetrahedron formed by four fully connected nodes corresponds to a 3-simplex, and so forth. The resulting complex, often called a clique complex or flag complex, encodes all complete subgraphs of the original graph as simplices. For notational simplicity, we continue to denote this extended simplicial complex by G.
To analyze the topology of G, we build a filtration by extending the function g from the vertices to all simplices using the maximum rule. Specifically, for any simplex (viewed as a simplicial complex), we define This extension ensures monotonicity; if , then . As a result, the sublevel sets of g form valid simplicial complexes, which allows us to define a filtration. The resulting Lower-Star filtration is defined as a sequence of nested subcomplexes where the filtration parameter t increases over the range of values taken by g. In this construction, a simplex enters the filtration at the smallest threshold t such that all its vertices satisfy . Because the value on a simplex depends on the maximum among its vertices, this method provides a consistent, hierarchical way to explore the topological features of the graph.
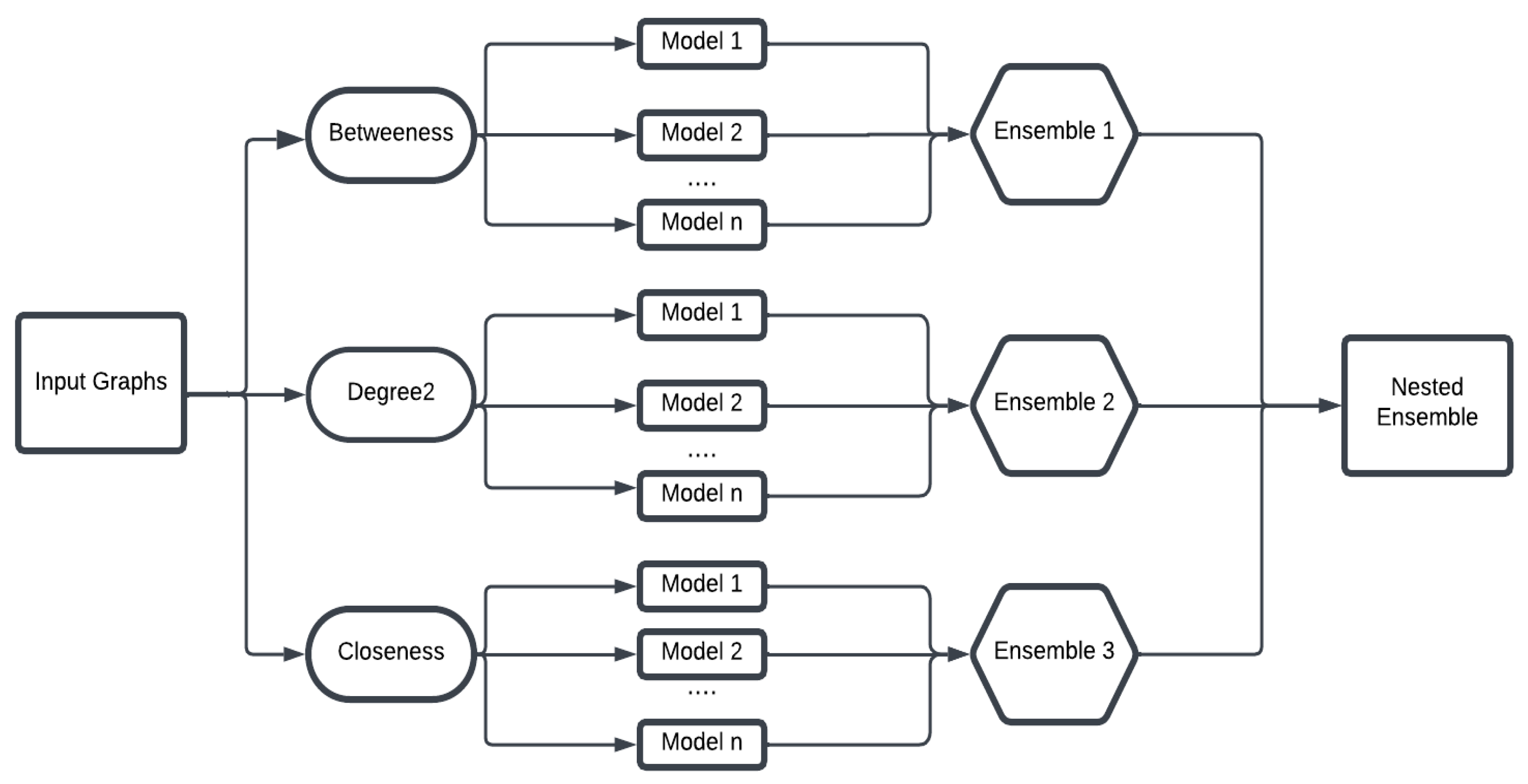
In the Lower-Star filtration, the choice of function values assigned to the vertices is critical, as it directly determines how the filtration unfolds and what topological features are captured. These function values act as filtration parameters, deciding the exact stage at which each simplex enters the filtration, ultimately shaping the topological summary we obtain. In this work, we define the vertex function g based on three types of node attributes: closeness centrality, betweenness centrality, and a custom measure we call degree2 centrality. Closeness centrality measures how close a node is, on average, to all other nodes in the network, based on the shortest-path distances. When analyzing unweighted graphs, each edge is treated as having unit length, and distances between nodes are computed as the number of edges along the shortest path. This node-level measure reflects the overall accessibility or influence of a node within the graph. Betweenness centrality measures how often a node lies on the shortest paths between other nodes, highlighting nodes that serve as bridges or bottlenecks within the graph. The degree2 centrality metric is defined as the count of unique nodes connected to a given node through paths of length one or two. By incorporating nodes at a distance of two hops, this measure provides a more extensive characterization of a node’s local neighborhood structure than that offered by standard degree centrality.
Each of these filter functions induces a distinct ordering of simplices, which governs how connected components, cycles, and other topological features appear and persist across scales. Applying persistent homology to these filtrations yields a persistence diagram that records the birth and death of each topological feature as the threshold t varies. These diagrams provide compact, multi-scale summaries of the graph’s topological structure relative to the chosen function g. (See Example 2 for an illustration of how lower-star filtrations can be used to compute persistence diagrams.)
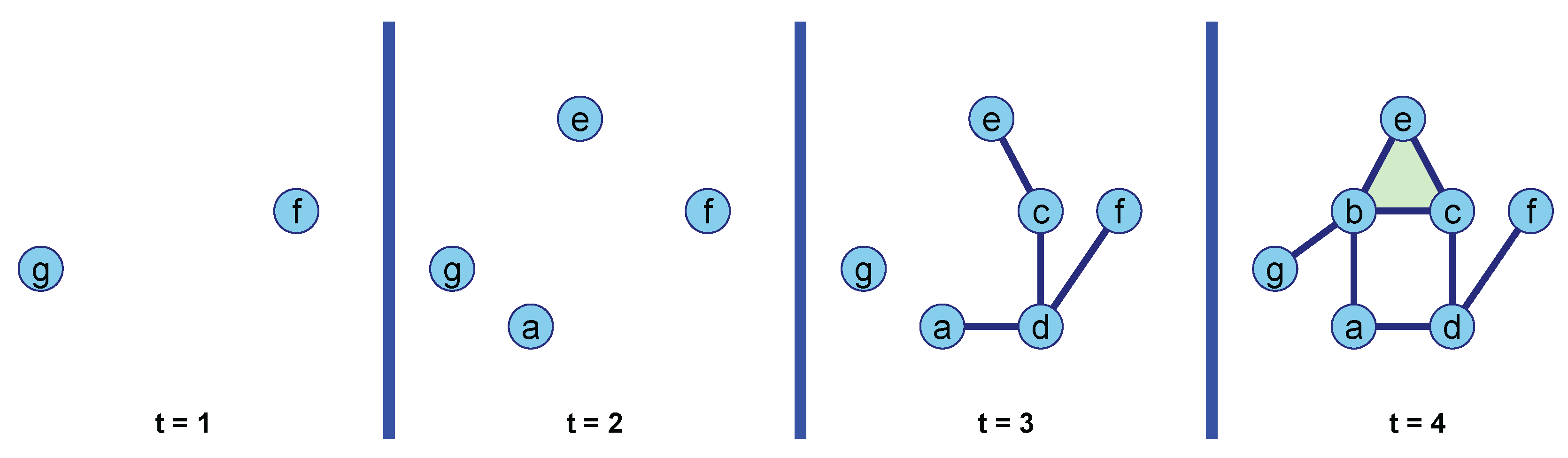
Example 2. Consider a graph with vertex set and edge set . This graph can be viewed as a 1-dimensional simplicial complex with seven vertices (0-simplices) and eight edges (1-simplices). We extend this complex by adding a 2-simplex, the triangle , as shown in Figure 3. To analyze the topological structure of this complex, we define a filter function based on the degree of each vertex, such that , where denotes the number of edges incident to vertex v. The degree values for each vertex are: We construct the lower-star filtration of the graph step by step. At each filtration step t, we include all simplices such that .
Step : We begin with an empty complex . At a filtration value of 1, we add vertices f and g since . No edges are included at this stage. Thus, the complex is .
Step : We include vertices a and e, as their degree values are . No edges are added because all neighboring vertices have higher degree values. The complex is now .
Step : We add vertices c and d, with degree values of 3. At this step, several edges are also included: , , , and , as the maximum degree of their endpoints is at most 3. The complex becomes .
Step
: Finally, we add vertex
b and all remaining simplices. All edges with a maximum vertex degree of 4—
,
,
, and
—are added. The 2-simplex, the triangle
, is also added since its maximum vertex degree is 4. The final complex is
.
Figure 4 visually demonstrates how the scalar function
g induces a lower-star filtration on the graph
G, as described above.
To compute the PD, we observe how topological features evolve as the filtration parameter
t increases. At
, two connected components are born; one persists indefinitely, while the other dies at
when it merges with another component through the formation of an edge (a 1-simplex). At
, two more connected components are born; both of these die at
as they merge with existing components via edge formation. At
, four more edges and a triangle (2-simplex) are added, which results in the birth of a loop. Since this loop is never filled in by a triangle at any later stage in the filtration, it persists indefinitely and is assigned an infinite death time in the PD. The PD resulting from the lower-star filtration is visualized in
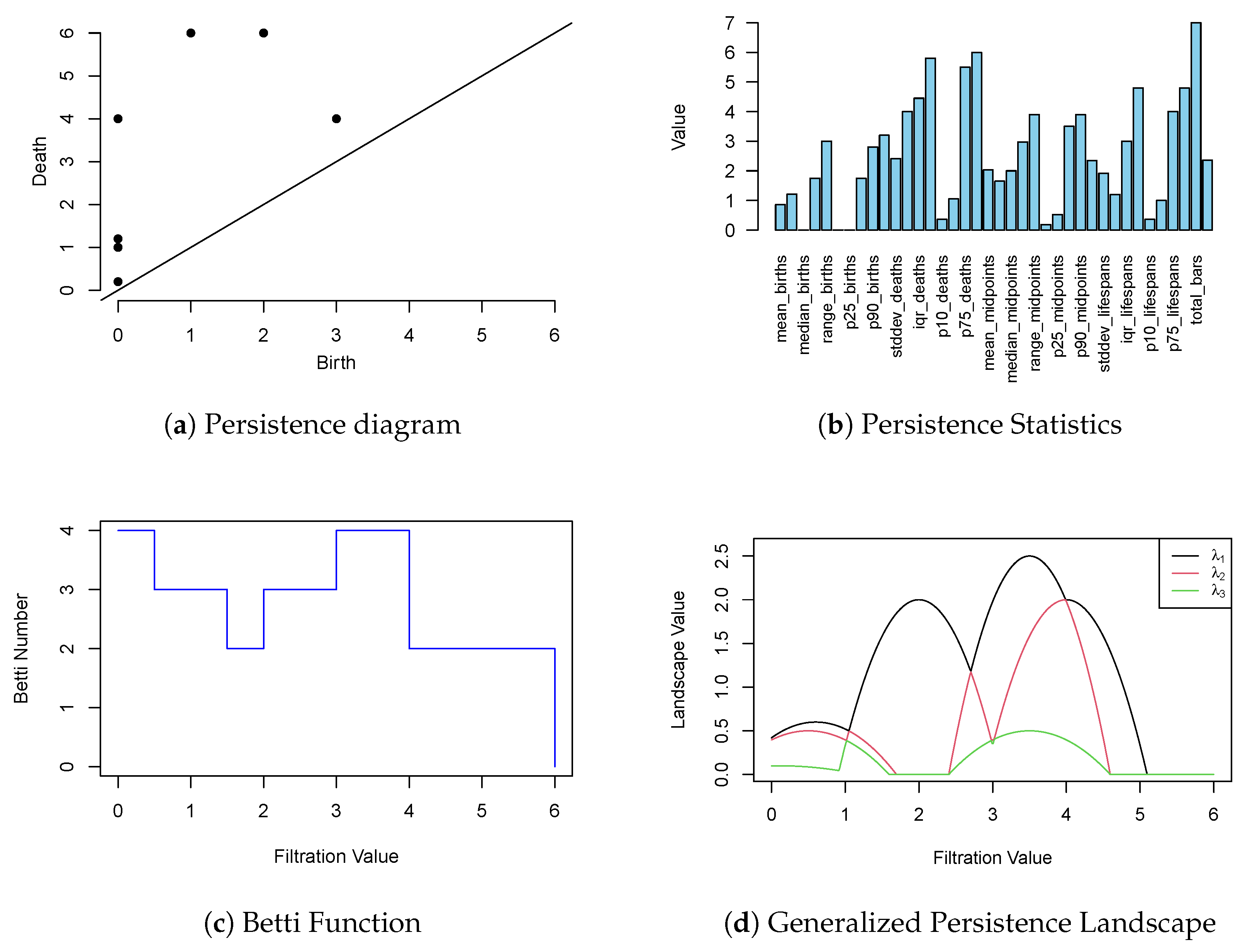
Figure 5 and detailed numerically in
Table 1. In practical applications, features with infinite death values—representing topological structures that never disappear—are handled differently depending on the problem at hand. They are often either excluded from the diagram or assigned a suitable finite constant to facilitate analysis and computation. However, in scenarios where the PD is used directly (e.g., with kernel methods or distance-based comparisons), it is often preferable to retain infinite death values, as they may carry meaningful topological information about essential or dominant features of the space.
Figure 5.
Visual representation of the persistence diagram shown in
Table 1, where each point corresponds to a topological feature characterized by its birth and death values.
Figure 5.
Visual representation of the persistence diagram shown in
Table 1, where each point corresponds to a topological feature characterized by its birth and death values.
Table 1.
Persistence diagram for the lower-star filtration of the graph in
Figure 3. Each row represents a topological feature with its dimension, birth time, and death time. Infinite death values indicate features that persist throughout the entire filtration.
Table 1.
Persistence diagram for the lower-star filtration of the graph in
Figure 3. Each row represents a topological feature with its dimension, birth time, and death time. Infinite death values indicate features that persist throughout the entire filtration.
| Dimension | Birth | Death |
|---|
| 0 | 1 | ∞ |
| 0 | 1 | 4 |
| 0 | 2 | 3 |
| 0 | 2 | 3 |
| 1 | 4 | ∞ |