Counting Polynomials in Chemistry II
Abstract
1. Introduction
2. μ-Polynomial
3. F-Polynomial
4. β-Polynomial
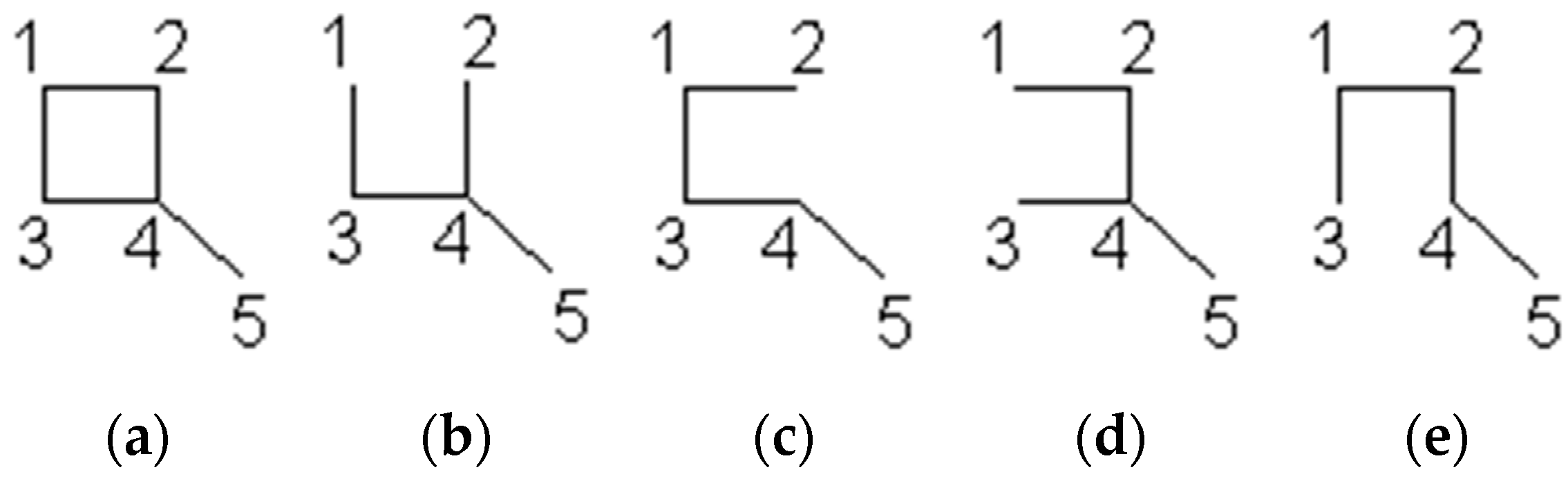
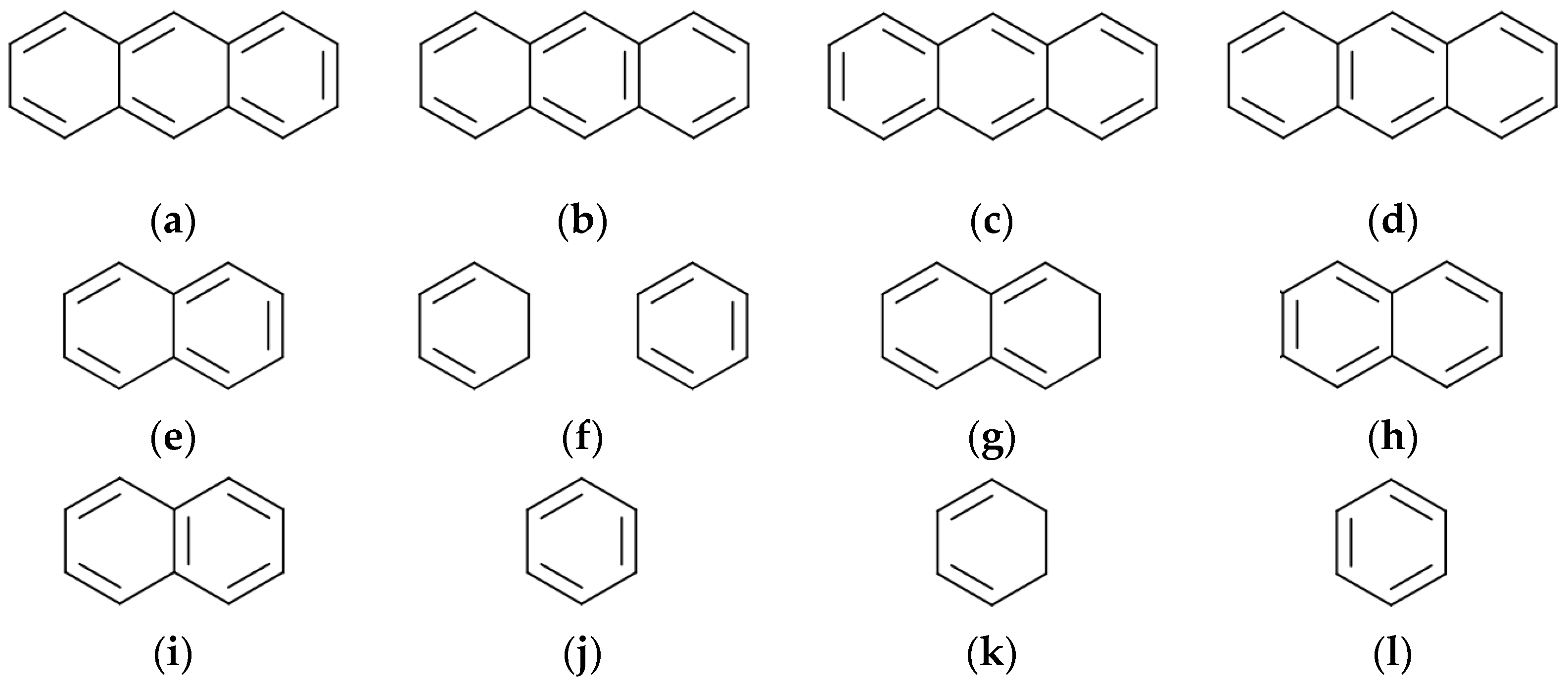
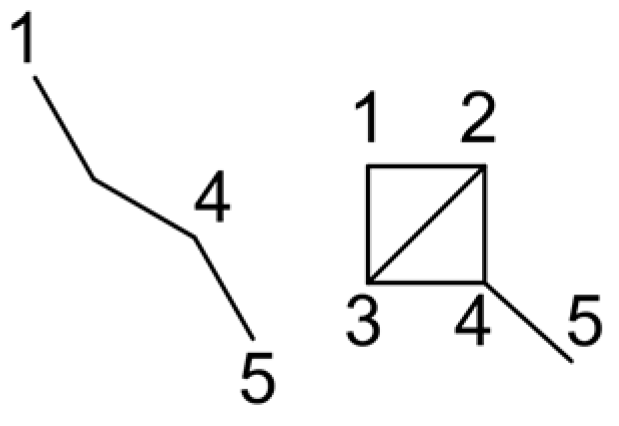
5. Cyclic Polynomial (Knop and Trinajstić)
- for m ≥ 0, and k > 1, it is not possible to find more than one disconnected conjugated ring in graph G2. Therefore, what is left is k = 1 and to describe the cases of m = 1, 2, and 3;
- for m = 3 and k = 1, n(G − Cy3; 1) = 1, as the result is the empty graph after eliminating the 14-atom ring.
- As such, .
6. Chromatic Polynomial
7. Tutte Polynomial
8. Other Polynomials
8.1. Information Polynomial
8.2. Orbit Polynomial
8.3. B-Polynomial
8.4. Clar Covering Polynomial
8.5. Forcing and Anti-Forcing Polynomials
8.6. Some More Polynomials
9. Discussion
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Jäntschi, L.; Bolboacă, S.-D. Characteristic Polynomial. In New Frontiers in Nanochemistry: Concepts, Theories, and Trends; Putz, M.V., Ed.; Apple Academic Press: Burlington, NJ, USA, 2020; Volume 2, pp. 95–118. ISBN 978-1-77188-780-9. [Google Scholar]
- Jäntschi, L.; Bolboacă, S.-D. Counting Polynomials. In New Frontiers in Nanochemistry: Concepts, Theories, and Trends; Putz, M.V., Ed.; Apple Academic Press: Burlington, NJ, USA, 2020; Volume 2, pp. 141–148. ISBN 978-1-77188-780-9. [Google Scholar]
- Brezovnik, S.; Dehmer, M.; Tratnik, N.; Žigert Pleteršek, P. Szeged and Mostar Root-Indices of Graphs. Appl. Math. Comput. 2023, 442, 127736. [Google Scholar] [CrossRef]
- Joița, D.-M.; Tomescu, M.A.; Jäntschi, L. Counting Polynomials in Chemistry: Past, Present, and Perspectives. Symmetry 2023, 15, 1815. [Google Scholar] [CrossRef]
- Jäntschi, L.; Bolboacă, S.-D. Informational Entropy of B-Ary Trees after a Vertex Cut. Entropy 2008, 10, 576–588. [Google Scholar] [CrossRef]
- Jäntschi, L.; Diudea, M.V. Subgraphs of Pair Vertices. J. Math. Chem. 2009, 45, 364–371. [Google Scholar] [CrossRef]
- Jäntschi, L.; Bălan, M.C.; Bolboacă, S.-D. Counting Polynomials on Regular Iterative Structures. Appl. Med. Inform. 2009, 24, 67–95. [Google Scholar]
- Bolboacă, S.-D.; Jäntschi, L. Counting Distance and Szeged (on Distance) Polynomials in Dodecahedron Nano-Assemblies. In Distance, Symmetry, and Topology in Carbon Nanomaterials; Ashrafi, A.R., Diudea, M.V., Eds.; Springer International Publishing: Cham, Switzerland, 2016; Volume 9, pp. 391–408. ISBN 978-3-319-31582-9. [Google Scholar]
- Putz, M.V.; Mirica, M.C. (Eds.) Sustainable Nanosystems Development, Properties, and Applications. In Advances in Chemical and Materials Engineering; IGI Global: Hershey, PA, USA, 2017; ISBN 978-1-5225-0492-4. [Google Scholar]
- Joiţa, D.-M.; Jäntschi, L. Extending the Characteristic Polynomial for Characterization of C20 Fullerene Congeners. Mathematics 2017, 5, 84. [Google Scholar] [CrossRef]
- Jäntschi, L. The Eigenproblem Translated for Alignment of Molecules. Symmetry 2019, 11, 1027. [Google Scholar] [CrossRef]
- Jäntschi, L. Characteristic and Counting Polynomials of Nonane Isomers; AcademicDirect: Cluj-Napoca, Romania, 2007. [Google Scholar]
- Bolboacă, S.-D.; Jäntschi, L. How Good Can the Characteristic Polynomial Be for Correlations? Int. J. Mol. Sci. 2007, 8, 335–345. [Google Scholar] [CrossRef]
- Jäntschi, L.; Bolboacă, S.-D.; Furdui, C.M. Characteristic and Counting Polynomials: Modelling Nonane Isomers Properties. Mol. Simul. 2009, 35, 220–227. [Google Scholar] [CrossRef]
- Harary, F. Graph Theory; Addison-Wesley Series in Mathematics; Addison-Wesley: Reading, MA, USA, 1964. [Google Scholar]
- Bonchev, D.; Rouvray, D.H. (Eds.) Chemical Graph Theory: Introduction and Fundamentals; Mathematical Chemistry; Abacus Press: New York, NY, USA, 1991; ISBN 978-0-85626-454-2. [Google Scholar]
- Joiţa, D.-M.; Tomescu, M.A.; Bàlint, D.; Jäntschi, L. An Application of the Eigenproblem for Biochemical Similarity. Symmetry 2021, 13, 1849. [Google Scholar] [CrossRef]
- Jäntschi, L.; Bálint, D.; Pruteanu, L.L.; Bolboacă, S.-D. Elemental Factorial Study on One-Cage Pentagonal Face Nanostructure Congeners. Mater. Discov. 2016, 5, 14–21. [Google Scholar] [CrossRef]
- Jäntschi, L.; Bolboacă, S.-D. Nanoporous Carbon. In New Frontiers in Nanochemistry; Putz, M.V., Ed.; Apple Academic Press: New York, NY, USA, 2020; pp. 313–325. ISBN 978-0-429-02294-4. [Google Scholar]
- Janežič, D.; Jäntschi, L.; Bolboacă, S.-D. Sugars and Sweeteners: Structure, Properties and In Silico Modeling. CMC 2020, 27, 5–22. [Google Scholar] [CrossRef] [PubMed]
- Gutman, I.; Polansky, O.E. Cyclic Conjugation and the Hückel Molecular Orbital Model. Theoret. Chim. Acta 1981, 60, 203–226. [Google Scholar] [CrossRef]
- Farrell, E.J. On a General Class of Graph Polynomials. J. Comb. Theory B 1979, 26, 111–122. [Google Scholar] [CrossRef]
- Gutman, I. Difficulties with Topological Resonance Energy. Chem. Phys. Lett. 1979, 66, 595–597. [Google Scholar] [CrossRef]
- Farrell, E.J.; Wahid, S.A. Some General Classes of Comatching Graphs. Int. J. Math. Sci. 1987, 10, 519–524. [Google Scholar] [CrossRef]
- Radenković, S.; Đurđević, J.; Gutman, I. Quantitative Study of the PCP Effect. Chem. Phys. Lett. 2009, 475, 289–292. [Google Scholar] [CrossRef]
- Knop, J.V.; Trinajstić, N. Chemical Graph Theory. II. On the Graph Theoretical Polynomials of Conjugated Structures. Int. J. Quantum Chem. 2009, 18, 503–520. [Google Scholar] [CrossRef]
- Diudea, M.V.; Gutman, I.; Jäntschi, L. Molecular Topology, 1st ed.; Nova Science Publishers: Hauppauge, NY, USA, 2001. [Google Scholar]
- Gutman, I. General Theory of Cycle-Dependence of Total Pi-Electron Energy. Iran. J. Math. Chem. 2018, 9, 9–16. [Google Scholar] [CrossRef]
- Alsharafi, M.S.Y.; Alameri, A.Q. The F-Index and Coindex of V-Phenylenic Nanotubes and Nanotorus and Their Molecular Complement Graphs. Nanosyst. Phys. Chem. Math. 2021, 12, 263–270. [Google Scholar] [CrossRef]
- Kauffman, L.H. An Invariant of Regular Isotopy. Trans. Amer. Math. Soc. 1990, 318, 417–471. [Google Scholar] [CrossRef]
- Salman, M.; Ali, F.; Rehman, M.U.; Khalid, I. Some Valency Oriented Molecular Invariants of Certain Networks. CCHTS 2022, 25, 462–475. [Google Scholar] [CrossRef]
- Ahila Jeyanthi, D.; Selvarajan, T.M. Enhanced Mesh Network Using Novel Forgotten Polynomial Algorithm for Pharmaceutical Design. Intell. Autom. Soft Comput. 2022, 33, 669–680. [Google Scholar] [CrossRef]
- Zhang, H.-P.; Zhang, F. The Clar Covering Polynomial of Hexagonal Systems I. Discret. Appl. Math. 1996, 69, 147–167. [Google Scholar] [CrossRef]
- Rosenfeld, V.R. The Cycle (Circuit) Polynomial of a Graph with Double and Triple Weights of Edges and Cycles. ejgta 2019, 7, 189–205. [Google Scholar] [CrossRef]
- Awan, J.; Bernardi, O. Tutte Polynomials for Directed Graphs. J. Comb. Theory B 2020, 140, 192–247. [Google Scholar] [CrossRef]
- Aihara, J. Resonance Energies of Benzenoid Hydrocarbons. J. Am. Chem. Soc. 1977, 99, 2048–2053. [Google Scholar] [CrossRef]
- Li, X.; Zhao, H.; Wang, L. A Complete Solution to a Conjecture on the β-Polynomials of Graphs. J. Math. Chem. 2003, 33, 189–193. [Google Scholar] [CrossRef]
- Mizoguchi, N. Unified Rule for Stability of Hueckel-Type and Moebius-Type Systems. J. Phys. Chem. 1988, 92, 2754–2756. [Google Scholar] [CrossRef]
- Herges, R. Topology in Chemistry: Designing Möbius Molecules. Chem. Rev. 2006, 106, 4820–4842. [Google Scholar] [CrossRef]
- Gutman, I. Cycle Energy and Its Size Dependence. Discret. Appl. Math. 2020, 284, 534–537. [Google Scholar] [CrossRef]
- Heilbronner, E. Some More Difficulties with Topological Resonance Energy. Chem. Phys. Lett. 1982, 85, 377–380. [Google Scholar] [CrossRef]
- Gutman, I.; Mohar, B. More Difficulties with Topological Resonance Energy. Chem. Phys. Lett. 1981, 77, 567–570. [Google Scholar] [CrossRef]
- Nikolić, S.; Randić, M.; Klein, D.J.; Plavšić, D.; Trinajstić, N. The Conjugated-Circuit Model: Application to Benzenoid Hydrocarbons. J. Mol. Struct. 1989, 198, 223–237. [Google Scholar] [CrossRef]
- Rosenfeld, V.R.; Klein, D.J. Enumeration of Substitutional Isomers with Restrictive Mutual Positions of Ligands: I. Overall Counts. Overall Counts. J. Math. Chem. 2013, 51, 21–37. [Google Scholar] [CrossRef]
- Rosenfeld, V.R. The Circuit Polynomial of the Restricted Rooted Product G (Γ) of Graphs with a Bipartite Core G. Discret. Appl. Math. 2008, 156, 500–510. [Google Scholar] [CrossRef]
- Zhang, J. An Introduction to Chromatic Polynomials. 2018. Available online: https://math.mit.edu/~apost/courses/18.204_2018/Julie_Zhang_paper.pdf (accessed on 14 October 2024).
- Perrett, T. Chromatic Polynomial Calculator. Available online: https://tjperr.github.io/chromatic-polynomial/ (accessed on 14 October 2024).
- Farrell, E.J.; Rosenfeld, V.R. Block and Articulation Node Polynomials of the Generalized Rooted Product of Graphs. J. Math. Sci. 2000, 10, 35–47. [Google Scholar]
- Bollobas, B.; Gehring, F.W.; Halmos, P.R. Modern Graph Theory; Springer: New York, NY, USA, 2013; ISBN 978-1-4612-0619-4. [Google Scholar]
- Tutte, W.T. A Contribution to the Theory of Chromatic Polynomials. Can. J. Math. 1954, 6, 80–91. [Google Scholar] [CrossRef]
- Chbili, N.; Alderai, N.; Ali, R.; AlQedra, R. Tutte Polynomials and Graph Symmetries. Symmetry 2022, 14, 2072. [Google Scholar] [CrossRef]
- Hubai, T. The Chromatic Polynomial; Eotvos Lorand University: Budapest, Hungary, 2009. [Google Scholar]
- Ellis-Monaghan, J.A.; Merino, C. Graph Polynomials and Their Applications I: The Tutte Polynomial. In Structural Analysis of Complex Networks; Dehmer, M., Ed.; Birkhäuser Boston: Boston, MA, USA, 2011; pp. 219–255. ISBN 978-0-8176-4788-9. [Google Scholar]
- Farr, G.; Oxley, J. The Contributions of W.T. Tutte to Matroid Theory. In 2017 MATRIX Annals; De Gier, J., Praeger, C.E., Tao, T., Eds.; MATRIX Book Series; Springer International Publishing: Cham, Switzerland, 2019; Volume 2, pp. 343–361. ISBN 978-3-030-04160-1. [Google Scholar]
- Younger, D.H. William Thomas Tutte. 14 May 1917–2 May 2002. Biogr. Mems Fell. R. Soc. 2012, 58, 283–297. [Google Scholar] [CrossRef]
- Nafaa, C. Graph Polynomials and Symmetries. arXiv 2018, arXiv:1802.08487. [Google Scholar]
- Björklund, A.; Husfeldt, T.; Kaski, P.; Koivisto, M. Computing the Tutte Polynomial in Vertex-Exponential Time. In Proceedings of the 2008 49th Annual IEEE Symposium on Foundations of Computer Science, Philadelphia, PA, USA, 25–28 October 2008; IEEE: Philadelphia, PA, USA, 2008; pp. 677–686. [Google Scholar]
- Fath-Tabar, G.; Gholam-Rezaei, Z.; Ashrafi, A.R. On the Tutte Polynomial of Benzenoid Chains. Iran. J. Math. Chem. 2012, 3, 113–119. [Google Scholar] [CrossRef]
- Cadavid Muñoz, J.J. Using Tutte Polynomials to Analyze the Structure of the Benzodiazepines. In Independent Component Analyses, Compressive Sampling, Wavelets, Neural Net, Biosystems, and Nanoengineering XII; Szu, H.H., Dai, L., Eds.; SPIE: Baltimore, MD, USA, 2014; p. 911815. [Google Scholar]
- Gong, H.; Jin, X.; Zhang, F. Tutte Polynomials for Benzenoid Systems with One Branched Hexagon. J. Math. Chem. 2016, 54, 1057–1071, Erratum in J. Math. Chem. 2016, 54, 1748–1749. [Google Scholar] [CrossRef]
- Ren, H.; Xu, D.; Yang, W. The Tutte Polynomials of Catacondensed Benzenoid Systems. J. Math. Chem. 2021, 59, 529–541. [Google Scholar] [CrossRef]
- Ma, T.; Jin, X.; Zhang, F. Tutte Polynomials of Fan-like Graphs with Applications in Benzenoid Systems. Appl. Math. Comput. 2021, 411, 126496. [Google Scholar] [CrossRef]
- Chen, H.; Guo, Q. Tutte Polynomials of Alternating Polycyclic Chains. J. Math. Chem. 2019, 57, 2248–2260. [Google Scholar] [CrossRef]
- Chen, H.; Li, C. The Tutte Polynomial of Phenylene Systems with given Number of Branching Hexagons. Int. J. Quantum. Chem. 2022, 122, e26959. [Google Scholar] [CrossRef]
- Chen, H. Tutte Polynomials for Some Chemical Polycyclic Graphs. J. Math. Chem. 2023. [CrossRef]
- Dehmer, M.; Mueller, L.A.J.; Graber, A. New Polynomial-Based Molecular Descriptors with Low Degeneracy. PLoS ONE 2010, 5, e11393. [Google Scholar] [CrossRef]
- Dehmer, M.; Chen, Z.; Emmert-Streib, F.; Mowshowitz, A.; Varmuza, K.; Feng, L.; Jodlbauer, H.; Shi, Y.; Tao, J. The Orbit-Polynomial: A Novel Measure of Symmetry in Networks. IEEE Access 2020, 8, 36100–36112. [Google Scholar] [CrossRef]
- Rosenfeld, V.R.; Diudea, M.V. The Block–Polynomials and Block–Spectra of Dendrimers. Internet Electron. J. Mol. Des. 2002, 1, 142–156. [Google Scholar]
- Zhang, H.-P. The Clar Covering Polynomials of S,T-Isomers. Match 1993, 29, 189–197. [Google Scholar]
- Zhang, H.-P. The Clar Covering Polynomial of Hexagonal Systems with an Application to Chromatic Polynomials. Discret. Math. 1997, 172, 163–173. [Google Scholar] [CrossRef]
- Fu-Ji, Z.; He-Ping, Z.; Yu-Ting, L. The Clar Covering Polynomial of Hexagonal Systems II. An Application to Resonance Energy of Condensed Aromatic Hydrocarbons. Chin. J. Chem. 2010, 14, 321–325. [Google Scholar] [CrossRef]
- Witek, H.A.; Kang, J.-S. ZZ Polynomials for Isomers of (5,6)-Fullerenes Cn with n = 20–50. Symmetry 2020, 12, 1483. [Google Scholar] [CrossRef]
- Li, P.; Bian, H.; Yu, H.; Dou, Y. Clar Covering Polynomials of Polycyclic Aromatic Hydrocarbons. AIMS Math. 2024, 9, 13385–13409. [Google Scholar] [CrossRef]
- Klein, D.J.; Rosenfeld, V. Forcing, Freedom, & Uniqueness in Graph Theory & Chemistry. Croat. Chem. Acta 2014, 87, 49–59. [Google Scholar] [CrossRef]
- Zhao, S.; Zhang, H. Forcing Polynomials of Benzenoid Parallelogram and Its Related Benzenoids. Appl. Math. Comput. 2016, 284, 209–218. [Google Scholar] [CrossRef]
- Deng, K.; Zhang, H. Anti-Forcing Spectrum of Any Cata-Condensed Hexagonal System Is Continuous. Front. Math. China 2017, 12, 325–337. [Google Scholar] [CrossRef]
- Zhao, S.; Zhang, H. Anti-Forcing Polynomials for Benzenoid Systems with Forcing Edges. Discret. Appl. Math. 2018, 250, 342–356. [Google Scholar] [CrossRef]
- Zhao, S.; Zhang, H. Forcing and Anti-Forcing Polynomials of Perfect Matchings for Some Rectangle Grids. J. Math. Chem. 2019, 57, 202–225. [Google Scholar] [CrossRef]
- Zhao, S. Matching Forcing Polynomials of Constructable Hexagonal Systems. Match 2022, 88, 767–797. [Google Scholar] [CrossRef]
- Zhao, S.; Fan, X. Forcing Polynomial of Double Hexagonal Chains. Polycycl. Aromat. Compd. 2023, 43, 4055–4069. [Google Scholar] [CrossRef]
- Zhao, S. Matching Anti-Forcing Polynomials of Catacondensed Hexagonal Systems. Discret. Appl. Math. 2023, 337, 54–67. [Google Scholar] [CrossRef]
- Deng, K.; Liu, S.; Zhou, X. Forcing and Anti–Forcing Polynomials of Perfect Matchings of a Pyrene System. Match 2021, 85, 27–46. [Google Scholar]
- Deng, K.; Lü, H.; Wu, T. Forcing and Anti-Forcing Polynomials of a Type of Polyomino Graphs. Comp. Appl. Math. 2023, 42, 91. [Google Scholar] [CrossRef]
- Randić, M. In Search of Structural Invariants. J. Math. Chem. 1992, 9, 97–146. [Google Scholar] [CrossRef]
- Tsukamoto, T.; Kambe, T.; Imaoka, T.; Yamamoto, K. Modern Cluster Design Based on Experiment and Theory. Nat. Rev. Chem. 2021, 5, 338–347. [Google Scholar] [CrossRef]
- Feher, M.; Schmidt, J.M. Fuzzy Clustering as a Means of Selecting Representative Conformers and Molecular Alignments. J. Chem. Inf. Comput. Sci. 2003, 43, 810–818. [Google Scholar] [CrossRef]
- Sastry, G.M.; Dixon, S.L.; Sherman, W. Rapid Shape-Based Ligand Alignment and Virtual Screening Method Based on Atom/Feature-Pair Similarities and Volume Overlap Scoring. J. Chem. Inf. Model. 2011, 51, 2455–2466. [Google Scholar] [CrossRef]
- Jäntschi, L. Structure–Property Relationships for Solubility of Monosaccharides. Appl. Water Sci. 2019, 9, 38. [Google Scholar] [CrossRef]
- Schaeffer, S.E. Graph Clustering. Comput. Sci. Rev. 2007, 1, 27–64. [Google Scholar] [CrossRef]
- Kanj, I.; Komusiewicz, C.; Sorge, M.; van Leeuwen, E.J. Parameterized Algorithms for Recognizing Monopolar and 2-Subcolorable Graphs. J. Comput. Syst. Sci. 2018, 92, 22–47. [Google Scholar] [CrossRef]
- Ahmadi, R.; Nami, S. Investigation of Entanglement Entropy in Cyclic Bipartite Graphs Using Computer Software. Pramana-J. Phys. 2021, 95, 39. [Google Scholar] [CrossRef]
- Hosseinian, S.; Butenko, S. Polyhedral Properties of the Induced Cluster Subgraphs. Discret. Appl. Math. 2021, 297, 80–96. [Google Scholar] [CrossRef]
- Nakamura, K.; Araki, T. Partitioning Vertices into In- and out-Dominating Sets in Digraphs. Discret. Appl. Math. 2020, 285, 43–54. [Google Scholar] [CrossRef]
- Barbato, M.; Bezzi, D. Monopolar Graphs: Complexity of Computing Classical Graph Parameters. Discret. Appl. Math. 2021, 291, 277–285. [Google Scholar] [CrossRef]
- Füredi, Z.; Jiang, T.; Kostochka, A.; Mubayi, D.; Verstraëte, J. Partitioning Ordered Hypergraphs. J. Comb. Theory A 2021, 177, 105300. [Google Scholar] [CrossRef]
- Hellmann, T. Pairwise Stable Networks in Homogeneous Societies with Weak Link Externalities. Eur. J. Oper. Res. 2021, 291, 1164–1179. [Google Scholar] [CrossRef]
- McDiarmid, C.; Yolov, N. Recognition of Unipolar and Generalised Split Graphs. Algorithms 2015, 8, 46–59. [Google Scholar] [CrossRef]
- Golumbic, M.C.; Lewenstein, M. New Results on Induced Matchings. Discret. Appl. Math. 2000, 101, 157–165. [Google Scholar] [CrossRef]
- Hosoya, H.; Motoyama, A. An Effective Algorithm for Obtaining Polynomials for Dimer Statistics. Application of Operator Technique on the Topological Index to Two- and Three-dimensional Rectangular and Torus Lattices. J. Math. Phys. 1985, 26, 157–167. [Google Scholar] [CrossRef]
- Eschen, E.M.; Wang, X. Algorithms for Unipolar and Generalized Split Graphs. Discret. Appl. Math. 2014, 162, 195–201. [Google Scholar] [CrossRef]
- Adoni, W.Y.H.; Nahhal, T.; Krichen, M.; El byed, A.; Assayad, I. DHPV: A Distributed Algorithm for Large-Scale Graph Partitioning. J. Big Data 2020, 7, 76. [Google Scholar] [CrossRef]
- Zhang, T.; Gao, Y.; Zheng, B.; Chen, L.; Wen, S.; Guo, W. Towards Distributed Node Similarity Search on Graphs. World Wide Web 2020, 23, 3025–3053. [Google Scholar] [CrossRef]
- Schaudt, O. On Weighted Efficient Total Domination. J. Discret. Algorithms 2012, 10, 61–69. [Google Scholar] [CrossRef]
- Tratch, S.S.; Zefirov, N.S. From Substituted Derivatives to Substitution Profiles, Some New Possibilities of the Polya’s Enumeration Techniques. Adv. Chem. Model 2015, 5, 179. [Google Scholar]
- Yousaf, A.; Nadeem, M. An Efficient Technique to Construct Certain Counting Polynomials and Related Topological Indices for 2D-Planar Graphs. Polycycl. Aromat. Compd. 2022, 42, 4328–4342. [Google Scholar] [CrossRef]
- de Freitas, R.; Dias, B.; Maculan, N.; Szwarcfiter, J. On Distance Graph Coloring Problems. Intl. Trans. Op. Res. 2021, 28, 1213–1241. [Google Scholar] [CrossRef]
- Slamin, S.; Adiwijaya, N.O.; Hasan, M.A.; Dafik, D.; Wijaya, K. Local Super Antimagic Total Labeling for Vertex Coloring of Graphs. Symmetry 2020, 12, 1843. [Google Scholar] [CrossRef]
- Šurimová, M.; Lužar, B.; Madaras, T. A Dynamic Coloring of Graphs. Discret. Appl. Math. 2020, 284, 224–233. [Google Scholar] [CrossRef]
- Dokeroglu, T.; Sevinc, E. Memetic Teaching–Learning-Based Optimization Algorithms for Large Graph Coloring Problems. Eng. Appl. Artif. Intell. 2021, 102, 104282. [Google Scholar] [CrossRef]
- Zaker, M. A New Vertex Coloring Heuristic and Corresponding Chromatic Number. Algorithmica 2020, 82, 2395–2414. [Google Scholar] [CrossRef]
- Lehner, F.; Smith, S.M. On Symmetries of Edge and Vertex Colourings of Graphs. Discret. Math. 2020, 343, 111959. [Google Scholar] [CrossRef]
- Ahmadi, B.; Alinaghipour, F.; Shekarriz, M.H. Number of Distinguishing Colorings and Partitions. Discret. Math. 2020, 343, 111984. [Google Scholar] [CrossRef]
- Buluç, A.; Meyerhenke, H.; Safro, I.; Sanders, P.; Schulz, C. Recent Advances in Graph Partitioning. In Algorithm Engineering; Lecture Notes in Computer Science; Kliemann, L., Sanders, P., Eds.; Springer International Publishing: Cham, Switzerland, 2016; Volume 9220, pp. 117–158. ISBN 978-3-319-49486-9. [Google Scholar]
- Choi, D.; Han, J.; Lim, J.; Han, J.; Bok, K.; Yoo, J. Dynamic Graph Partitioning Scheme for Supporting Load Balancing in Distributed Graph Environments. IEEE Access 2021, 9, 65254–65265. [Google Scholar] [CrossRef]
- Miyazawa, F.K.; Moura, P.F.S.; Ota, M.J.; Wakabayashi, Y. Partitioning a Graph into Balanced Connected Classes: Formulations, Separation and Experiments. Eur. J. Oper. Res. 2021, 293, 826–836. [Google Scholar] [CrossRef]
- Miyazawa, F.K.; Moura, P.F.S.; Ota, M.J.; Wakabayashi, Y. Cut and Flow Formulations for the Balanced Connected K-Partition Problem. In Combinatorial Optimization; Lecture Notes in Computer Science; Baïou, M., Gendron, B., Günlük, O., Mahjoub, A.R., Eds.; Springer International Publishing: Cham, Switzerland, 2020; Volume 12176, pp. 128–139. ISBN 978-3-030-53261-1. [Google Scholar]
- Bruglieri, M.; Cordone, R. Metaheuristics for the Minimum Gap Graph Partitioning Problem. Comput. Oper. Res. 2021, 132, 105301. [Google Scholar] [CrossRef]
- Bok, K.; Kim, J.; Yoo, J. Dynamic Partitioning Supporting Load Balancing for Distributed RDF Graph Stores. Symmetry 2019, 11, 926. [Google Scholar] [CrossRef]
- Zheng, Z.; Wang, C.; Ding, Y.; Li, L.; Li, D. Research on Partitioning Algorithm Based on RDF Graph. Concurr. Comput. Pr. Exper 2021, 33, e5612. [Google Scholar] [CrossRef]
- Wagner, K. Über eine Eigenschaft der ebenen Komplexe. Math. Ann. 1937, 114, 570–590. [Google Scholar] [CrossRef]
- Bodlaender, H.L. A Tourist Guide through Treewidth. Acta Cybern 1993, 11, 1–21. [Google Scholar]
- Ateskan, E.R.; Erciyes, K.; Dalkilic, M.E. Parallelization of Network Motif Discovery Using Star Contraction. Parallel Comput. 2021, 101, 102734. [Google Scholar] [CrossRef]
- Pothen, A.; Simon, H.D.; Liou, K.-P. Partitioning Sparse Matrices with Eigenvectors of Graphs. SIAM J. Matrix Anal. Appl. 1990, 11, 430–452. [Google Scholar] [CrossRef]
- Gupta, A. Fast and Effective Algorithms for Graph Partitioning and Sparse-Matrix Ordering. IBM J. Res. Dev. 1997, 41, 171–183. [Google Scholar] [CrossRef]
- Gilbert, J.R.; Miller, G.L.; Teng, S.-H. Geometric Mesh Partitioning: Implementation and Experiments. SIAM J. Sci. Comput. 1998, 19, 2091–2110. [Google Scholar] [CrossRef]
- Karypis, G.; Kumar, V. A Fast and High Quality Multilevel Scheme for Partitioning Irregular Graphs. SIAM J. Sci. Comput. 1998, 20, 359–392. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Joița, D.-M.; Jäntschi, L. Counting Polynomials in Chemistry II. Int. J. Topol. 2024, 1, 13-26. https://doi.org/10.3390/ijt1010003
Joița D-M, Jäntschi L. Counting Polynomials in Chemistry II. International Journal of Topology. 2024; 1(1):13-26. https://doi.org/10.3390/ijt1010003
Chicago/Turabian StyleJoița, Dan-Marian, and Lorentz Jäntschi. 2024. "Counting Polynomials in Chemistry II" International Journal of Topology 1, no. 1: 13-26. https://doi.org/10.3390/ijt1010003
APA StyleJoița, D.-M., & Jäntschi, L. (2024). Counting Polynomials in Chemistry II. International Journal of Topology, 1(1), 13-26. https://doi.org/10.3390/ijt1010003






