Cell-Surface Glycan Labeling and Sensing
Abstract
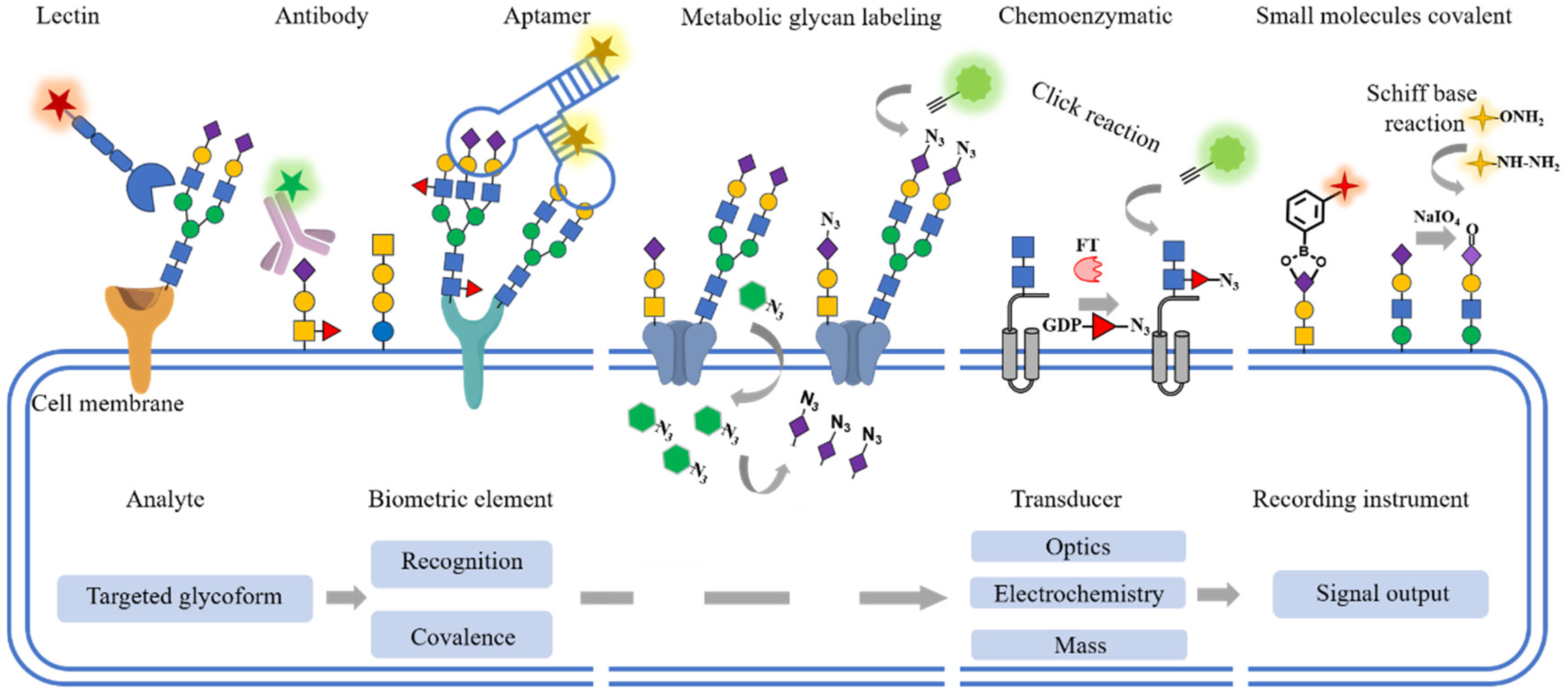
:1. Introduction
2. Glycan Recognition
2.1. Lectins
2.2. Antibodies
2.3. Aptamers
3. Glycan Covalent Labeling
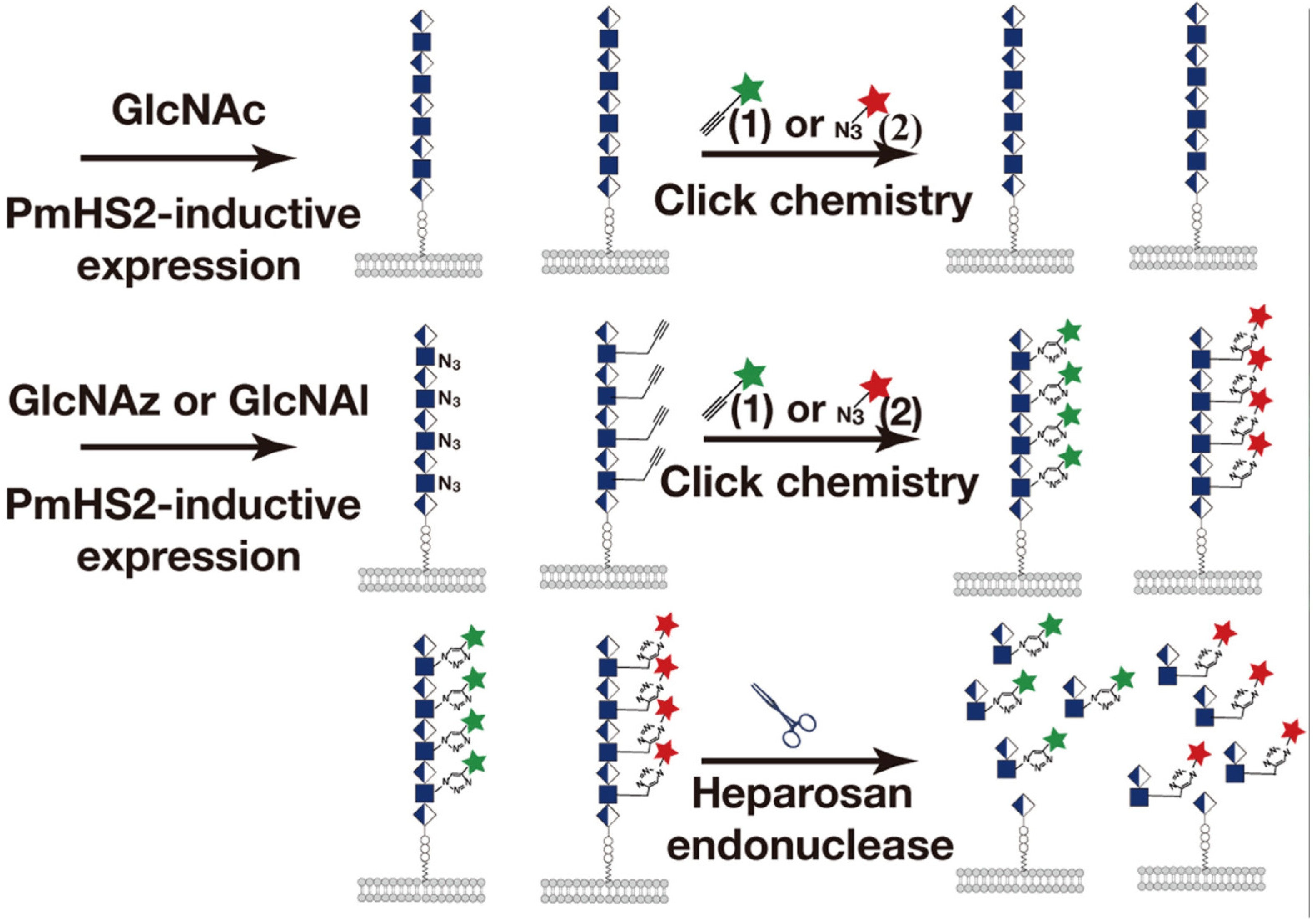
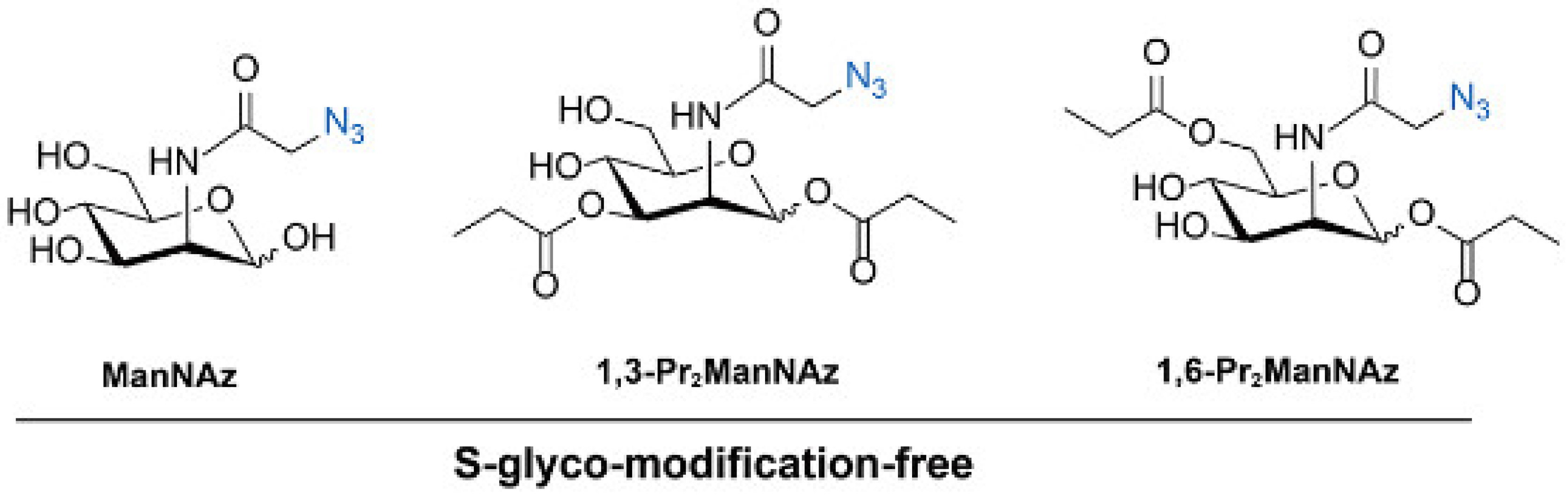
3.1. Metabolic Glycan Labeling (MGL)
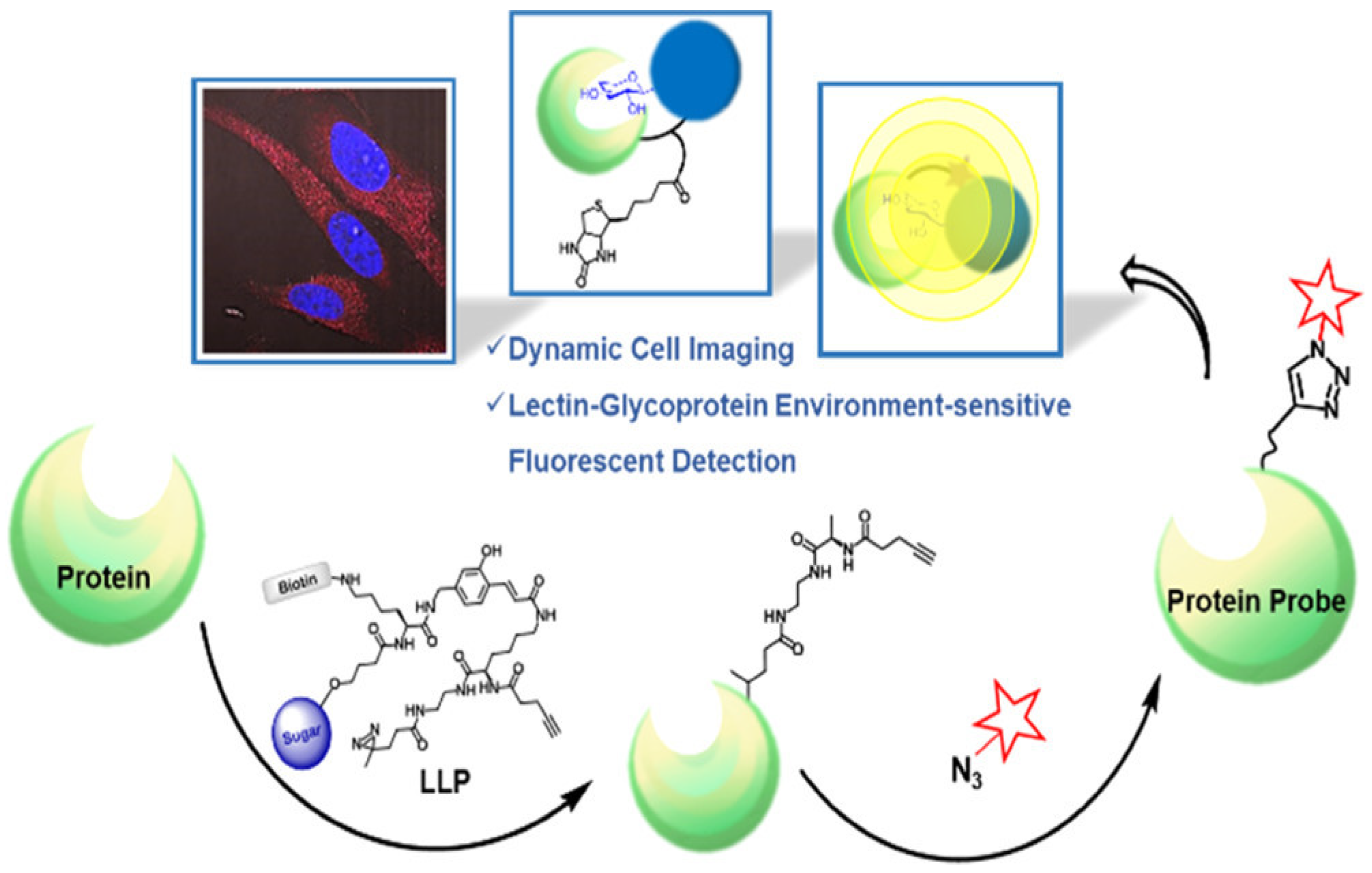
3.2. Chemoenzymatic Glycan Labeling (CeGL)
3.3. Chemical Covalent Labeling
4. Glycan-Specific Sensing
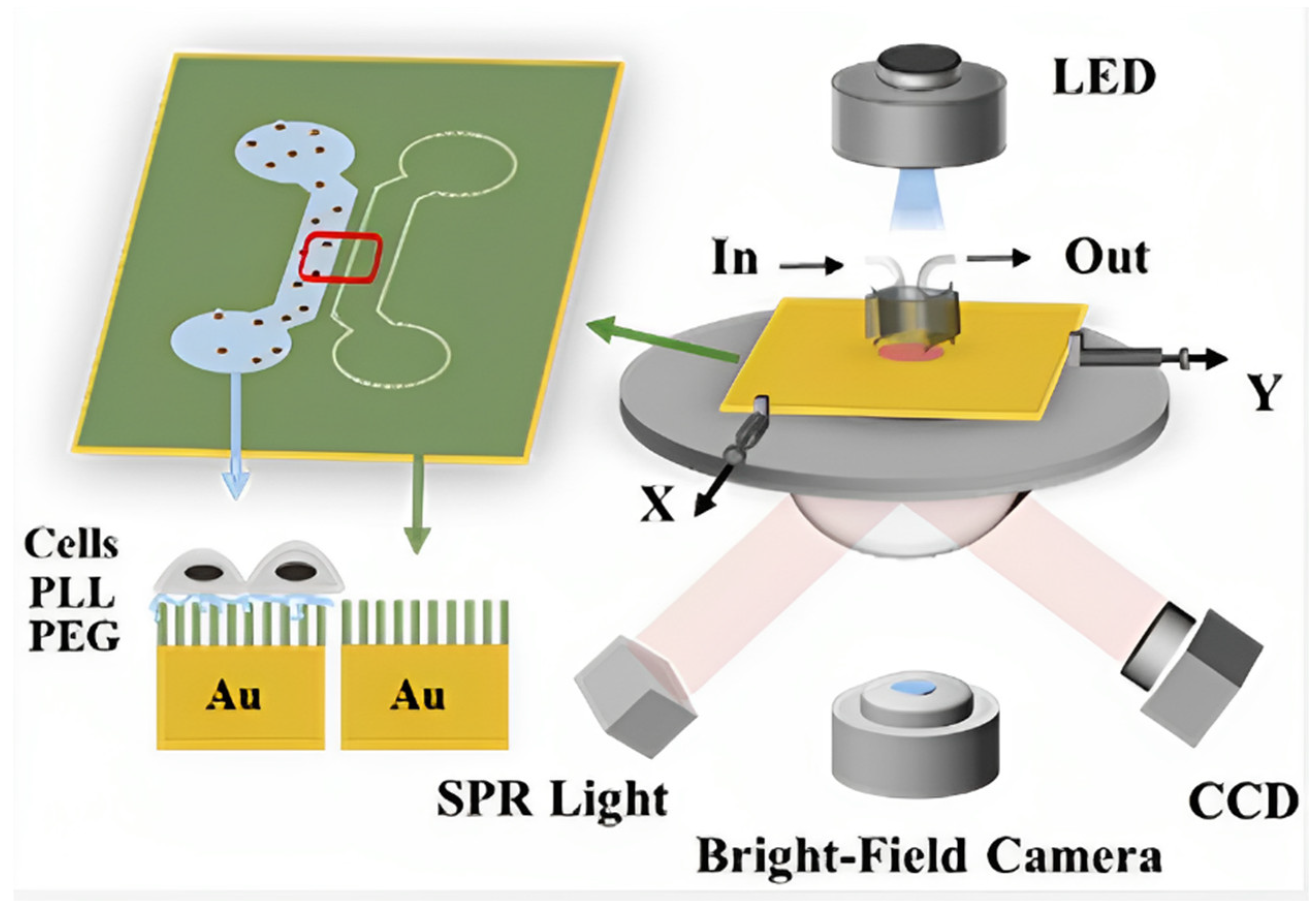
4.1. Surface Plasmon Resonance
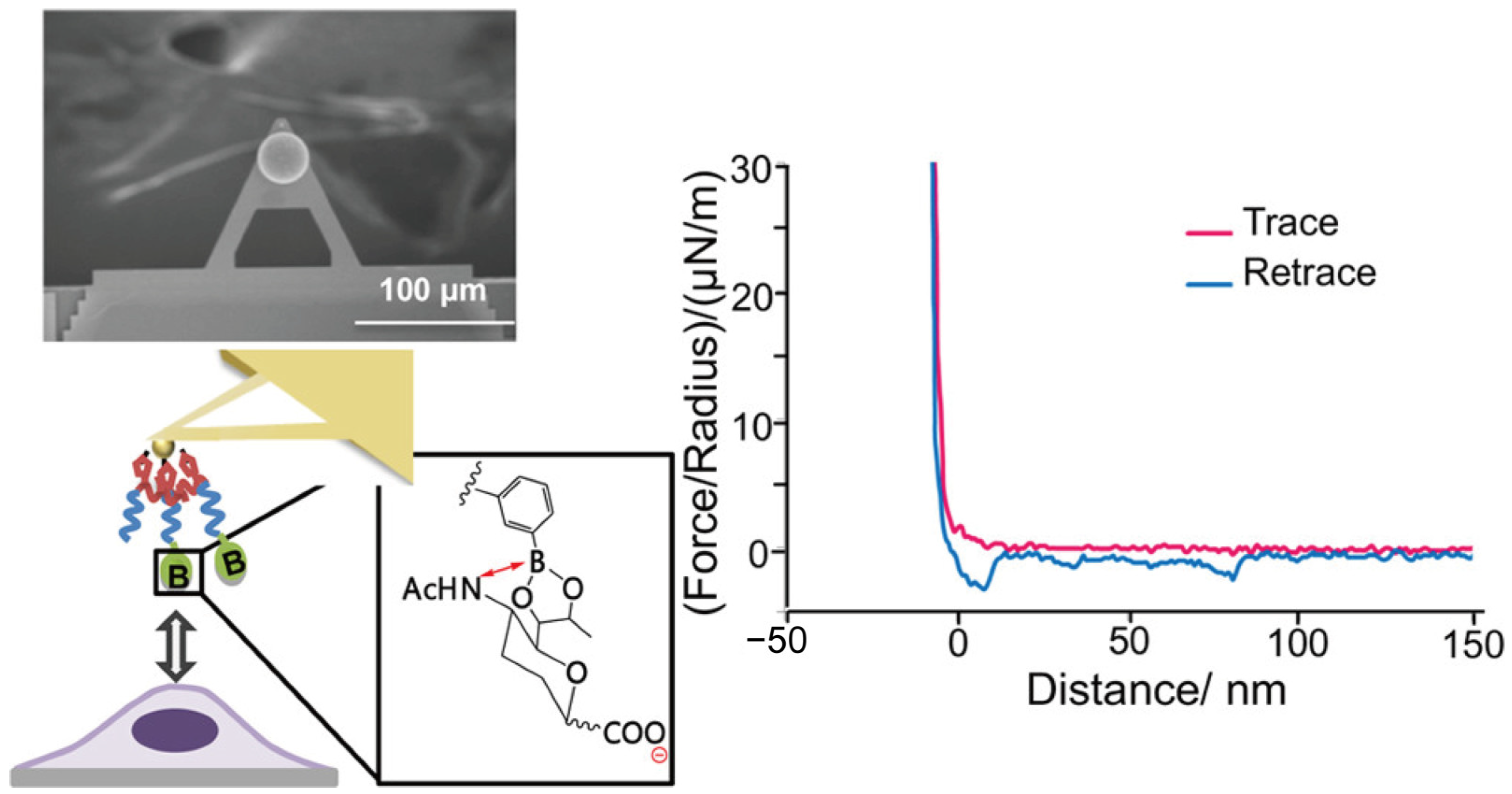
4.2. Atomic Force Microscope (AFM)
4.3. Optical Detection
4.3.1. Fluorescence
4.3.2. Surface-Enhanced Raman Scattering (SERS)
4.3.3. Other Optical Methods
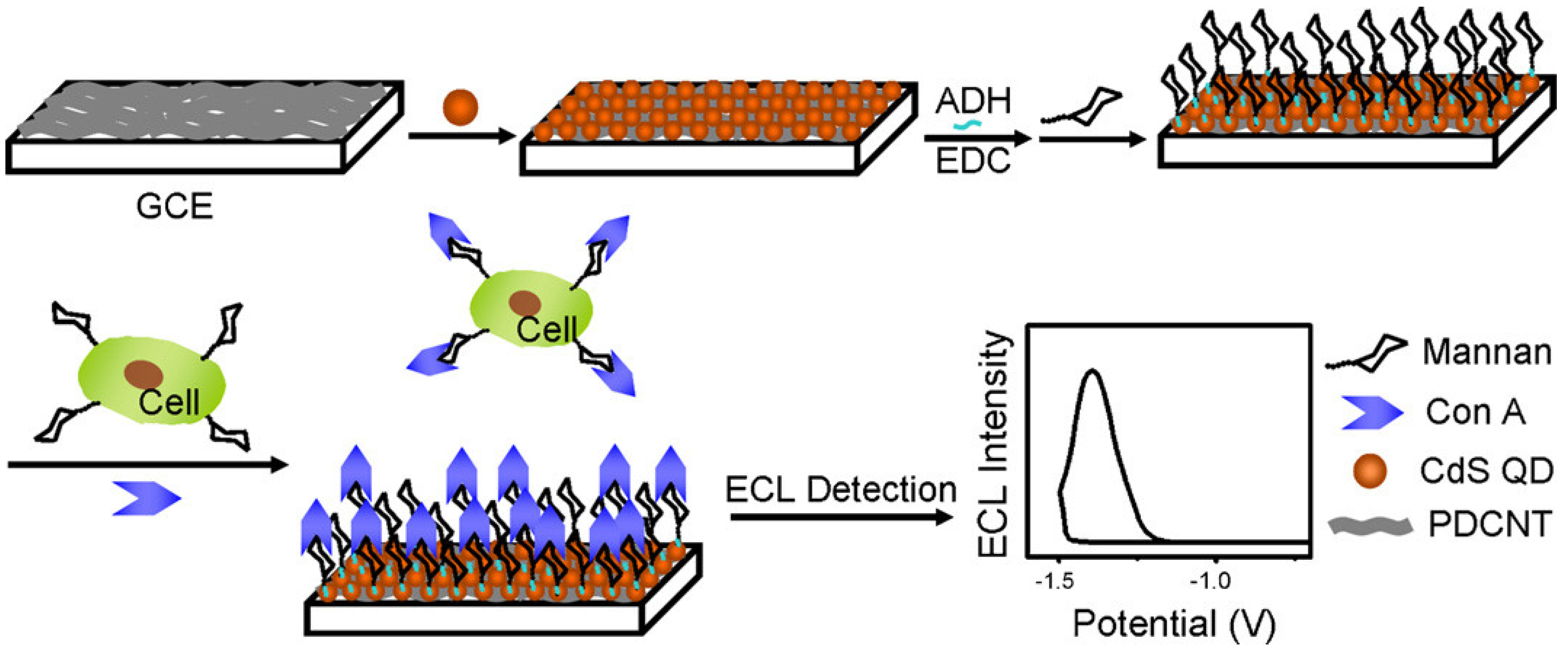
4.4. Electrochemical Detection
4.5. Mass Spectrometry (MS)
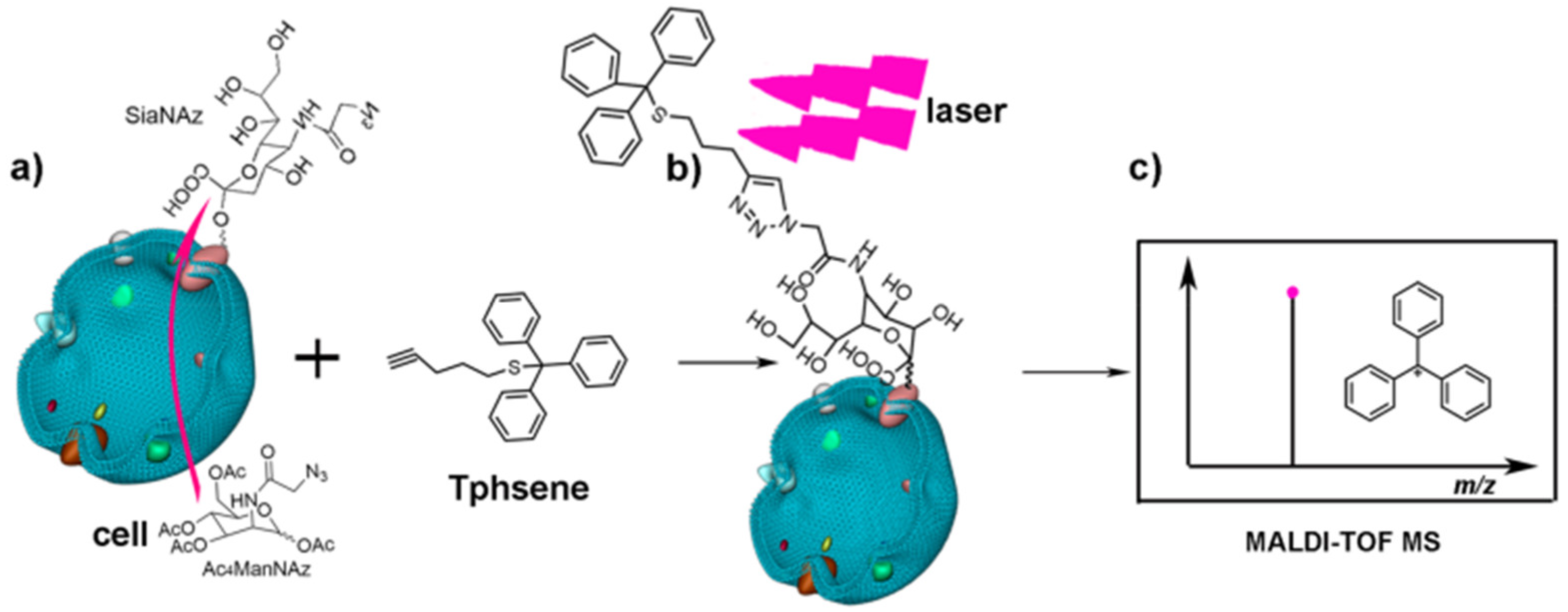
4.5.1. Laser-Cleavage Assisted In Situ Detection
4.5.2. Enzyme-Catalyzed Assisted In Situ Detection

| Recognition Molecule | Name | Specificity | Labeling Strategy | Application | Ref. |
|---|---|---|---|---|---|
| Lectin | AAL | Fuc | Fluorescent nanodiamonds | Glycan targeting therapy | [23] |
| ECA SBA BPL | Gal GalNAc GlcNAc | Fluorescent molecule | Cellular localization of glycan residues | [24] | |
| NPA | Man | Cell-selective glycoform analysis | [25] | ||
| SSA | Sia | ||||
| LTL | Fuc | Label-free | Detection of tumor cell vesicles | [26] | |
| hMGL | Tn/sTn (GalNAcα1-Ser/Thr, Siaα2-3GalNAcα1-Ser/Thr) | Label-free | Assessment of glycan–protein interactions | [30] | |
| RCA120 | Gal, Lac | Fluorescent molecule | [31] | ||
| Siglec-2-Fc | Sia | ||||
| WGA | GlcNAc | Label-free | [112] | ||
| UEA I | Fuc | DNA-fluorescence conjugate | Analysis of exosomal glycans of tumor cells | [125] | |
| SNA | Sia | ||||
| Jacalin | Gal | ||||
| PNA | Gal | ||||
| HPA | GalNAc | ||||
| LCA | Man | Nanoparticles | Quantification of cell-surface glycans | [139] | |
| WGA | GlcNAc | [142] | |||
| Con A | Man | Nanoparticles, Mass tag | Quantification of cell-surface glycans | [164,165,169,171,176,177] | |
| Antibody | S0G0-GlcNAc2-Qβ | GlcNAc | - | Antibody screening | [33] |
| Anti CA19-9 | CA19-9 | Composite nanomaterial | Detection of CA19-9 | [35] | |
| Anti-SSEA-1 | Lewis x | Technetium-99m | - | [38] | |
| Anti-TF | TF (Galβ1-3GalNAcα1-Ser/Thr) | - | - | [40] | |
| Anti-α-Gal (27H8) | Galili (Gal-α1,3-Gal) | - | - | [41,42] | |
| IgM to the Forssman disaccharide | Forssman (GalNAc-α1,3-GalNAc) | - | - | [42,43] | |
| Aptamer | SL-11 | Sialyllactose | - | - | [47] |
| Apt9 | Sialyllactose | Nanoparticles | - | [48] | |
| Clone 5 RNA aptamer | Sialyl Lewis X | - | Inhibition of cell adhesion | [49] | |
| TGP4 | Biantennary digalactosylated disialylated N-glycan A2G2S2 | Fluorescent molecule | Quantification of cell membrane glycans | [51] |
| Covalent Labeling Method | Probe | Glycan | Application | Ref. |
|---|---|---|---|---|
| Metabolic glycan labeling | Ac4Fuc7Alk | Fuc | Fuc imaging | [57] |
| 1,3-Pr2GalNAz | GalNAc | Identification of protein glycosylation | [58] | |
| Ac4ManAz | Sia | Sia imaging | [59] | |
| GlcNAz, GlcNAl | Polysaccharide E. coli K5 | In vivo bacterial detection | [60] | |
| ManNAz 1,3-Pr2ManNAz 1,6-Pr2ManNAz | Sia | Avoiding S-glyco-modification and SA imaging | [62,63,64] | |
| Chemoenzymatic glycan labeling | ST3Gal1 | Sia (donor sugar) | Probing of the binding specificities of siglecs | [73] |
| ST6Gal1 | Sia (donor sugar) | Capturing glycan–protein interactions | [74] | |
| CgtA | GalNAc (donor sugar) | Investigation of the effects of pathogen invasion | [75] | |
| WbwK | Fuc (donor sugar) | TF detection and imaging | [76] | |
| Galactose oxidase | Terminal Gal/GalNAc | Gal/GalNAc quantification and imaging | [77,78,79,80,81,82,83,84,85,86,87] | |
| Chemical covalent labeling | PBA | Sia | Sia quantification and imaging | [88,89,90,91,92] |
| Wulff-type PBA | Lipopolysaccharide | Gram-negative bacteria detection | [93,94] | |
| BOB | Terminal Gal | Gal detection | [95,96] | |
| NaIO4 | Sia | Sia quantification and imaging | [97,98,99] |
| Method | Target | Application | Sensitivity | Linear Range | Ref. |
|---|---|---|---|---|---|
| Optics | Sia | Quantification of Sia on PD-L1 of cell surface | - | - | [119] |
| Terminal Gal, Sia | Quantification of Sia and Gal of sperm | - | - | [120] | |
| Integrin-αxβ2/EGFR/TGF-β receptor-specific Sia | Imaging glycans on specific proteins (FRET) | - | - | [126] | |
| IL-36R-specific Sia | Imaging glycans on specific proteins (FRET) | - | - | [127] | |
| EGFR-specific Sia | Imaging glycans on specific proteins (FRET) | - | - | [129] | |
| EpCAM-specific Sia | Imaging glycans on specific proteins (SERS) | - | - | [130] | |
| Man | In situ and dynamically evaluating N-glycans in live cells (colorimetric and fluorescence) | Colorimetric method: 33 cells/mL; fluorescence method: 26 cells/mL | Colorimetric method: 100 to 1.0 × 107 cells/mL; fluorescence method: 80–5.0 × 107 cells/mL | [131] | |
| Glu | Quantitative glucose detection (SERS) | 1.8 mM | 0~25 mM | [136] | |
| Glu | Detection of glucose in urine (SERS) | - | - | [137] | |
| Man, Sia, GlcNAc | Quantification of multiple glycans on intact cell surfaces (SERS) | - | - | [139] | |
| Sia | Imaging Sia at single-cell level (SERS) | - | - | [140] | |
| Man, Sia, Gal, Fuc | In situ monitoring of glycans on the surface of cells in body fluids (SERS) | - | - | [142] | |
| Electrochemistry | Sia | Highly specific and sensitive detection in serum (DPV) | 10 pg/mL | 10–500 pg/mL | [146] |
| Man | Monitoring of glycans on living cells in response to drugs (DPV) | 3000 cells/mL | 1 × 104~1 × 107 cells/mL | [147] | |
| Sia | PSA detection (LSV) | 0.2 pg/mL | 0.5~200 pg/mL | [148] | |
| Recombinant glycoproteins | Accurate and sensitive determination (DPV) | 5.3 pg/mL | 0.01–50 ng/mL | [149] | |
| Cell-surface N-glycans | Evaluation of surface N-glycans (DPV) | 10 cells | 1.0 × 102~5.0 × 104 cells/mL | [150] | |
| Cell-surface N-glycans | Evaluation of surface N-glycans (I-t curve) | 3 cells/mL | 1 × 102~1 × 106 cells/mL | [151] | |
| AFP | Evaluating the AFP N-glycans and discriminating AFP between healthy and cancer patient’s serum (EIS) | 0.1 ng/L | 1–100 ng/L | [152] | |
| Man | Monitoring of glycans on living cells (DPV) | 1.5 × 103 cell/mL | 2.0 × 103~2.0 × 106 cell/mL | [164] | |
| Man | Label-free analysis of cell-surface glycans (electrochemiluminescence) | 1.2 × 103 cells/mL | 2 × 103~1 × 107 cells/mL | [165] | |
| Man | Monitoring the dynamic variation of glycans in cancer cells (electrochemiluminescence) | 100 cells/mL | 1 × 102~1 × 106 cells/mL | [167] | |
| N-glycans | Dynamically evaluating cell-surface N-glycans (electrochemiluminescence) | 600 cells/mL | 1 × 103~1 × 107 cells/mL | [169] | |
| N-glycans | Evaluation of cancer cell-surface glycans (electrochemiluminescence) | 38 cells/mL | 1.0 × 102~1.0 × 105 cells/mL | [170] | |
| Mass | MUC1-specific Sia | Quantification of MUC1-specific Sia in breast cells (LC–MS/MS) | 50 pM | 50 pM~10 nM | [101] |
| Man, Sia, GlcNAc | Fast and in situ multiplexed glycan detection (LDI-MS) | 200 cells/mL (Man) | 5 × 102~2 × 105 cells/mL (Man) | [172] | |
| Man, Gal | Imaging glycomic alterations in cancer cells (MALDI-TOF MS) | - | - | [176] | |
| Sialoglycoconjugates | In situ detection of cell-surface sialoglycoconjugates (LDI-MS) | 5 fmol | 100 fmol~100 pmol | [178] |
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Reily, C.; Stewart, T.J.; Renfrow, M.B.; Novak, J. Glycosylation in health and disease. Nat. Rev. Nephrol. 2019, 15, 346–366. [Google Scholar] [CrossRef] [PubMed]
- Pinho, S.S.; Reis, C.A. Glycosylation in cancer: Mechanisms and clinical implications. Nat. Rev. Cancer 2015, 15, 540–555. [Google Scholar] [CrossRef] [PubMed]
- Rudd, P.M.; Elliott, T.; Cresswell, P.; Wilson, I.A.; Dwek, R.A. Glycosylation and the immune system. Science 2001, 291, 2370–2376. [Google Scholar] [CrossRef] [PubMed]
- Parekh, R.; Dwek, R.; Sutton, B.; Fernandes, D.; Leung, A.; Stanworth, D.; Rademacher, T.; Mizuochi, T.; Taniguchi, T.; Matsuta, K. Association of rheumatoid arthritis and primary osteoarthritis with changes in the glycosylation pattern of total serum IgG. Nature 1985, 316, 452–457. [Google Scholar] [CrossRef] [PubMed]
- Marth, J.D.; Grewal, P.K. Mammalian glycosylation in immunity. Nat. Rev. Immunol. 2008, 8, 874–887. [Google Scholar] [CrossRef] [PubMed]
- Goto, Y.; Uematsu, S.; Kiyono, H. Epithelial glycosylation in gut homeostasis and inflammation. Nat. Immunol. 2016, 17, 1244–1251. [Google Scholar] [CrossRef]
- Reticker-Flynn, N.E.; Bhatia, S.N. Aberrant glycosylation promotes lung cancer metastasis through adhesion to galectins in the metastatic niche. Cancer Discov. 2015, 5, 168–181. [Google Scholar] [CrossRef]
- Thomas, D.; Rathinavel, A.K.; Radhakrishnan, P. Altered glycosylation in cancer: A promising target for biomarkers and therapeutics. Biochim. Biophys. Acta Rev. Cancer 2021, 1875, 188464. [Google Scholar] [CrossRef]
- Durand, G.; Seta, N. Protein glycosylation and diseases: Blood and urinary oligosaccharides as markers for diagnosis and therapeutic monitoring. Clin. Chem. 2000, 46, 795–805. [Google Scholar] [CrossRef]
- Theodoratou, E.; Campbell, H.; Ventham, N.T.; Kolarich, D.; Pučić-Baković, M.; Zoldoš, V.; Fernandes, D.; Pemberton, I.K.; Rudan, I.; Kennedy, N.A. The role of glycosylation in IBD. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 588–600. [Google Scholar] [CrossRef]
- Costa, A.F.; Campos, D.; Reis, C.A.; Gomes, C. Targeting glycosylation: A new road for cancer drug discovery. Trends Cancer 2020, 6, 757–766. [Google Scholar] [CrossRef] [PubMed]
- Mereiter, S.; Balmaña, M.; Campos, D.; Gomes, J.; Reis, C.A. Glycosylation in the era of cancer-targeted therapy: Where are we heading? Cancer Cell 2019, 36, 6–16. [Google Scholar] [CrossRef] [PubMed]
- Dotz, V.; Haselberg, R.; Shubhakar, A.; Kozak, R.P.; Falck, D.; Rombouts, Y.; Reusch, D.; Somsen, G.W.; Fernandes, D.L.; Wuhrer, M. Mass spectrometry for glycosylation analysis of biopharmaceuticals. TrAC-Trends Anal. Chem. 2015, 73, 1–9. [Google Scholar] [CrossRef]
- Dwek, R.A.; Butters, T.D.; Platt, F.M.; Zitzmann, N. Targeting glycosylation as a therapeutic approach. Nat. Rev. Drug Discov. 2002, 1, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Weis, W.I.; Drickamer, K. Structural basis of lectin-carbohydrate recognition. Annu. Rev. Biochem. 1996, 65, 441–473. [Google Scholar] [CrossRef] [PubMed]
- Lis, H.; Sharon, N. Lectins as molecules and as tools. Annu. Rev. Biochem. 1986, 55, 35–67. [Google Scholar] [CrossRef]
- Kilpatrick, D.C. Animal lectins: A historical introduction and overview. Biochim. Biophys. Acta 2002, 1572, 187–197. [Google Scholar] [CrossRef]
- Kaji, H.; Saito, H.; Yamauchi, Y.; Shinkawa, T.; Taoka, M.; Hirabayashi, J.; Kasai, K.-i.; Takahashi, N.; Isobe, T. Lectin affinity capture, isotope-coded tagging and mass spectrometry to identify N-linked glycoproteins. Nat. Biotechnol. 2003, 21, 667–672. [Google Scholar] [CrossRef]
- Wu, D.; Li, J.; Struwe, W.B.; Robinson, C.V. Probing N-glycoprotein microheterogeneity by lectin affinity purification-mass spectrometry analysis. Chem. Sci. 2019, 10, 5146–5155. [Google Scholar] [CrossRef]
- Regnier, F.E.; Jung, K.; Hooser, S.B.; Wilson, C.R. Glycoproteomics based on lectin affinity chromatographic selection of glycoforms. In Lectins: Analytical Technologies; Elsevier Science: Amsterdam, The Netherlands, 2007; pp. 193–212. [Google Scholar]
- Safina, G. Application of surface plasmon resonance for the detection of carbohydrates, glycoconjugates, and measurement of the carbohydrate-specific interactions: A comparison with conventional analytical techniques. A critical review. Anal. Chim. Acta 2012, 712, 9–29. [Google Scholar] [CrossRef]
- Hendrickson, O.D.; Zherdev, A.V. Analytical application of lectins. Crit. Rev. Anal. Chem. 2018, 48, 279–292. [Google Scholar] [CrossRef] [PubMed]
- Fard, M.G.; Khabir, Z.; Reineck, P.; Cordina, N.M.; Abe, H.; Ohshima, T.; Dalal, S.; Gibson, B.C.; Packer, N.H.; Parker, L.M. Targeting cell surface glycans with lectin-coated fluorescent nanodiamonds. Nanoscale Adv. 2022, 4, 1551–1564. [Google Scholar] [CrossRef] [PubMed]
- Gewaily, M.S.; Kassab, M.; Aboelnour, A.; Almadaly, E.A.; Noreldin, A.E. Comparative cellular localization of sugar residues in bull (Bos taurus) and donkey (Equus asinus) testes using lectin histochemistry. Microsc. Microanal. 2021, 27, 1529–1538. [Google Scholar] [CrossRef] [PubMed]
- Nagai-Okatani, C.; Nagai, M.; Sato, T.; Kuno, A. An improved method for cell type-selective glycomic analysis of tissue sections assisted by fluorescence laser microdissection. Int. J. Mol. Sci. 2019, 20, 700. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, T.; Sato, K.; Wakahara, S.; Mitamura, K.; Taga, A. A method for detecting tumor cells derived from colorectal cancer by targeting cell surface glycosylation with affinity capillary electrophoresis. J. Pharm. Biomed. Anal. 2020, 182, 113138. [Google Scholar] [CrossRef] [PubMed]
- Geijtenbeek, T.B.H.; Gringhuis, S.I. C-type lectin receptors in the control of T helper cell differentiation. Nat. Rev. Immunol. 2016, 16, 433–448. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.H.; Floyd, H.; Olson, N.E.; Magaletti, D.; Li, C.; Draves, K.; Clark, E.A. Dendritic-cell-associated C-type lectin 2 (DCAL-2) alters dendritic-cell maturation and cytokine production. Blood 2006, 107, 1459–1467. [Google Scholar] [CrossRef]
- Tommasone, S.; Allabush, F.; Tagger, Y.K.; Norman, J.; Köpf, M.; Tucker, J.H.R.; Mendes, P.M. The challenges of glycan recognition with natural and artificial receptors. Chem. Soc. Rev. 2019, 48, 5488–5505. [Google Scholar] [CrossRef]
- Beckwith, D.M.; FitzGerald, F.G.; Rodriguez Benavente, M.C.; Mercer, E.R.; Ludwig, A.K.; Michalak, M.; Kaltner, H.; Kopitz, J.; Gabius, H.J.; Cudic, M. Calorimetric analysis of the interplay between synthetic Tn antigen-presenting MUC1 glycopeptides and human macrophage galactose-type lectin. Biochemistry 2021, 60, 547–558. [Google Scholar] [CrossRef]
- Li, P.J.; Anwar, M.T.; Fan, C.Y.; Juang, D.S.; Lin, H.Y.; Chang, T.C.; Kawade, S.K.; Chen, H.J.; Chen, Y.J.; Tan, K.T.; et al. Fluorescence “turn-on” lectin sensors fabricated by ligand-assisted labeling probes for detecting protein–glycoprotein interactions. Biomacromolecules 2020, 21, 815–824. [Google Scholar] [CrossRef]
- Xie, Y.X.; Sheng, Y.; Li, Q.Y.; Ju, S.; Reyes, J.; Lebrilla, C.B. Determination of the glycoprotein specificity of lectins on cell membranes through oxidative proteomics. Chem. Sci. 2020, 11, 9501–9512. [Google Scholar] [CrossRef] [PubMed]
- Donahue, T.C.; Zong, G.H.; O’Brien, N.A.; Ou, C.; Gildersleeve, J.C.; Wang, L.X. Synthesis and mmunological tudy of N-glycan-bacteriophage Qβ conjugates reveal dominant antibody responses to the conserved chitobiose core. Bioconjugate Chem. 2022, 33, 1350–1362. [Google Scholar] [CrossRef] [PubMed]
- Koprowski, H.; Steplewski, Z.; Mitchell, K.; Herlyn, M.; Herlyn, D.; Fuhrer, P. Colorectal carcinoma antigens detected by hybridoma antibodies. Somat. Cell Mol. Genet. 1979, 5, 957–971. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Li, Y.X.; Li, C.C.; Yang, J.H.; Liu, D.L.; Wang, H.B.; Xu, R.; Zhang, Y.; Wei, Q. An ultrasensitive split-type electrochemical immunosensor based on controlled-release strategy for detection of CA19-9. Biosens. Bioelectron. 2023, 227, 115180. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Bamlet, W.R.; Oberg, A.L.; Chaffee, K.G.; Donahue, G.; Cao, X.J.; Chari, S.; Garcia, B.A.; Petersen, G.M.; Zaret, K.S. Detection of early pancreatic ductal adenocarcinoma with thrombospondin-2 and CA19-9 blood markers. Sci. Transl. Med. 2017, 9, eaah5583. [Google Scholar] [CrossRef]
- Partyka, K.; Maupin, K.A.; Brand, R.E.; Haab, B.B. Diverse monoclonal antibodies against the CA 19-9 antigen show variation in binding specificity with consequences for clinical interpretation. Proteomics 2012, 12, 2212–2220. [Google Scholar] [CrossRef]
- Thakur, M.L.; Marcus, C.S.; Henneman, P.; Butler, J.; Sinow, R.; Diggles, L.; Minami, C.; Mason, G.; Klein, S.; Rhodes, B. Imaging inflammatory diseases with neutrophil-specific technetium-99m-labeled monoclonal antibody anti-SSEA-1. J. Nucl. Med. 1996, 37, 1789–1795. [Google Scholar]
- Briard, J.G.; Jiang, H.; Moremen, K.W.; Macauley, M.S.; Wu, P. Cell-based glycan arrays for probing glycan–glycan binding protein interactions. Nat. Commun. 2018, 9, 880. [Google Scholar] [CrossRef]
- Kurtenkov, O.; Innos, K.; Sergejev, B.; Klaamas, K. The Thomsen-Friedenreich Antigen-Specific Antibody Signatures in Patients with Breast Cancer. BioMed Res. Int. 2018, 2018, 9579828. [Google Scholar] [CrossRef]
- Kreft, L.; Schepers, A.; Hils, M.; Swiontek, K.; Flatley, A.; Janowski, R.; Mirzaei, M.K.; Dittmar, M.; Charkrapani, N.; Desai, M.S.; et al. A novel monoclonal IgG1 antibody specific for Galactose-alpha-1,3-galactose questions alpha-Gal epitope expression by bacteria. Front. Immunol. 2022, 13, 958952. [Google Scholar] [CrossRef]
- Kappler, K.; Hennet, T. Emergence and significance of carbohydrate-specific antibodies. Genes Immun. 2020, 21, 224–239. [Google Scholar] [CrossRef] [PubMed]
- Butler, D.L.; Imberti, L.; Quaresima, V.; Fiorini, C.; Gildersleeve, J.C.; NIAID COVID-19 Consortium. Abnormal antibodies to self-carbohydrates in SARS-CoV-2-infected patients. PNAS Nexus 2022, 1, pgac062. [Google Scholar] [CrossRef] [PubMed]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef]
- Zhao, L.H.; Qi, X.Y.; Yan, X.C.; Huang, Y.F.; Liang, X.G.; Zhang, L.; Wang, S.; Tan, W.H. Engineering aptamer with enhanced affinity by triple helix-based terminal fixation. J. Am. Chem. Soc. 2019, 141, 17493–17497. [Google Scholar] [CrossRef] [PubMed]
- Kawakami, J.; Kawase, Y.; Sugimoto, N. In vitro selection of aptamers that recognize a monosaccharide. Anal. Chim. Acta 1998, 365, 95–100. [Google Scholar] [CrossRef]
- Mehedi Masud, M.; Kuwahara, M.; Ozaki, H.; Sawai, H. Sialyllactose-binding modified DNA aptamer bearing additional functionality by SELEX. Bioorg. Med. Chem. 2004, 12, 1111–1120. [Google Scholar] [CrossRef]
- Chen, J.; Chen, X.; Zhang, Y.; Wang, X.; Zhou, N. Screening of a Sialyllactose-Specific Aptamer and Engineering a Pair of Recognition Elements with Unique Fluorescent Characteristics for Sensitive Detection of Sialyllactose. J. Agric. Food Chem. 2023, 71, 2628–2636. [Google Scholar] [CrossRef]
- Jeong, S.; Eom, T.Y.; Kim, S.J.; Lee, S.W.; Yu, J. In vitro selection of the RNA aptamer against the Sialyl Lewis X and its inhibition of the cell adhesion. Biochem. Biophys. Res. Commun. 2001, 281, 237–243. [Google Scholar] [CrossRef]
- Ma, Y.; Li, X.; Li, W.; Liu, Z. Glycan-imprinted magnetic nanoparticle-based SELEX for efficient screening of glycoprotein-binding aptamers. ACS Appl. Mater. Interfaces 2018, 10, 40918–40926. [Google Scholar] [CrossRef]
- Li, W.; Ma, Y.; Guo, Z.; Xing, R.; Liu, Z. Efficient screening of glycan-specific aptamers using a glycosylated peptide as a scaffold. Anal. Chem. 2021, 93, 956–963. [Google Scholar] [CrossRef]
- Sminia, T.J.; Zuilhof, H.; Wennekes, T. Getting a grip on glycans: A current overview of the metabolic oligosaccharide engineering toolbox. Carbohydr. Res. 2016, 435, 121–141. [Google Scholar] [CrossRef] [PubMed]
- Cheng, B.; Tang, Q.; Zhang, C.; Chen, X. Glycan labeling and analysis in cells and in vivo. Annu. Rev. Anal. Chem. 2021, 14, 363–387. [Google Scholar] [CrossRef] [PubMed]
- Bertozzi, C.R.; Kiessling, L.L. Chemical glycobiology. Science 2001, 291, 2357–2364. [Google Scholar] [CrossRef] [PubMed]
- Dube, D.H.; Bertozzi, C.R. Metabolic oligosaccharide engineering as a tool for glycobiology. Curr. Opin. Chem. Biol. 2003, 7, 616–625. [Google Scholar] [CrossRef] [PubMed]
- Goon, S.; Bertozzi, C.R. Metabolic substrate engineering as a tool for glycobiology. J. Carbohydr. Chem. 2002, 21, 943–977. [Google Scholar] [CrossRef]
- Kizuka, Y.; Funayama, S.; Shogomori, H.; Nakano, M.; Nakajima, K.; Oka, R.; Kitazume, S.; Yamaguchi, Y.; Sano, M.; Korekane, H.; et al. High-sensitivity and low-toxicity fucose probe for glycan imaging and biomarker discovery. Cell Chem. Biol. 2016, 23, 782–792. [Google Scholar] [CrossRef]
- Hao, Y.; Fan, X.; Shi, Y.; Zhang, C.; Sun, D.-E.; Qin, K.; Qin, W.; Zhou, W.; Chen, X. Next-generation unnatural monosaccharides reveal that ESRRB O-GlcNAcylation regulates pluripotency of mouse embryonic stem cells. Nat. Commun. 2019, 10, 4065. [Google Scholar] [CrossRef]
- Chen, D.X.; Lin, Y.Y.; Li, A.; Luo, X.J.; Yang, C.Y.; Gao, J.H.; Lin, H.Y. Bio-orthogonal metabolic fluorine labeling enables deep-tissue visualization of tumor cells in vivo by 19F magnetic resonance imaging. Anal. Chem. 2022, 94, 16614–16621. [Google Scholar] [CrossRef]
- Wang, Y.J.; Li, L.; Yu, J.; Hu, H.Y.; Liu, Z.X.; Jiang, W.J.; Xu, W.; Guo, X.P.; Wang, F.S.; Sheng, J.Z. Imaging of Escherichia coli K5 and glycosaminoglycan precursors via targeted metabolic labeling of capsular polysaccharides in bacteria. Sci. Adv. 2023, 9, eade4770. [Google Scholar] [CrossRef]
- Parle, D.R.; Bulat, F.; Fouad, S.; Zecchini, H.; Brindle, K.M.; Neves, A.A.; Leeper, F.J. Metabolic glycan labeling of cancer cells using variably acetylated monosaccharides. Bioconjug. Chem. 2022, 33, 1467–1473. [Google Scholar] [CrossRef]
- Qin, W.; Qin, K.; Fan, X.; Peng, L.; Hong, W.; Zhu, Y.; Lv, P.; Du, Y.; Huang, R.; Han, M.; et al. Artificial cysteine S-glycosylation induced by per-O-acetylated unnatural monosaccharides during metabolic glycan labeling. Angew. Chem. Int. Ed. 2018, 57, 1817–1820. [Google Scholar] [CrossRef] [PubMed]
- Qin, K.; Zhang, H.; Zhao, Z.; Chen, X. Protein S-glyco-modification through an elimination–addition mechanism. J. Am. Chem. Soc. 2020, 142, 9382–9388. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.Y.; Huang, Z.M.; Du, Y.F.; Lv, P.; Fan, X.Q.; Dai, P.; Chen, X. Metabolic glycan labeling in primary neurons enabled by unnatural sugars with no S-glyco-modification. ACS Chem. Biol. 2023, 18, 1416–1424. [Google Scholar] [CrossRef] [PubMed]
- Lopez Aguilar, A.; Briard, J.G.; Yang, L.; Ovryn, B.; Macauley, M.S.; Wu, P. Tools for studying glycans: Recent advances in chemoenzymatic glycan labeling. ACS Chem. Biol. 2017, 12, 611–621. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.R.; Mo, T.; Huang, W.Q.; Xu, X.P.; Tian, W.S.; Wang, Y.X.; Song, Y.L.; Li, J.; Shi, S.; Liu, X.; et al. Glycosylation of aromatic glycosides by a promiscuous glycosyltransferase UGT71BD1 from Cistanche tubulosa. J. Nat. Prod. 2022, 85, 1826–1836. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.D.; Hou, X.D.; Zhang, Y.; Deng, Z.W.; Ping, Q.; Fu, K.; Yuan, Z.B.; Rao, Y.J. Highly efficient production of rebaudioside D enabled by structure-guided engineering of bacterial glycosyltransferase YojK. Front. Bioeng. Biotechnol. 2022, 10, 985826. [Google Scholar] [CrossRef]
- Zhong, R.Q.; Phillips, D.R.; Adams, E.R.; Ye, Z.H. An Arabidopsis family GT106 glycosyltransferase is essential for xylan biosynthesis and secondary wall deposition. Planta 2023, 257, 43. [Google Scholar] [CrossRef]
- Tan, Y.M.; Zhang, X.; Feng, Y.; Yang, G.Y. Directed evolution of glycosyltransferases by a single-cell ultrahigh-throughput FACS-based screening method. Methods Mol. Biol. 2022, 2461, 211–224. [Google Scholar]
- Deng, J.Q.; Lu, Z.; Liu, J.; Zhao, Y.; Hou, X.B.; Guo, X.P.; Jiang, W.J.; Wang, F.S.; Sheng, J.Z. Heparosan oligosaccharide synthesis using engineered single-function glycosyltransferases. Catal. Sci. Technol. 2022, 12, 3793–3803. [Google Scholar] [CrossRef]
- Cid, E.; Yamamoto, M.; Yamamoto, F. Mixed-up sugars: Glycosyltransferase cross-reactivity in cancerous tissues and their therapeutic targeting. ChemBioChem 2022, 23, e202100460. [Google Scholar] [CrossRef]
- Hirata, T.; Harada, Y.; Hirosawa, K.M.; Tokoro, Y.; Suzuki, K.G.; Kizuka, Y. N-acetylglucosaminyltransferase-V (GnT-V)-enriched small extracellular vesicles mediate N-glycan remodeling in recipient cells. iScience 2023, 26, 105747. [Google Scholar] [CrossRef] [PubMed]
- Büll, C.; Nason, R.; Sun, L.; Van Coillie, J.; Madriz Sørensen, D.; Moons, S.J.; Yang, Z.; Arbitman, S.; Fernandes, S.M.; Furukawa, S.; et al. Probing the binding specificities of human Siglecs by cell-based glycan arrays. Proc. Natl. Acad. Sci. USA 2021, 118, e2026102118. [Google Scholar] [CrossRef] [PubMed]
- Babulic, J.L.; Capicciotti, C.J. Exo-enzymatic cell-surface glycan labeling for capturing glycan–protein interactions through photo-cross-linking. Bioconjug. Chem. 2022, 33, 773–780. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Ye, J.; Chao, Y.; Dong, S.; Niu, M.; Wang, Y.; Liu, Z.; Chen, W.; Ge, N.; Lu, S.; et al. Chemoenzymatic labeling pathogens containing terminal N-acetylneuraminic acid-α (2-3)-galactose glycans. ACS Infect. Dis. 2022, 8, 657–664. [Google Scholar] [CrossRef]
- Li, Q.; Li, Z.; Duan, X.; Yi, W. A tandem enzymatic approach for detecting and imaging tumorassociated Thomsen-Friedenreich antigen disaccharide. J. Am. Chem. Soc. 2014, 136, 12536–12539. [Google Scholar] [CrossRef]
- Whittaker, J.W. Free radical catalysis by galactose oxidase. Chem. Rev. 2003, 103, 2347–2364. [Google Scholar] [CrossRef]
- Rannes, J.B.; Ioannou, A.; Willies, S.C.; Grogan, G.; Behrens, C.; Flitsch, S.L.; Turner, N.J. Glycoprotein labeling using engineered variants of galactose oxidase obtained by directed evolution. J. Am. Chem. Soc. 2011, 133, 8436–8439. [Google Scholar] [CrossRef]
- Mattey, A.P.; Birmingham, W.R.; Both, P.; Kress, N.; Huang, K.; van Munster, J.M.; Bulmer, G.S.; Parmeggiani, F.; Voglmeir, J.; Martinez, J.E.R.; et al. Selective oxidation of N-glycolylneuraminic acid using an engineered galactose oxidase variant. ACS Catal. 2019, 9, 8208–8212. [Google Scholar] [CrossRef]
- Ramya, T.N.C.; Weerapana, E.; Cravatt, B.F.; Paulson, J.C. Glycoproteomics enabled by tagging sialic acid- or galactose-terminated glycans. Glycobiology 2013, 23, 211–221. [Google Scholar] [CrossRef]
- Han, E.; Ding, L.; Qian, R.C.; Bao, L.; Ju, H.X. Sensitive chemiluminescent imaging for chemoselective analysis of glycan expression on living cells using a multifunctional nanoprobe. Anal. Chem. 2012, 84, 1452–1458. [Google Scholar] [CrossRef]
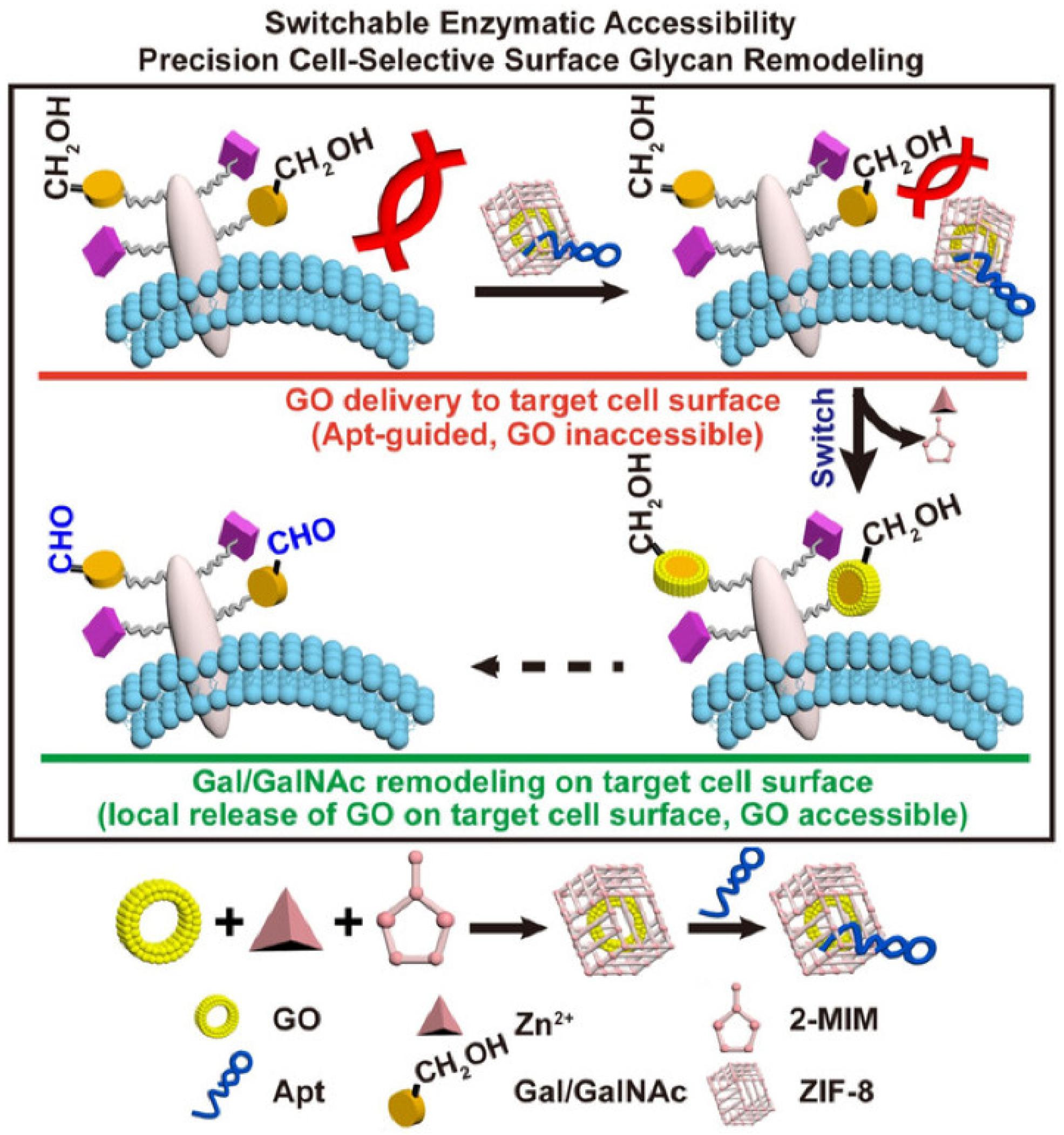
- Zhang, P.W.; Li, Y.R.; Yu, X.F.; Ju, H.X.; Ding, L. Switchable enzymatic accessibility for precision cell-selective surface glycan remodeling. Chem. Eur. J. 2019, 25, 10505–10510. [Google Scholar] [CrossRef]
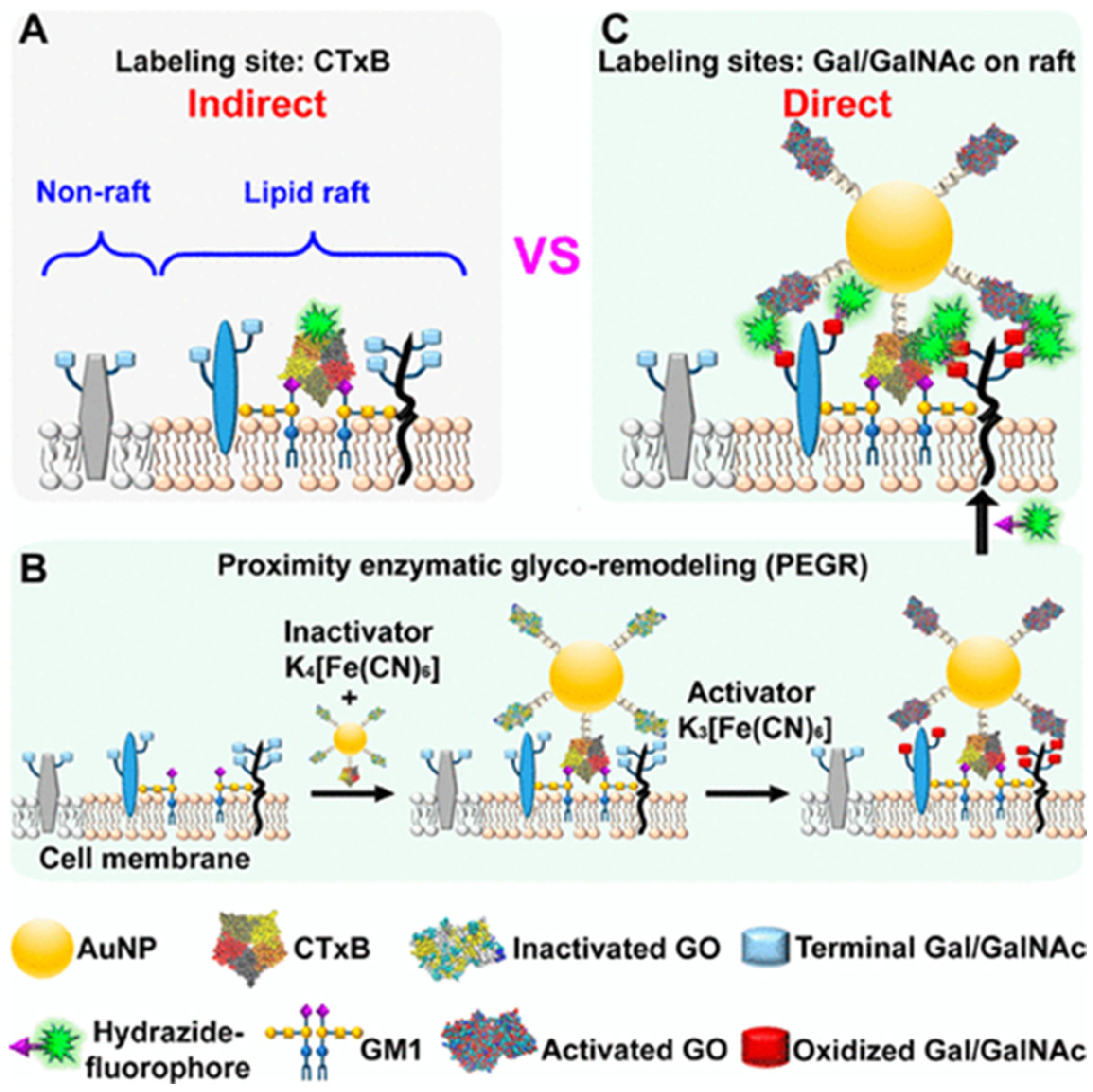
- Tao, J.; Yu, X.F.; Guo, Y.N.; Wang, G.Y.; Ju, H.X.; Ding, L. Proximity enzymatic glyco-remodeling enables direct and highly efficient lipid raft imaging on live cells. Anal. Chem. 2020, 92, 7232–7239. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.F.; Shi, H.F.; Li, Y.R.; Guo, Y.N.; Zhang, P.W.; Wang, G.Y.; Li, L.; Chen, X.; Ding, L.; Ju, H.X. Thermally triggered, cell-specific enzymatic glyco-editing: In situ regulation of lectin recognition and immune response on target cells. ACS Appl. Mater. Interfaces 2020, 12, 54387–54398. [Google Scholar] [CrossRef] [PubMed]
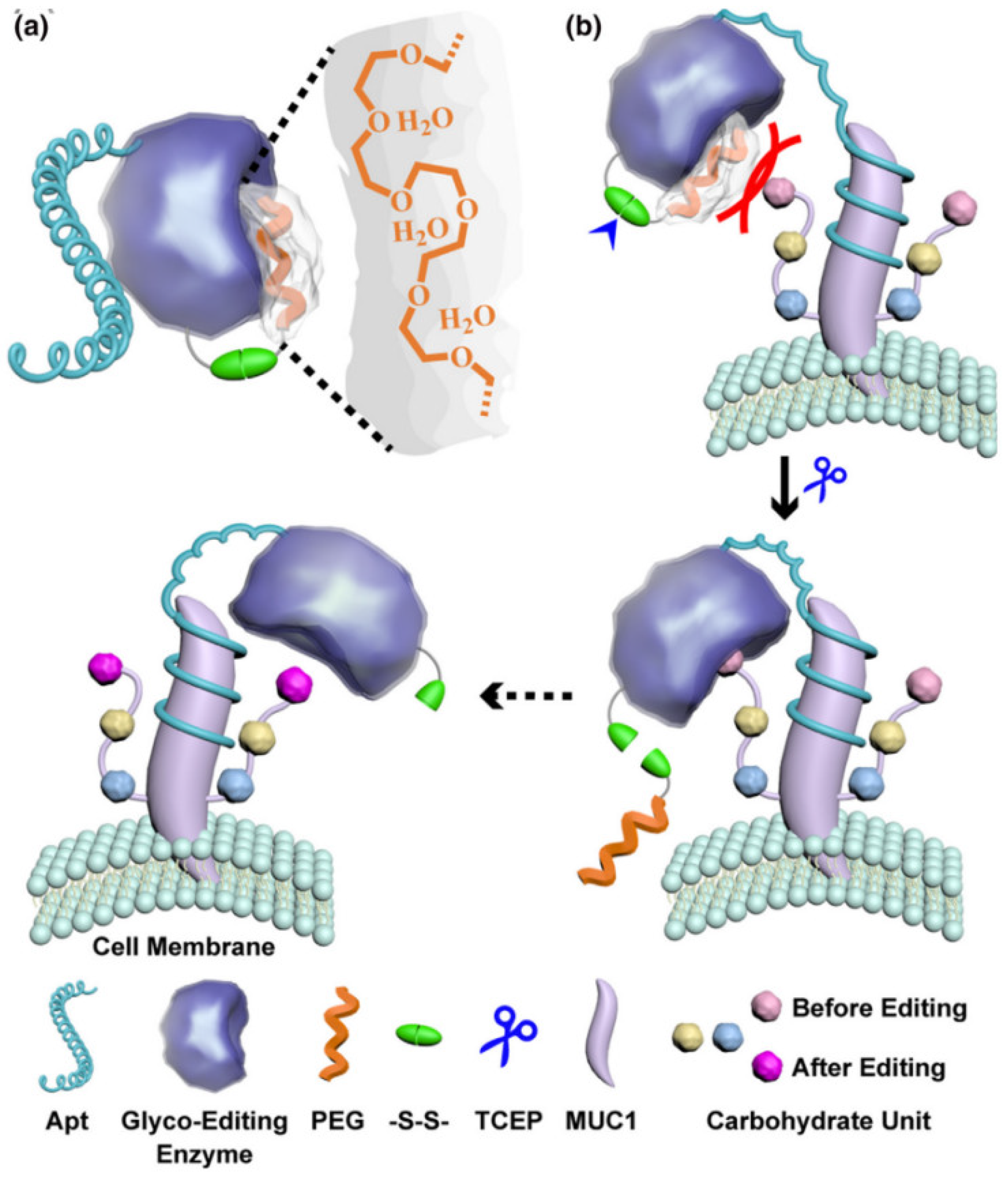
- Li, S.Q.; Mao, A.W.; Huo, F.; Wang, X.J.; Guo, Y.N.; Liu, L.; Yan, C.; Ding, L.; Ju, H.X. A localized glyco-editing probe for revelation of protein-specific glycan function. Mater. Today 2021, 49, 85–96. [Google Scholar] [CrossRef]
- Guo, Y.N.; Tao, J.; Li, Y.R.; Feng, Y.M.; Ju, H.X.; Wang, Z.F.; Ding, L. Quantitative localized analysis reveals distinct exosomal protein-specific glycosignatures: Implications in cancer cell subtyping, exosome biogenesis, and function. J. Am. Chem. Soc. 2020, 142, 7404–7412. [Google Scholar] [CrossRef] [PubMed]
- Mao, A.W.; Zhang, Y.; Wang, G.Y.; Zhong, T.; Chen, X.Y.; Wang, H.Q.; Xie, R.; Wang, X.J.; Ding, L.; Ju, H.X. Aglycone sterics-selective enzymatic glycan remodeling. iScience 2022, 25, 104578. [Google Scholar] [CrossRef]
- Otsuka, H.; Uchimura, E.; Koshino, H.; Okano, T.; Kataoka, K. Anomalous binding profile of phenylboronic acid with N-acetylneuraminic acid (Neu5Ac) in aqueous solution with varying pH. J. Am. Chem. Soc. 2003, 125, 3493–3502. [Google Scholar] [CrossRef]
- Matsumoto, A.; Sato, N.; Kataoka, K.; Miyahara, Y. Noninvasive sialic acid detection at cell membrane by using phenylboronic acid modified self-assembled monolayer gold electrode. J. Am. Chem. Soc. 2009, 131, 12022–12023. [Google Scholar] [CrossRef]
- Li, T.; Liu, Y. Self-assembled nanorods of phenylboronic acid functionalized pyrene for in situ two-photon imaging of cell surface sialic acids and photodynamic therapy. Anal. Chem. 2021, 93, 7029–7036. [Google Scholar] [CrossRef]
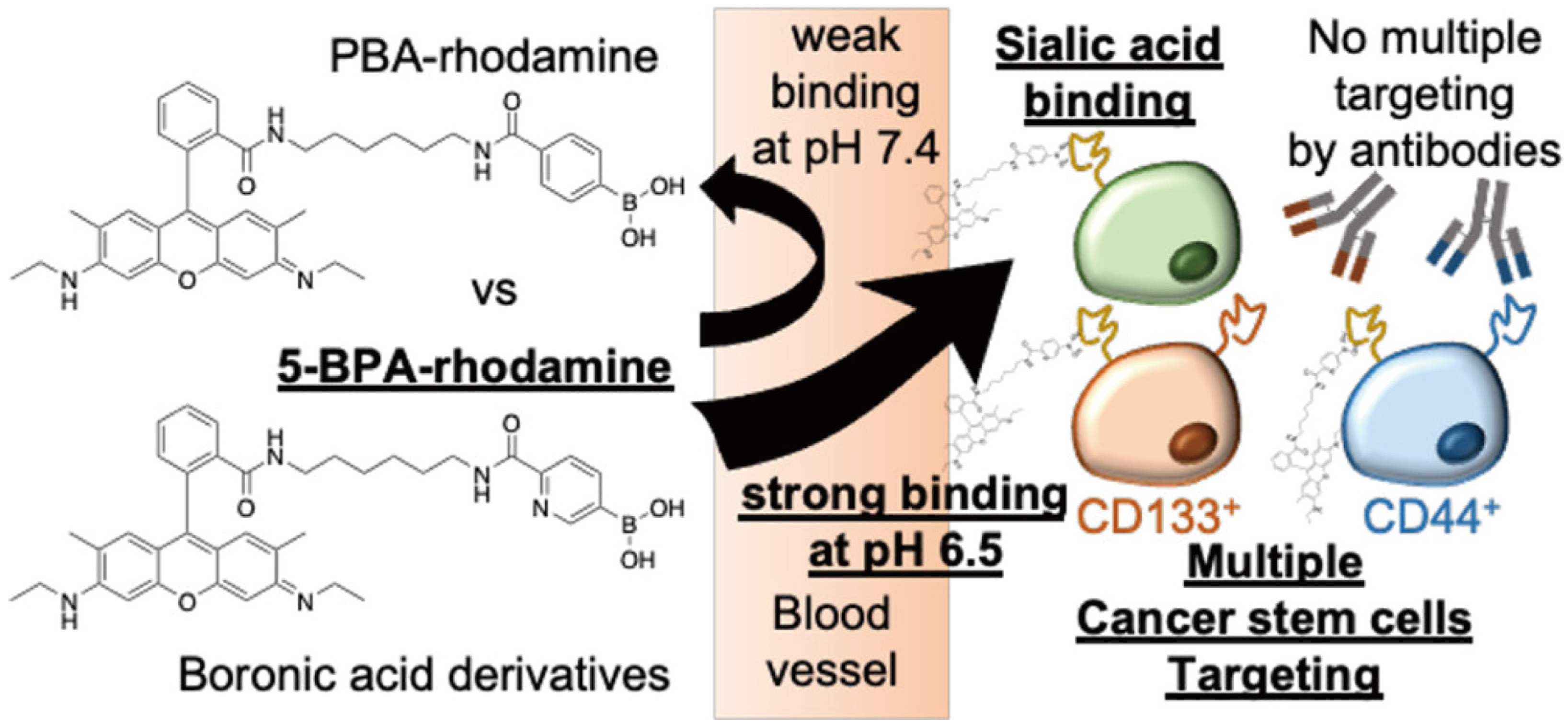
- Miyazaki, T.; Khan, T.; Tachihara, Y.; Itoh, M.; Miyazawa, T.; Suganami, T.; Miyahara, Y.; Cabral, H.; Matsumoto, A. Boronic acid ligands can target multiple subpopulations of pancreatic cancer stem cells via pH-dependent glycan-terminal sialic acid recognition. ACS Appl. Bio. Mater. 2021, 4, 6647–6651. [Google Scholar] [CrossRef]
- Qualls, M.L.; Hagewood, H.; Lou, J.; Mattern-Schain, S.I.; Zhang, X.Y.; Mountain, D.J.; Best, M.D. Bis-boronic acid liposomes for carbohydrate recognition and cellular delivery. ChemBioChem 2022, 23, e202200402. [Google Scholar] [CrossRef] [PubMed]
- Wulff, G.; Lauer, M.; Böhnke, H. Rapid proton transfer as cause of an unusually large neighboring group effect. Angew. Chem. Int. Ed. 1984, 23, 741–742. [Google Scholar] [CrossRef]
- Ye, S.; Han, T.; Cheng, M.; Dong, L. Wulff-type boronic acid-functionalized quantum dots for rapid and sensitive detection of Gram-negative bacteria. Sens. Actuators B Chem. 2022, 356, 131332. [Google Scholar] [CrossRef]
- Dowlut, M.; Hall, D.G. An improved class of sugar-binding boronic acids, soluble and capable of complexing glycosides in neutral water. J. Am. Chem. Soc. 2006, 128, 4226–4227. [Google Scholar] [CrossRef] [PubMed]
- Sorensen, M.D.; Martins, R.; Hindsgaul, O. Assessing the terminal glycosylation of a glycoprotein by the naked eye. Angew. Chem. Int. Ed. 2007, 46, 2403–2407. [Google Scholar] [CrossRef] [PubMed]
- Lindstedt, G. Periodate oxidation of sugars in neutral phosphate buffer. Nature 1945, 156, 448–449. [Google Scholar] [CrossRef]
- Zeng, Y.; Ramya, T.; Dirksen, A.; Dawson, P.E.; Paulson, J.C. High-efficiency labeling of sialylated glycoproteins on living cells. Nat. Methods 2009, 6, 207–209. [Google Scholar] [CrossRef]
- Chen, B.B.; Wang, X.Y.; Qian, R.C. Rolling “wool-balls”: Rapid live-cell mapping of membrane sialic acids via poly-p-benzoquinone/ethylenediamine nanoclusters. Chem. Commun. 2019, 55, 9681–9684. [Google Scholar] [CrossRef]
- Yang, Y.Q.; Qian, X.P.; Zhang, L.H.; Miao, W.J.; Ming, D.M.; Jiang, L.; Huang, H. Enhanced imaging of glycan expressing cancer cells using poly (glycidyl methacrylate)-grafted silica nanospheres labeled with quantum dots. Anal. Chim. Acta 2020, 1095, 138–145. [Google Scholar] [CrossRef]
- Liu, L.; Chen, X.Y.; Sun, B. Construction of a recyclable DNAzyme motor for MUC1-specific glycoform in situ quantification. Anal. Chem. 2022, 94, 13745–13752. [Google Scholar] [CrossRef]
- Fernandez-Poza, S.; Padros, A.; Thompson, R.; Butler, L.; Islam, M.; Mosely, J.A.; Scrivens, J.H.; Rehman, M.F.; Akram, M.S. Tailor-Made Recombinant Prokaryotic Lectins for Characterisation of Glycoproteins. Anal. Chim. Acta 2021, 1155, 338352. [Google Scholar] [CrossRef] [PubMed]
- Tamura, T.; Omura, Y.; Kotera, K.; Ito, R.; Ohno, S.; Manabe, N.; Yamaguchi, Y.; Tamura, J. Synthesis of the Matriglycan Hexasaccharide, -3xylα1-3glcaβ1-Trimer and Its Interaction with Laminin. Org. Biomol. Chem. 2022, 20, 8489–8500. [Google Scholar] [CrossRef] [PubMed]
- Lim, B.; Kydd, L.; Jaworski, J. A Peptide-Lectin Fusion Strategy for Developing a Glycan Probe for Use in Various Assay Formats. Chemosensors 2019, 7, 55. [Google Scholar] [CrossRef] [PubMed]
- Kelly, S.; Hansen, S.B.; Rübsam, H.; Saake, P.; Pedersen, E.B.; Gysel, K.; Madland, E.; Wu, S.L.; Wawra, S.; Reid, D.; et al. A Glycan Receptor Kinase Facilitates Intracellular Accommodation of Arbuscular Mycorrhiza and Symbiotic Rhizobia in the Legume Lotus japonicus. PLoS Biol. 2023, 21, e3002127. [Google Scholar] [CrossRef] [PubMed]
- Sáez, F.J.; Madrid, J.F.; Cardoso, S.; Gómez, L.; Hernández, F. Glycoconjugates of the Urodele Amphibian Testis Shown by Lectin Cytochemical Methods. Microsc. Res. Tech. 2004, 64, 63–76. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.J.; Zhu, M.Z.; Qiu, Y.N.; Zhong, Y.G.; Yan, H.; Wang, Q.; Bian, H.J.; Li, Z. Analysis of Glycan-Related Genes Expression and Glycan Profiles in Mice with Liver Fibrosis. J. Proteome Res. 2012, 11, 5277–5285. [Google Scholar] [CrossRef] [PubMed]
- Hirakawa, T.; Nasu, K.; Kai, K.; Aoyagi, Y.; Ishii, T.; Uemura, T.; Yano, M.; Narahara, H. Wisteria Floribunda Agglutinin-Binding Glycan Expression Is Decreased in Endometriomata. Reprod. Biol. Endocrinol. 2014, 12, 100. [Google Scholar] [CrossRef]
- Jones, C.J.P.; Wilsher, S.; Russo, G.; Aplin, J.D. Lectin Histochemistry Reveals Two Cytotrophoblast Differentiation Pathways During Placental Development in the Feline (Felis catus). Placenta 2023, 134, 30–38. [Google Scholar] [CrossRef]
- Duk, M.; Lisowska, E.; Wu, J.H.; Wu, A.M. The biotin/avidin-mediated microtiter plate lectin assay with the use of chemically modified glycoprotein ligand. Anal. Biochem. 1994, 221, 266–272. [Google Scholar] [CrossRef]
- Mislovičová, D.; Katrlík, J.; Paulovičová, E.; Gemeiner, P.; Tkac, J. Comparison of three distinct ELLA protocols for determination of apparent affinity constants between Con A and glycoproteins. Colloids Surf. B 2012, 94, 163–169. [Google Scholar] [CrossRef]
- Han, C.W.; Dong, T.B.; Wang, P.C.; Zhou, F.M. Microfluidically partitioned dual channels for accurate background subtraction in cellular binding studies by surface plasmon resonance microscopy. Anal. Chem. 2022, 94, 17303–17311. [Google Scholar] [CrossRef] [PubMed]
- Dong, T.B.; Han, C.W.; Liu, X.; Wang, Z.C.; Wang, Y.H.; Kang, Q.; Wang, P.C.; Zhou, F.M. Live cells versus fixated cells: Kinetic measurements of biomolecular interactions with the ligand tracer method and surface plasmon resonance microscopy. Mol. Pharm. 2023, 20, 2094–2104. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.W.; Wang, Y.M.; Xu, Y.; Wang, X.; Huang, Z.J.; Chen, J.M.; Li, Y.X.; Duan, Y.X. Ultrasensitive U-shaped fiber optic LSPR cytosensing for label-free and in situ evaluation of cell surface N-glycan expression. Sens. Actuators B Chem. 2019, 284, 582–588. [Google Scholar] [CrossRef]
- Koehler, M.; Aravamudhan, P.; Guzman-Cardozo, C.; Dumitru, A.C.; Yang, J.; Gargiulo, S.; Soumillion, P.; Dermody, T.S.; Alsteens, D. Glycan-mediated enhancement of reovirus receptor binding. Nat. Commun. 2019, 10, 4460. [Google Scholar] [CrossRef]
- Koehler, M.; Delguste, M.; Sieben, C.; Gillet, L.; Alsteens, D. Initial step of virus entry: Virion binding to cell-surface glycans. Annu. Rev. Virol. 2020, 7, 143–165. [Google Scholar] [CrossRef]
- Petitjean, S.J.L.; Chen, W.; Koehler, M.; Jimmidi, R.; Yang, J.; Mohammed, D.; Juniku, B.; Stanifer, M.L.; Boulant, S.; Vincent, S.P. Multivalent 9-O-Acetylated-sialic acid glycoclusters as potent inhibitors for SARS-CoV-2 infection. Nat. Commun. 2022, 13, 2564. [Google Scholar] [CrossRef]
- Osawa, S.; Matsumoto, A.; Maejima, Y.; Suzuki, T.; Miyahara, Y.; Otsuka, H. Direct observation of cell surface sialylation by atomic force microscopy employing boronic acid–sialic acid reversible interaction. Anal. Chem. 2020, 92, 11714–11720. [Google Scholar] [CrossRef]
- Fu, Y.; Qian, H.; Zhou, X.; Wu, Y.; Song, L.; Chen, K.; Bai, D.; Yang, Y.; Li, J.; Xie, G. Proximity ligation assay mediated rolling circle amplification strategy for in situ amplified imaging of glycosylated PD-L1. Anal. Bioanal. Chem. 2021, 413, 6929–6939. [Google Scholar] [CrossRef]
- Xu, L.; Zhong, T.; Zhao, W.; Yao, B.; Ding, L.; Ju, H. Chemoselective labeling-based spermatozoa glycan imaging reveals abnormal glycosylation in oligoasthenotspermia. Chin. Chem. Lett. 2023, 34, 108760. [Google Scholar] [CrossRef]
- Elgavish, S.; Shaanan, B. Lectin-carbohydrate interactions: Different folds, common recognition principles. Trends Biochem. Sci. 1997, 22, 462–467. [Google Scholar] [CrossRef]
- Hirabayashi, J.; Yamada, M.; Kuno, A.; Tateno, H. Lectin microarrays: Concept, principle and applications. Chem. Soc. Rev. 2013, 42, 4443–4458. [Google Scholar] [CrossRef] [PubMed]
- Hsu, K.L.; Pilobello, K.T.; Mahal, L.K. Analyzing the dynamic bacterial glycome with a lectin microarray approach. Nat. Chem. Biol. 2006, 2, 153–157. [Google Scholar] [CrossRef] [PubMed]
- Pilobello, K.T.; Slawek, D.E.; Mahal, L.K. A ratiometric lectin microarray approach to analysis of the dynamic mammalian glycome. Proc. Natl. Acad. Sci. USA 2007, 104, 11534–11539. [Google Scholar] [CrossRef] [PubMed]
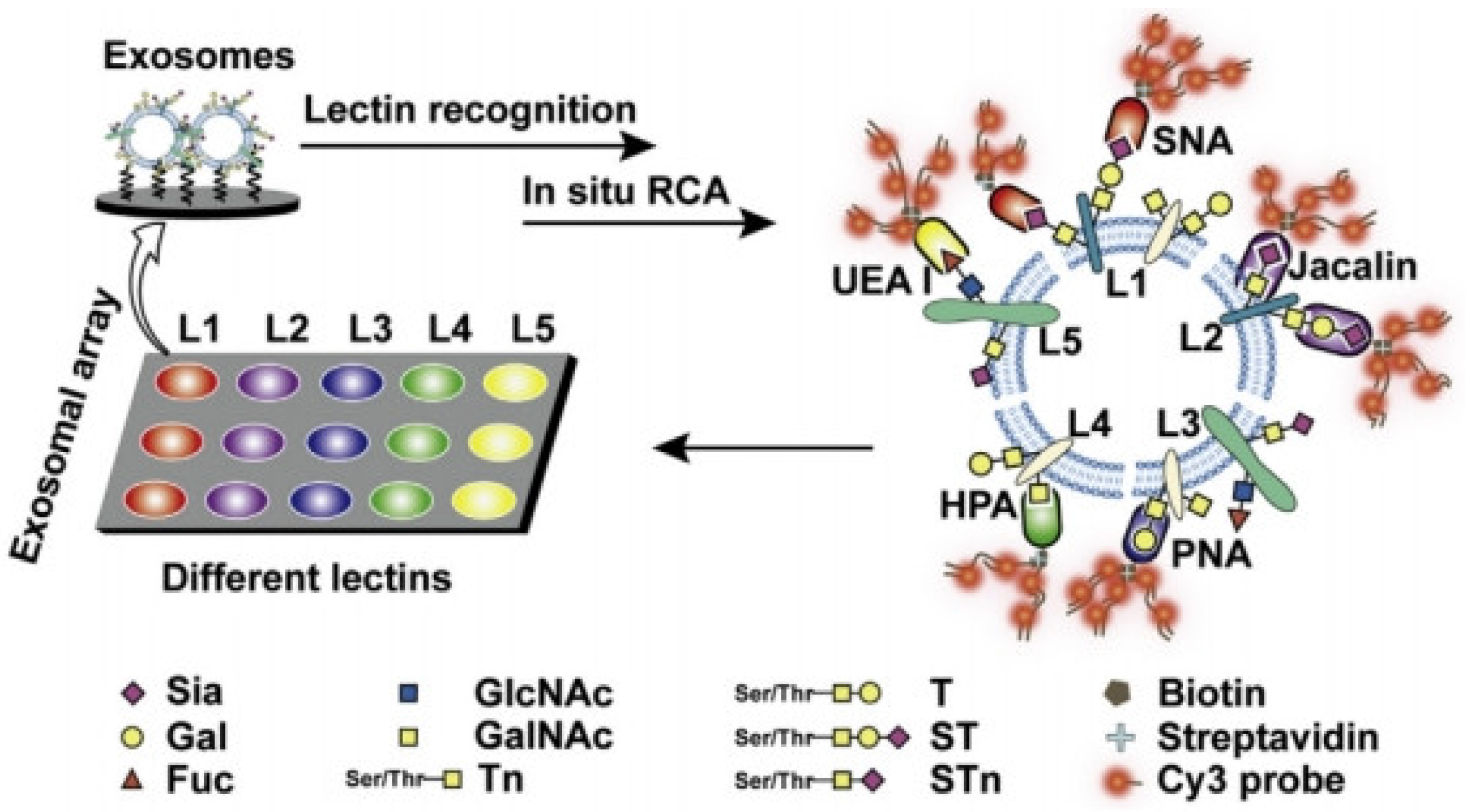
- Feng, Y.M.; Guo, Y.N.; Li, Y.R.; Tao, J.; Ding, L.; Wu, J.; Ju, H.X. Lectin-mediated in situ rolling circle amplification on exosomes for probing cancer-related glycan pattern. Anal. Chim. Acta 2018, 1039, 108–115. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Du, Y.F.; Zhu, Y.T.; Chen, X. A cis-membrane FRET-based method for protein-specific imaging of cell-surface glycans. J. Am. Chem. Soc. 2014, 136, 679–687. [Google Scholar] [CrossRef]
- Li, N.; Zhang, W.; Lin, L.; Shah, S.N.A.; Li, Y.; Lin, J.M. Nongenetically encoded and erasable imaging strategy for receptor-specific glycans on live cells. Anal. Chem. 2019, 91, 2600–2604. [Google Scholar] [CrossRef]
- Belardi, B.; de la Zerda, A.; Spiciarich, D.R.; Maund, S.L.; Peehl, D.M.; Bertozzi, C.R. Imaging the glycosylation state of cell surface glycoproteins by two-photon fluorescence lifetime imaging microscopy. Angew. Chem. Int. Ed. 2013, 52, 14045–14049. [Google Scholar] [CrossRef]
- Haga, Y.; Ishii, K.; Hibino, K.; Sako, Y.; Ito, Y.; Taniguchi, N.; Suzuki, T. Visualizing specific protein glycoforms by transmembrane fluorescence resonance energy transfer. Nat. Commun. 2012, 3, 907. [Google Scholar] [CrossRef]
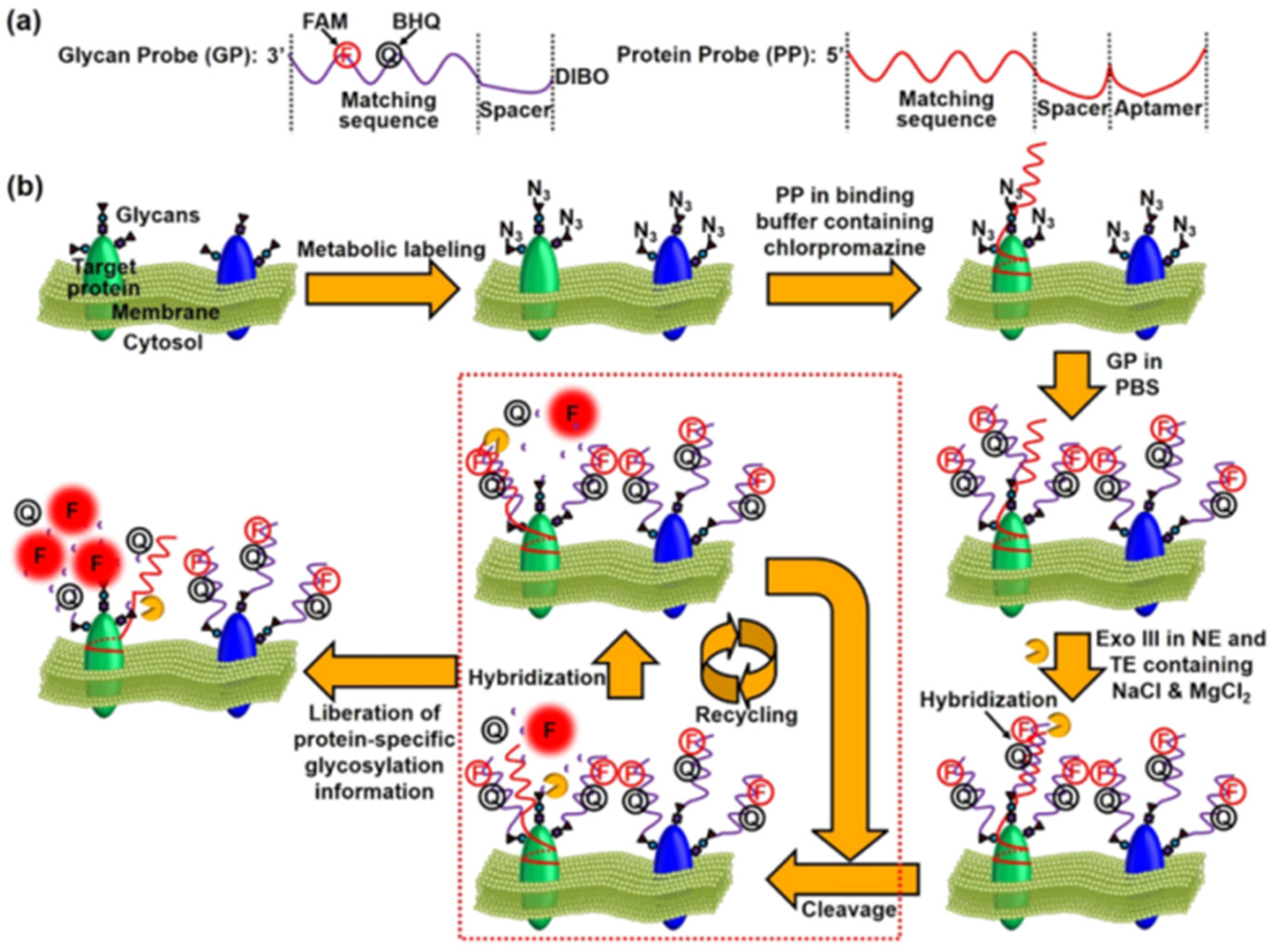
- Chen, Y.; Ding, L.; Song, W.; Yang, M.; Ju, H.X. Liberation of protein-specific glycosylation information for glycan analysis by exonuclease III-aided recycling hybridization. Anal. Chem. 2016, 88, 2923–2928. [Google Scholar] [CrossRef]
- Liang, L.L.; Lan, F.F.; Li, L.; Ge, S.; Yu, J.H.; Ren, N.; Liu, H.Y.; Yan, M. Paper analytical devices for dynamic evaluation of cell surface N-glycan expression via a bimodal biosensor based on multibranched hybridization chain reaction amplification. Biosens. Bioelectron. 2016, 86, 756–763. [Google Scholar] [CrossRef]
- Swierczewska, M.; Liu, G.; Lee, S.; Chen, X. High-sensitivity nanosensors for biomarker detection. Chem. Soc. Rev. 2012, 41, 2641–2655. [Google Scholar] [CrossRef] [PubMed]
- Reguera, J.; Langer, J.; Jimenez de Aberasturi, D.; Liz-Marzan, L.M. Anisotropic metal nanoparticles for surface enhanced Raman scattering. Chem. Soc. Rev. 2017, 46, 3866–3885. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.Y.; Zong, S.F.; Wu, L.; Zhu, D.; Cui, Y.P. SERS-activated platforms for immunoassay: Probes, encoding methods, and applications. Chem. Rev. 2017, 117, 7910–7963. [Google Scholar] [CrossRef] [PubMed]
- Vangala, K.; Yanney, M.; Hsiao, C.T.; Wu, W.W.; Shen, R.F.; Zou, S.; Sygula, A.; Zhang, D. Sensitive carbohydrate detection using surface enhanced Raman tagging. Anal. Chem. 2010, 82, 10164–10171. [Google Scholar] [CrossRef] [PubMed]
- Shafer-Peltier, K.E.; Haynes, C.L.; Glucksberg, M.R.; Van Duyne, R.P. Toward a glucose biosensor based on surface-enhanced Raman scattering. J. Am. Chem. Soc. 2003, 125, 588–593. [Google Scholar] [CrossRef]
- Sun, D.; Qi, G.H.; Xu, S.P.; Xu, W.Q. Construction of highly sensitive surface-enhanced Raman scattering (SERS) nanosensor aimed for the testing of glucose in urine. RSC Adv. 2016, 6, 53800–53803. [Google Scholar] [CrossRef]
- Sun, F.; Bai, T.; Zhang, L.; Ella-Menye, J.R.; Liu, S.J.; Nowinski, A.K.; Jiang, S.Y.; Yu, Q.M. Sensitive and fast detection of fructose in complex media via symmetry breaking and signal amplification using surface-enhanced Raman spectroscopy. Anal. Chem. 2014, 86, 2387–2394. [Google Scholar] [CrossRef]
- Chen, Y.L.; Ding, L.; Xu, J.Q.; Song, W.Y.; Yang, M.; Hu, J.J.; Ju, H.X. Micro-competition system for Raman quantification of multiple glycans on intact cell surface. Chem. Sci. 2015, 6, 3769–3774. [Google Scholar] [CrossRef]
- Di, H.; Liu, H.; Li, M.; Li, J.; Liu, D. High-precision profiling of sialic acid expression in cancer cells and tissues using background-free surface-enhanced raman scattering tags. Anal. Chem. 2017, 89, 5874–5881. [Google Scholar] [CrossRef]
- Ye, J.; Chen, Y.; Liu, Z. A boronate affinity sandwich assay: An appealing alternative to immunoassays for the determination of glycoproteins. Angew. Chem. Int. Ed. 2014, 53, 10386–10389. [Google Scholar] [CrossRef]
- Cordina, N.M.; Zhang, W.; Packer, N.H.; Wang, Y. Rapid and sensitive glycan targeting by lectin-SERS assay. Mol. Omics 2020, 16, 339–344. [Google Scholar] [CrossRef] [PubMed]
- Miao, X.R.; Wen, S.P.; Su, Y.; Fu, J.J.; Luo, X.J.; Wu, P.; Cai, C.X.; Jelinek, R.; Jiang, L.P.; Zhu, J.J. Graphene quantum dots wrapped gold nanoparticles with integrated enhancement mechanisms as sensitive and homogeneous substrates for surface-enhanced Raman spectroscopy. Anal. Chem. 2019, 91, 7295–7303. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Liang, Y.; Cao, W.; Gao, W.; Tang, B. Proximity-induced hybridization chain reaction-based photoacoustic imaging system for amplified visualization protein-specific glycosylation in mice. Anal. Chem. 2021, 93, 8915–8922. [Google Scholar] [CrossRef] [PubMed]
- Akiba, U.; Anzai, J.I. Recent progress in electrochemical biosensors for glycoproteins. Sensors 2016, 16, 2045. [Google Scholar] [CrossRef] [PubMed]
- Shah, A.K.; Hill, M.M.; Shiddiky, M.J.; Trau, M. Electrochemical detection of glycan and protein epitopes of glycoproteins in serum. Analyst 2014, 139, 5970–5976. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.D.; Ding, L.; Lei, J.P.; Ju, H.X. A simple electrochemical lectin-probe for in situ homogeneous cytosensing and facile evaluation of cell surface glycan. Biosens. Bioelectron. 2010, 26, 169–174. [Google Scholar] [CrossRef]
- Xia, N.; Cheng, C.; Liu, L.; Peng, P.Z.; Liu, C.; Chen, J.X. Electrochemical glycoprotein aptasensors based on the in-situ aggregation of silver nanoparticles induced by 4-mercaptophenylboronic acid. Microchim. Acta 2017, 184, 4393–4400. [Google Scholar] [CrossRef]
- Chang, Y.; Liu, G.; Li, S.; Liu, L.; Song, Q.J. Biorecognition element-free electrochemical detection of recombinant glycoproteins using metal-organic frameworks as signal tags. Anal. Chim. Acta 2023, 1273, 341540. [Google Scholar] [CrossRef]
- Chen, X.J.; Wang, Y.Z.; Zhang, Y.Y.; Chen, Z.H.; Liu, Y.; Li, Z.L.; Li, J.H. Sensitive electrochemical aptamer biosensor for dynamic cell surface N-glycan evaluation featuring multivalent recognition and signal amplification on a dendrimer-graphene electrode interface. Anal. Chem. 2014, 86, 4278–4286. [Google Scholar] [CrossRef]
- Liu, J.X.; Bao, N.; Luo, X.; Ding, S.N. Nonenzymatic amperometric aptamer cytosensor for ultrasensitive detection of circulating tumor cells and dynamic evaluation of cell surface N-glycan expression. ACS Omega 2018, 3, 8595–8604. [Google Scholar] [CrossRef]
- Yang, H.Y.; Li, Z.J.; Wei, X.M.; Huang, R.; Qi, H.L.; Gao, Q.; Li, C.Z.; Zhang, C.X. Detection and discrimination of alpha-fetoprotein with a label-free electrochemical impedance spectroscopy biosensor array based on lectin functionalized carbon nanotubes. Talanta 2013, 111, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.A.; Teng, Y.Q.; Fu, Y.; Zhang, S.P.; Wang, T.; Wang, C.G.; Jin, L.; Zhang, W. Lectin-based electrochemical biosensor constructed by functionalized carbon nanotubes for the competitive assay of glycan expression on living cancer cells. Chem. Sci. 2011, 2, 2353–2360. [Google Scholar] [CrossRef]
- Rivnay, J.; Inal, S.; Salleo, A.; Owens, R.M.; Berggren, M.; Malliaras, G.G. Organic electrochemical transistors. Nat. Rev. Mater. 2018, 3, 17086. [Google Scholar] [CrossRef]
- Chen, L.Z.; Wang, N.X.; Wu, J.; Yan, F.; Ju, H.X. Organic electrochemical transistor for sensing of sialic acid in serum samples. Anal. Chim. Acta 2020, 1128, 231–237. [Google Scholar] [CrossRef] [PubMed]
- Liao, C.; Mak, C.; Zhang, M.; Chan, H.L.; Yan, F. Flexible organic electrochemical transistors for highly selective enzyme biosensors and used for saliva testing. Adv. Mater. 2015, 27, 676–681. [Google Scholar] [CrossRef]
- Chen, L.Z.; Fu, Y.; Wang, N.X.; Yang, A.N.; Li, Y.Z.; Wu, J.; Ju, H.; Yan, F. Organic electrochemical transistors for the detection of cell surface glycans. ACS Appl. Mater. Interfaces 2018, 10, 18470–18477. [Google Scholar] [CrossRef]
- Chen, L.Z.; Wu, J.; Yan, F.; Ju, H.X. Monose-modified organic electrochemical transistors for cell surface glycan analysis via competitive recognition to enzyme-labeled lectin. Microchim. Acta 2021, 188, 252. [Google Scholar] [CrossRef]
- Chen, L.Z.; Wu, J.; Yan, F.; Ju, H.X. A facile strategy for quantitative sensing of glycans on cell surface using organic electrochemical transistors. Biosens. Bioelectron. 2021, 175, 112878. [Google Scholar] [CrossRef]
- Zhang, J.J.; Cheng, F.F.; Zheng, T.T.; Zhu, J.J. Versatile aptasensor for electrochemical quantification of cell surface glycan and naked-eye tracking glycolytic inhibition in living cells. Biosens. Bioelectron. 2017, 89, 937–945. [Google Scholar] [CrossRef]
- Qian, R.C.; Ding, L.; Bao, L.; He, S.J.; Ju, H.X. In situ electrochemical assay of cell surface sialic acids featuring highly efficient chemoselective recognition and a dual-functionalized nanohorn probe. Chem. Commun. 2012, 48, 3848–3850. [Google Scholar] [CrossRef]
- Hu, Q.; Wan, J.W.; Liang, Z.W.; Li, S.Q.; Feng, W.X.; Liang, Y.Y.; Luo, Y.L.; Cao, X.J.; Ma, Y.M.; Han, D.X.; et al. Dually amplified electrochemical aptasensor for endotoxin detection via target-assisted electrochemically mediated ATRP. Anal. Chem. 2023, 95, 5463–5469. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.A.; Lu, W.J.; Shen, J.Z.; Jiang, Y.X.; Han, E.; Dong, X.Y.; Huang, J.L. Carbohydrate derivative-functionalized biosensing toward highly sensitive electrochemical detection of cell surface glycan expression as cancer biomarker. Biosens. Bioelectron. 2015, 74, 291–298. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Ji, Q.J.; Qian, R.C.; Cheng, W.; Ju, H.X. Lectin-based nanoprobes functionalized with enzyme for highly sensitive electrochemical monitoring of dynamic carbohydrate expression on living cells. Anal. Chem. 2010, 82, 1292–1298. [Google Scholar] [CrossRef] [PubMed]
- Han, E.; Ding, L.; Jin, S.; Ju, H.X. Electrochemiluminescent biosensing of carbohydrate-functionalized CdS nanocomposites for in situ label-free analysis of cell surface carbohydrate. Biosens. Bioelectron. 2011, 26, 2500–2505. [Google Scholar] [CrossRef] [PubMed]
- Han, E.; Ding, L.; Lian, H.; Ju, H.X. Cytosensing and dynamic monitoring of cell surface carbohydrate expression by electrochemiluminescence of quantum dots. Chem. Commun. 2010, 46, 5446–5448. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Yang, Y.Y.; Li, C.X.; Yu, B.; Zhang, S.S. Enhanced iridium complex electrochemiluminescence cytosensing and dynamic evaluation of cell-surface carbohydrate expression. Chem. Eur. J. 2014, 20, 14736–14743. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Z.; Chen, Z.H.; Liu, Y.; Li, J.H. A functional glycoprotein competitive recognition and signal amplification strategy for carbohydrate-protein interaction profiling and cell surface carbohydrate expression evaluation. Nanoscale 2013, 5, 7349–7355. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.H.; Liu, Y.; Wang, Y.Z.; Zhao, X.; Li, J.H. Dynamic evaluation of cell surface N-glycan expression via an electrogenerated chemiluminescence biosensor based on concanavalin A-integrating gold-nanoparticle-modified Ru(bpy)32+-doped silica nanoprobe. Anal. Chem. 2013, 85, 4431–4438. [Google Scholar] [CrossRef]
- Chen, X.J.; He, Y.; Zhang, Y.Y.; Liu, M.L.; Liu, Y.; Li, J.H. Ultrasensitive detection of cancer cells and glycan expression profiling based on a multivalent recognition and alkaline phosphatase-responsive electrogenerated chemiluminescence biosensor. Nanoscale 2014, 6, 11196–11203. [Google Scholar] [CrossRef]
- Ma, W.; Xu, S.; Liu, H.W.; Bai, Y. Mass spectrometry methods for in situ analysis of clinical biomolecules. Small Methods 2020, 4, 1900407. [Google Scholar] [CrossRef]
- Ma, W.; Xu, S.T.; Nie, H.G.; Hu, B.Y.; Bai, Y.; Liu, H.W. Bifunctional cleavable probes for in situ multiplexed glycan detection and imaging using mass spectrometry. Chem. Sci. 2019, 10, 2320–2325. [Google Scholar] [CrossRef] [PubMed]
- Dressman, J.W.; McDowell, C.T.; Lu, X.; Angel, P.M.; Drake, R.R.; Mehta, A.S. Development of an antibody-based platform for the analysis of immune cell-specific N-linked glycosylation. Anal. Chem. 2023, 95, 10289–10297. [Google Scholar] [CrossRef] [PubMed]
- Powers, T.W.; Jones, E.E.; Betesh, L.R.; Romano, P.R.; Gao, P.; Copland, J.A.; Mehta, A.S.; Drake, R.R. Matrix assisted laser desorption ionization imaging mass spectrometry workflow for spatial profiling analysis of N-linked glycan expression in tissues. Anal. Chem. 2013, 85, 9799–9806. [Google Scholar] [CrossRef]
- Dai, C.; Cazares, L.H.; Wang, L.; Chu, Y.; Wang, S.L.; Troyer, D.A.; Semmes, O.J.; Drake, R.R.; Wang, B. Using boronolectin in MALDI-MS imaging for the histological analysis of cancer tissue expressing the sialyl Lewis X antigen. Chem. Commun. 2011, 47, 10338–10340. [Google Scholar] [CrossRef] [PubMed]
- He, Z.Y.; Chen, Q.S.; Chen, F.M.; Zhang, J.; Li, H.F.; Lin, J.M. DNA-mediated cell surface engineering for multiplexed glycan profiling using MALDI-TOF mass spectrometry. Chem. Sci. 2016, 7, 5448–5452. [Google Scholar] [CrossRef]
- Han, J.; Huang, X.; Liu, H.H.; Wang, J.Y.; Xiong, C.Q.; Nie, Z.X. Laser cleavable probes for in situ multiplexed glycan detection by single cell mass spectrometry. Chem. Sci. 2019, 10, 10958–10962. [Google Scholar] [CrossRef]
- Sun, J.; Liu, H.; Zhan, L.; Xiong, C.; Huang, X.; Xue, J.; Nie, Z.X. Laser cleavable probes-based cell surface engineering for in situ sialoglycoconjugates profiling by laser desorption/Ionization mass spectrometry. Anal. Chem. 2018, 90, 6397–6402. [Google Scholar] [CrossRef]
- Sun, B.; Xu, F.F.; Zhang, Y.Y.; Hu, Y.C.; Chen, Y. Dual-probe approach for mass spectrometric quantification of MUC1-specific terminal Gal/GalNAc in situ. Anal. Chem. 2020, 92, 8340–8349. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Wang, L.; Ding, L.; Ju, H. Cell-Surface Glycan Labeling and Sensing. Targets 2024, 2, 1-31. https://doi.org/10.3390/targets2010001
Li Y, Wang L, Ding L, Ju H. Cell-Surface Glycan Labeling and Sensing. Targets. 2024; 2(1):1-31. https://doi.org/10.3390/targets2010001
Chicago/Turabian StyleLi, Yiran, Lele Wang, Lin Ding, and Huangxian Ju. 2024. "Cell-Surface Glycan Labeling and Sensing" Targets 2, no. 1: 1-31. https://doi.org/10.3390/targets2010001
APA StyleLi, Y., Wang, L., Ding, L., & Ju, H. (2024). Cell-Surface Glycan Labeling and Sensing. Targets, 2(1), 1-31. https://doi.org/10.3390/targets2010001








