Abstract
Chest radiography needs timely diseases diagnosis and reporting of potential findings in the images, as it is an important diagnostic imaging test in medical practice. A crucial step in radiology workflow is the fast, automated, and reliable detection of diseases created on chest radiography. To overcome this issue, an artificial intelligence-based algorithm such as deep learning (DL) are promising methods for automatic and fast diagnosis due to their excellent performance analysis of a wide range of medical images and visual information. This paper surveys the DL methods for lung disease detection from chest X-ray images. The common five attributes surveyed in the articles are data augmentation, transfer learning, types of DL algorithms, types of lung diseases and features used for detection of abnormalities, and types of lung diseases. The presented methods may prove extremely useful for people to ideate their research contributions in this area.
1. Introduction
On this planet Earth, respiratory diseases are the main health threat to humankind. Approximately, 450 million people, i.e., almost 7% of the world’s population are infected by pneumonia alone, resulting in nearly 4 million deaths every year [1]. The diagnosis of these respiratory diseases is performed through most common radiology methods such as chest X-ray (CXR) and chest radiography because they are easily accessible and low cost. Visual inspection of a large number of chest radiographs is carried out on a slice-by-slice basis globally. This method involves more concentration, a high degree of precision, and skill, and it is expensive, prone to operator bias, time-consuming, and difficult to extract the valuable information present in such large-scale data [2]. In many countries, the complexity of chest radiographs has resulted in the shortage of expert radiologists because it is a crucial task for them to discriminate the respiratory diseases. Hence, there is a necessity to develop an automated method for the computer-aided diagnosis of respiratory diseases based on chest radiography.
Deep learning (DL) methods have achieved tremendous growth in the last decade, in the area of various computer vision applications such as the classification of medical and natural images [3]. This led to the development of deep convolutional neural networks (CNNs) for the diagnosis of respiratory diseases based on chest radiography.
The computer-based diagnosis of respiratory diseases consists of the detection of pathological abnormalities, followed by their classification. The challenging task is automated abnormality detection on chest radiographs due to the diversity and complexity of respiratory diseases and their limited quality. On chest radiographs, manual marking of abnormal regions needs even more labor and time than labeling it. Therefore, in many chest radiography data, abnormalities are masked [4], leading to the computer-aided diagnosis solution to a weakly supervised problem by showing only the names of abnormalities in each radiograph without their locations.
To predict X-ray image diagnostic information, machine-learning-based methods are proposed by many researchers [5]. The use of computer science-based methods to control huge volumes of medical records can decrease medical costs for health and medical science applications. In recent years, the use of DL algorithms on medical images for respiratory disease detection has grown to great heights. DL is derived from machine learning in that its algorithms are inspired by the structure and function of the human brain. The identification, classification, and quantification of patterns in medical images [6] are supported by DL methods. DL is gaining much importance by improving performance in many medical applications. Figure 1 describes a generalized manner in which deep neural networks process data and classify images. In turn, these improvements support clinicians in the classification and detection of certain medical conditions in a more efficient way [7].
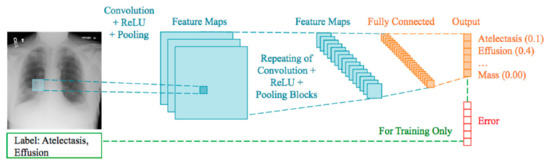
Figure 1.
Deep CNN learning paradigm of a multilabel classification task using the whole image.
The objective of this paper is to provide a comparison of the state-of-the-art DL-based respiratory disease detection methods and also identify the issues in this direction of research. Section 2 describes the taxonomy of various methods and respiratory issues that are considered for this study. Section 3 focuses on the issues that the authors observed during their research. The conclusion of the article is presented in Section 4.
2. Taxonomy of the State-of-the-Art DL Techniques for Respiratory Disorder from X-ray Images
This section discusses the state-of-the-art DL techniques used for the detection of lung diseases through CXR images. The main aim of this taxonomy is to provide a summarized and articulated view of the main focus points and major concepts related to the existing research carried out in this area. A total of five attributes that the authors found to be present commonly and imminent in the majority of the articles are identified and discussed in detail. These attributes are the types of DL algorithms, features used for detection of abnormalities, data augmentation, transfer learning, and types of lung diseases.
2.1. Features Extracted from Images
In the field of computer vision, a feature can be thought of as some form of numerical information that can be extracted from an image, which could prove beneficial in solving a certain set of problems [8]. Features could be certain structures in the image such as edges, shapes, and sizes of objects, or even specific points in the image.
The process of feature transformation relates to the generation of new features of an image based on the information extracted from existing ones. The newly generated features might have more powerful ways to represent the regions of importance in the image when viewed from a different dimension or space, compared with the original. This improved representation of the image has proven extremely beneficial when they are subjected to various machine learning algorithms. Some of the most prevalent extracted features in images include Gabor, local binary patterns (LBPs), edge histogram descriptor (EHD), color-and-edge direction descriptor (CEDD), color layout descriptor (CLD) [9], autocorrelation, scale-invariant feature transform (SIFT), edge frequency, and speeded up robust features (SURFs). The concept of histograms was also used to generate features in the form of a histogram of oriented gradients (HOGs), pyramid HOGs. intensity histograms (IHs), gradient magnitude histograms (GMs), and fuzzy color and texture histograms (FCTHs). From the recent literature, it has been observed that CNNs have the capability to automatically extract the relevant features without the need for explicit manual implementation of handpicked features from the images [10].
2.2. Data Augmentation
Having a large training dataset image in DL helps in improving the training accuracy. When compared with a strong algorithm with modest data, a weak algorithm on large-scale data can be more accurate. The presence of imbalanced classes is another obstacle that is encountered. The resulting model would be biased when the number of samples belonging to a particular class is larger than the other class during binary classification training. For the optimal performance of DL algorithms, the number of samples should be equal or balanced in each class. Image augmentation is a technique to increase the training dataset by creating variations of the original images, without obtaining new images. The variations are achieved by various processing methods such as flips, rotations, zooms, adding noise, and translations [11]. A few examples of augmented images are shown in Figure 2.
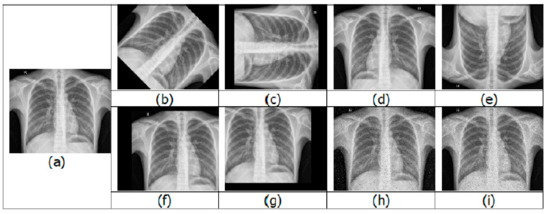
Figure 2.
Examples of image augmentation: (a) original; (b) 45° rotation; (c) 90° rotation; (d) horizontal flip; (e) vertical flip; (f) positive x and y translation; (g) negative x and y translation; (h) salt-and-pepper noise; (i) speckle noise.
Overfitting is also prevented during data augmentation, where the network tries to learn a very high variance function. Data augmentation addresses it by introducing the model with more diverse data, which decreases variance and improves the generalization of the model. Some of the disadvantages of data augmentation are its inability to overcome all biases in a small dataset [12], transformation computing costs, additional training time, and memory costs.
2.3. Types of DL Algorithm
CNNs are the common DL algorithm used to find patterns in images. A CNN consists of neurons with trainable weights and biases analogous to the neurons of the human brain. Several inputs are received by each neuron, which computes the weighted sum of its inputs, which is then activated to produce an output. CNNs have convolution layers, compared with other neural networks. Figure 3 shows a typical CNN architecture [13].
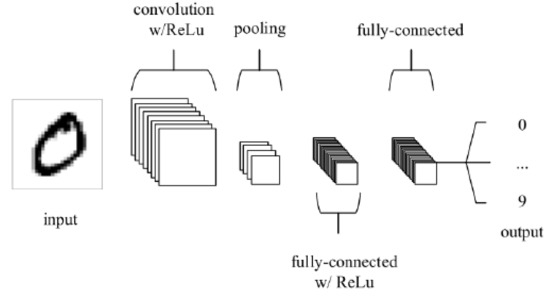
Figure 3.
CNN structure.
The two general stages during the learning phase of CNN are feature extraction and classification. In feature extraction, a kernel or a filter is used to perform convolution on the input data, which generates a feature map. The probability of the image belonging to a specific label/class is computed by the CNN in the classification stage. The main advantage of using CNN is that it automatically learns features for image classification and recognition without needing manual feature extraction. Transfer learning can be used to retrain CNN to be applied to a different domain [14], which is shown to produce better classification results.
Another DL algorithm that is a stack of restricted Boltzmann machines (RBMs) is DBN [15]. Except for the first and final layers of DBN, every other layer has two functions, that is, they serve as input layer for succeeding layer nodes and as a hidden layer for the preceding layer nodes. The design of the first RBM is made as accurate as possible to train a DBN. The output from the first RBM is used to train the second RBM by treating the first RBM’s hidden layer as the input layer. This process is iterated until all the network layer is trained. The model thus created during this initial training of the DBN can detect data patterns. DBN model finds applications in recognizing motion-capture data, objects in video sequences, and images [16].
2.4. Transfer Learning
Transfer learning is an emerging and popular method in computer vision, as it allows the building of accurate models. A model trained in a particular domain can be retrained using transfer learning to be used on a different domain. Transfer learning could be performed with or without a pretrained model.
A model that has been developed to solve an analogous task is called a pretrained model, which can be used as a starting point to solve the current task. The weights and architecture obtained by the pretrained models on large datasets can be applied to the current task. The main advantage in using such a pretrained model is a reduction in training costs for the new model [17], as it is sufficient to train and modify the weights related to the last few layers.
When the transfer learning approach is followed, there is a necessity to consider two main criteria. The first one is the selection of a pretrained model by ensuring that the model has worked on a similar dataset as the dataset under consideration. The second one is that the weights of the CNN have to be trained and fine-tuned with lower learning rates, such that they are not distorted and are expected to be relatively good [18].
2.5. Type of Disease
Applications of DL techniques for detection of most common causes of critical illness related to the lung [19] such as pneumonia and tuberculosis, as well as COVID-19, an ongoing pandemic, are discussed in the next few sections.
2.5.1. Tuberculosis
The bacteria that cause tuberculosis is Mycobacterium tuberculosis. A report by the WHO states that tuberculosis ranks in the 10 most common causes of death in the world. In 2017, tuberculosis caused a total death of around 1.6 million people out of 10 million people infected across the world. Therefore, in order to have an increase in chances of recovery, there is a need for early detection of tuberculosis [20].
Computer-aided detection for tuberculosis (CAD4TB) is a tool jointly developed by Delft Imaging Systems, University of Cape Town Lung Institute, and Radboud University, Nijmegen, for tuberculosis detection is used in two studies. The patient’s CXR images are obtained and analyzed via CAD4TB cloud server/computer, to display an abnormality score from 0 to 100 based on the heat map generated of the patient’s lung. Murphy et al. [21] verified that CAD4TB v6 accurately performs in comparison with data read by radiology experts. Melendez et al. [22] combined clinical information and X-ray score by computer-aided detection for automated tuberculosis screening, which improved accuracies and specificities, compared with when only either type of information is used alone.
Several studies carried out in the literature use CNNs to classify tuberculosis. Heo et al. proposed a technique to improve the performance of CNNs by incorporating demographical information—namely, gender, age, and weight. Results indicate that the proposed method has superior sensitivity and higher area under the receiver-operating characteristic curve (AUC) score than the CNN based on CXR images only. Pasa et al. proposed a simple CNN for tuberculosis detection with reduced computational requirements and memory without losing the performance in the classification that proved more efficient and accurate than previous models [23].
The use of transfer learning has also been researched by several authors. Hwang et.al claimed an accuracy level of more than 90%, along with an AUC value of 0.96, through the use of transfer learning of ImageNet, after they trained their network on more than 10,000 chest X-rays. Lakhani and Sundaram [24] used pretrained GoogLeNet and AlexNet for the classification of pulmonary tuberculosis. Their methodology displayed an AUC value of 0.97 and 0.98, respectively. A combination of SVMs, for classification, and pretrained VGGNet, ResNet, and GoogLeNet, for feature extraction, was used for the detection of X-ray images with tuberculosis by Lopes and Valiati. They obtained AUCs in the vicinity of 0.9–0.912 [25].
Some authors have also used the NIH-14 dataset instead of ImageNet for pretrained models. This dataset contains a wide variety of diseases and falls under the same modality as that of the dataset considered for tuberculosis. Models pretrained on this dataset have been shown to learn better features for classifying tuberculosis.
A variety of other methodologies such as k-nearest neighbors (kNN), sequential minimal optimization, and simple linear regression have also been adopted for the classification of X-ray images related to tuberculosis [26]. Another technique that has been attempted along with the previously mentioned methodologies is the multiple-instance learning-based approach. This method presents the advantage of requiring a lower labeling detail during optimization. Additionally, the previously optimized systems can easily be retained due to the minimal supervision required by this method. Moreover, COGNEX developed an industry-level DL-based image analysis software known as ViDi, which also displayed comparatively accurate detection of tuberculosis in chest X-ray images [27].
2.5.2. Pneumonia
Pneumonia is a condition in which alveoli of one or both lungs are filled by pus of fluid, which leads to difficulty in breathing. Symptoms of this lung infection include chest pain, severe shortness of breath, fever or fatigue, and cough. It is still a recurrent cause of mortality and morbidity. A majority of the computer-aided techniques used for detecting pneumonia are aligned toward the use of data augmentation and DL. Tobias et al. [28] used a direct and straightforward CNN technique to detect this condition. Stephen et al. used various data augmentation techniques, such as shear, flip, rescaling, zooming, rotation, and rescaling, to train their CNN from scratch [29].
Authors have also used a pretrained CNN architecture trained on augmented data for the detection of pneumonia. Rajpurkar et al. [30] used randomized horizontal flipping as data augmentation, while Ayan and Ünver [31] used flipping, rotation, and zooming to augment their data. Chouhan et al. [32] on the other hand, also incorporated adding noise to the images along with the other methods to obtain an augmented version of the data on which to train their architecture.
A deep Siamese CNN architecture was also adopted for the purpose of classification of X-ray images with pneumonia. It utilizes a symmetric architecture that takes two images as inputs, which consist of the X-ray being split into the left and right halves. The architecture compares the extent of infection that has spread across both regions in the image, which is claimed to make the classification system more robust, according to a study conducted by Ref. [30]. Ref. [33] proposed that CNNs present higher levels of accuracy when compared with methods such as random forest, AdaBoost, decision tree, KNN, and XGBoost.
2.5.3. COVID-19
Coronavirus disease 2019 (COVID-19) is a highly infectious disease that is caused by the very recently discovered coronavirus [34]. The most susceptible people are senior citizens and those who have a history of medical conditions such as chronic respiratory problems, cardiovascular disease, diabetes, and cancer.
Approaches to detect this disease through X-ray images have also been attempted through CNNs trained on augmented datasets. Ref. [35] used an InseptionV3 architecture for this purpose and used it as a feature extractor. Ref. [36] also followed similar approaches. Methodologies to classify X-ray images into normal, COVID-19, and pneumonia with the help of transfer learning have been investigated. Other authors have also used transfer learning with CNN architectures trained on datasets augmented with techniques such as rotation, scaling, flipping, translation, and shifting of image intensity for the three classes of classification of X-ray images [37].
Along with the classification of chest X-ray images for COVID-19, some authors have also modified CNN architectures to rather detect the disease. Sedik et.al [38] used an amalgam of CNN and LSTM, while Ahsan et al. [39] used an MLP–CNN model.
3. Issues Observed in the Present Area of Research
The main issues that were observed during this study were (a) data imbalance, (b) handling of images with large size, and (c) limitation of datasets that can be used.
- (a)
- Data imbalance: It is of paramount importance that there be a similar number of images for each class. If this condition is not satisfied, then the trained model would tend to be more biased toward the class that has more volume of data. This can be disastrous in medical applications.
- (b)
- Handling of images with large sizes: It is usually computationally expensive to work with high-resolution data. For this purpose, researchers tend to reduce the size of the original images to counter this problem. Even with the use of powerful GPUs, training a DL architecture is quite time-consuming.
- (c)
- Limitation of datasets: To obtain extremely accurate results, it would be desirable to have thousands of images for each class into which the images need to be classified. However, in real-life situations, the number of images available for training is less than ideal.
4. Conclusions
The presented study was an attempt to summarize and provide, in an organized manner, the key focus and concepts used for the detection of lung diseases through DL methodologies. A taxonomy of the state-of-the-art methodologies for this purpose was presented. Three main issues that might hinder the progress of research in this area were also put forward—namely, data imbalance, handling of images with large sizes, and limitation of datasets. The authors strongly believe that, in order for the research of this topic to progress in the right direction, such an investigative study would be helpful for other researchers who might be keen to contribute to this field.
Author Contributions
A.A.S.: Methodology, Formal Analysis; N.T.H.: Conceptualization, Investigation, Data Curation; A.C.V.: Resources, Validation; C.R.S.: Writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ruuskanen, O.; Lahti, E.; Jennings, L.C.; Murdoch, D.R. Viral pneumonia. Lancet 2011, 377, 1264–1275. [Google Scholar] [CrossRef]
- Wang, X.; Peng, Y.; Lu, L.; Lu, Z.; Bagheri, M.; Summers, R.M. Chestx-ray8: Hospital-scale chest X-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 3462–3471. [Google Scholar]
- Forum of International Respiratory Societies. The Global Impact of Respiratory Disease, 2nd ed.; European Respiratory Society: Sheffield, UK, 2017. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Wu, C.; Luo, C.; Xiong, N.; Zhang, W.; Kim, T.-H. A Greedy Deep Learning Method for Medical Disease Analysis. IEEE Access 2018, 6, 20021–20030. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [Green Version]
- Domingos, P. A few useful things to know about machine learning. Commun. ACM 2012, 55, 78–87. [Google Scholar] [CrossRef] [Green Version]
- Yao, L.; Poblenz, E.; Dagunts, D.; Covington, B.; Bernard, D.; Lyman, K. Learning to diagnose from scratch by exploiting dependencies among labels. arXiv 2017, arXiv:1710.10501. [Google Scholar]
- O’Mahony, N.; Campbell, S.; Carvalho, A.; Harapanahalli, S.; Hernandez, G.V.; Krpalkova, L.; Riordan, D.; Walsh, J. Deep Learning vs. Traditional Computer Vision. In Advances in Computer Vision; Springer International Publishing: Cham, Switzerland, 2019; pp. 128–144. [Google Scholar] [CrossRef] [Green Version]
- Mikołajczyk, A.; Grochowski, M. Data augmentation for improving deep learning in image classification problem. In Proceedings of the International Interdisciplinary Ph.D. Workshop (IIPh, DW), Swinoujscie, Poland, 9–12 May 2018; pp. 117–122. [Google Scholar] [CrossRef]
- Shorten, C.; Khoshgoftaar, T.M. A survey on Image Data Augmentation for Deep Learning. J. Big Data 2019, 6, 60. [Google Scholar] [CrossRef]
- Al-Ajlan, A.; Allali, A.E. CNN—MGP: Convolutional Neural Networks for Metagenomics Gene Prediction. Interdiscip. Sci. Comput. Life Sci. 2019, 11, 628–635. [Google Scholar] [CrossRef] [Green Version]
- O’Shea, K.; Nash, R. An Introduction to Convolutional Neural Networks. arXiv 2015, arXiv:1511.08458v2. [Google Scholar]
- Hinton, G.E.; Osindero, S.; The, Y.-W. A Fast Learning Algorithm for Deep Belief Nets. Neural Comput. 2006, 18, 1527–1554. [Google Scholar] [CrossRef]
- Nogueira, K.; Penatti, O.A.; dos Santos, J.A. Towards better exploiting convolutional neural networks for remote sensing scene classification. Pattern Recognit. 2017, 61, 539–556. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Chen, D.; Hao, L.; Liu, X.; Zeng, Y.; Chen, J.; Zhang, G. Pulmonary Image Classification Based on Inception-v3 Transfer Learning Model. IEEE Access 2019, 7, 146533–146541. [Google Scholar] [CrossRef]
- Cao, X.; Wipf, D.; Wen, F.; Duan, G.; Sun, J. A Practical Transfer Learning Algorithm for Face Verification. In Proceedings of the 2013 IEEE International Conference on Computer Vision, Sydney, Australia, 1–8 December 2013; pp. 3208–3215. [Google Scholar]
- Murphy, K.; Habib, S.S.; Zaidi, S.M.A.; Khowaja, S.; Khan, A.; Melendez, J.; Scholten, E.T.; Amad, F.; Schalekamp, S.; Verhagen, M.; et al. Computer aided detection of tuberculosis on chest radiographs: An evaluation of the CAD4TB v6 system. Sci. Rep. 2020, 10, 1–11. [Google Scholar] [CrossRef]
- Melendez, J.; Sánchez, C.I.; Philipsen, R.H.; Maduskar, P.; Dawson, R.; Theron, G.; Dheda, K.; Van Ginneken, B. An auto-mated tuberculosis screening strategy combining X-ray-based computer-aided detection and clinical information. Sci. Rep. 2016, 6, 1–8. [Google Scholar] [CrossRef]
- Heo, S.J.; Kim, Y.; Yun, S.; Lim, S.S.; Kim, J.; Nam, C.M.; Park, E.C.; Jung, I.; Yoon, J.H. Deep Learning Algorithms with De-mographic Information Help to Detect Tuberculosis in Chest Radiographs in Annual Workers’ Health Examination Data. Int. J. Environ. Res. Public Health 2019, 16, 250. [Google Scholar] [CrossRef] [Green Version]
- Vajda, S.; Karargyris, A.; Jaeger, S.; Santosh, K.C.; Candemir, S.; Xue, Z.; Antani, S.; Thoma, G. Feature Selection for Auto-matic Tuberculosis Screening in Frontal Chest Radiographs. J. Med. Syst. 2018, 42, 1–11. [Google Scholar] [CrossRef]
- Chauhan, A.; Chauhan, D.; Rout, C. Role of Gist and PHOG Features in Computer-Aided Diagnosis of Tuberculosis without Segmentation. PLoS ONE 2014, 9, e112980. [Google Scholar] [CrossRef]
- Jaeger, S.; Karargyris, A.; Candemir, S.; Folio, L.; Siegelman, J.; Callaghan, F.; Xue, Z.; Palaniappan, K.; Singh, R.K.; Antani, S.; et al. Automatic Tuberculosis Screening Using Chest Radiographs. IEEE Trans. Med. Imaging 2014, 33, 233–245. [Google Scholar] [CrossRef]
- Antony, B.; Nizar Banu, P.K. Lung tuberculosis detection using x-ray images. Int. J. Appl. Eng. Res. 2017, 12, 15196–15201. [Google Scholar]
- Pasa, F.; Golkov, V.; Pfeiffer, F.; Cremers, D.; Pfeiffer, D. Efficient Deep Network Architectures for Fast Chest X-Ray Tuber-culosis Screening and Visualization. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, Q.H.; Nguyen, B.P.; Dao, S.D.; Unnikrishnan, B.; Dhingra, R.; Ravichandran, S.R.; Satpathy, S.; Raja, P.N.; Chua, M.C.H. Deep Learning Models for Tuberculosis Detection from Chest X-ray Images. In Proceedings of the 2019 26th International Conference on Telecommunications (ICT), Hanoi, Vietnam, 8–10 April 2019; pp. 381–385. [Google Scholar]
- Moujahid, H.; Cherradi, B.; Gannour, O.E.; Bahatti, L.; Terrada, O.; Hamida, S. Convolutional Neural Network Based Classi-fication of Patients with Pneumonia using X-ray Lung Images. Adv. Sci. Technol. Eng. Syst. 2020, 5, 167–175. [Google Scholar] [CrossRef]
- Young, J.C.; Suryadibrata, A. Applicability of Various Pre-Trained Deep Convolutional Neural Networks for Pneumonia Classification based on X-Ray Images. Int. J. Adv. Trends Comput. Sci. Eng. 2020, 9, 2649–2654. [Google Scholar] [CrossRef]
- Stephen, O.; Sain, M.; Maduh, U.J.; Jeong, D.-U. An Efficient Deep Learning Approach to Pneumonia Classification in Healthcare. J. Health Eng. 2019, 2019, 4180949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kermany, D.S.; Goldbaum, M.; Cai, W.; Valentim, C.C.S.; Liang, H.; Baxter, S.L.; McKeown, A.; Yang, G.; Wu, X.; Yan, F.; et al. Identifying Medical Diagnoses and Treatable Diseases by Image-Based Deep Learning. Cell 2018, 172, 1122–1131.e9. [Google Scholar] [CrossRef] [PubMed]
- Wunderink, R.G.; Waterer, G. Advances in the causes and management of community acquired pneumonia in adults. BMJ 2017, 358, j2471. [Google Scholar] [CrossRef]
- Wardlaw, T.; Johansson, E.W.; Hodge, M. Pneumonia: The Forgotten Killer of Children; United Nations Children’s Fund (UNICEF): New York, NY, USA, 2006; p. 44. [Google Scholar]
- Rajpurkar, P.; Irvin, J.; Zhu, K.; Yang, B.; Mehta, H.; Duan, T.; Ding, D.; Bagul, A.; Ball, R.L.; Langlotz, C.; et al. CheXNet: Radiologist-Level Pneumonia Detection on Chest X-Rays with Deep Learning. arXiv 2017, arXiv:1711.05225v3. [Google Scholar]
- Zhu, J.; Shen, B.; Abbasi, A.; Hoshmand-Kochi, M.; Li, H.; Duong, T.Q. Deep transfer learning artificial intelligence accu-rately stages COVID-19 lung disease severity on portable chest radiographs. PLoS ONE 2020, 15, e0236621. [Google Scholar] [CrossRef]
- Narin, A.; Kaya, C.; Pamuk, Z. Automatic detection of coronavirus disease (COVID-19) using X-ray images and deep convolutional neural networks. Pattern Anal. Appl. 2021, 24, 1207–1220. [Google Scholar] [CrossRef]
- Li, L.; Qin, L.; Xu, Z.; Yin, Y.; Wang, X.; Kong, B.; Bai, J.; Lu, Y.; Fang, Z.; Song, Q.; et al. Artificial Intelligence Distinguishes COVID-19 from Community Acquired Pneumonia Based on Pulmonary CT: Evaluation of the Diagnostic Accuracy. Radiology 2020, 200905. [Google Scholar] [CrossRef]
- Sedik, A.; Iliyasu, A.M.; El-Rahiem, B.A.; Abdel Samea, M.E.; Abdel-Raheem, A.; Hammad, M.; Peng, J.; Abd El-Samie, F.E.; Abd El-Latif, A.A. Deploying machine and deep learning models for efficient data-augmented detection of COVID-19 infections. Viruses 2020, 12, 769. [Google Scholar] [CrossRef]
- Ahsan, M.; Alam, T.E.; Trafalis, T.; Huebner, P. Deep MLP-CNN Model Using Mixed-Data to Distinguish between COVID-19 and Non-COVID-19 Patients. Symmetry 2020, 12, 1526. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).