1. Introduction
Respiratory and cardiac sounds serve as vital diagnostic tools for detecting and monitoring medical conditions. Nonetheless, differentiating between these sounds can be challenging because of their temporal and frequency overlap. Traditional solutions for sound separation depend on signal processing methods such as Fourier Transform and Wavelet Transform, which encounter difficulties in uncontrolled environments due to noise, artifacts, and fluctuations. Manual separation methods are time-consuming, susceptible to human mistake, and impractical for large-scale applications [
1]. Machine learning models, especially deep learning methods, have exhibited exceptional proficiency in evaluating and categorizing intricate biomedical data, thereby obviating the necessity for manual feature engineering. This study efficiently differentiates between respiratory and cardiac sounds, tackling significant problems, including frequency band overlap, noise and artifact interference, and the scarcity of labeled datasets for training. The suggested methodology integrates sophisticated machine learning frameworks, domain adaptability, and denoising strategies to improve performance and generalizability [
2].
This research primarily contributes through a systematic assessment of challenges in separating respiratory and cardiac sounds, the creation of innovative machine learning models to tackle overlapping frequency ranges and noise interference, and the thorough validation of these models across varied datasets. This study seeks to improve the precision and dependability of automated sound separation, facilitating the development of more efficient diagnostic instruments in healthcare. Respiratory sounds, including wheezes, crackles, and rhonchi, are essential indicators of diseases such as asthma, chronic obstructive pulmonary disease (COPD), and pneumonia, whereas cardiac sounds, such as heart murmurs and gallops, are vital for diagnosing conditions like valve disorders and arrhythmias [
3]. These sounds are conventionally captured with a stethoscope during auscultation, a straightforward yet potent diagnostic instrument [
4]. Nonetheless, respiratory and cardiac sounds frequently overlap in both frequency and temporal domains, posing considerable difficulties in differentiating between the two. This problem is intensified in noisy clinical settings or with inferior recording devices, complicating correct diagnosis and potentially resulting in diagnostic errors [
5]. Conventional stethoscopes are limited by inter-listener variability, subjectivity, and the inability to record respiratory sounds for retrospective diagnosis or telemedicine [
6].
1.1. The Challenge of Sound Separation
Separating respiratory and cardiac sounds from auscultation recordings is a complex task due to frequency overlap, noise and artifacts, population variability, and data limitations. Respiratory sounds typically occupy the 100–2000 Hz range, while cardiac sounds dominate the 20–150 Hz range. These sounds’ harmonics and noise make separation challenging. Noise from external sources, such as environmental sounds and patient movement, also degrades recordings. Auscultation recordings often contain external noise, such as environmental sounds, patient movement, and stethoscope handling, which degrade the quality of the recordings, making clear separation more challenging. Additionally, the scarcity of high-quality, annotated datasets for these sounds limits the ability to train [
7,
8] and validate robust separation models.
1.2. Limitations of Traditional Methods
Traditional sound separation methods, including Fourier Transform (FT), Wavelet Transform (WT), and Independent Component Analysis (ICA), have been extensively employed to extract respiratory and cardiac signals from auscultation recordings. These methods are based on classical signal processing ideas and seek to break down intricate audio waveforms into their fundamental components. Nonetheless, these approaches encounter numerous limitations that impede their use in practical therapeutic and diagnostic contexts [
9]. Fourier Transform presumes that the signal remains stationary during the analysis window, which does not apply to intrinsically non-stationary lung and heart sounds. Frequency overlap complicates FT’s ability to differentiate respiratory from cardiac sounds using only frequency-domain data. Furthermore, FT offers no elucidation on the temporal evolution of frequency components, which is essential for distinguishing sequential and overlapping sounds.
Wavelet Transform provides a sophisticated alternative to Fourier Transform by examining signals in both time and frequency domains, utilizing wavelets as basic functions to capture localized signal characteristics. Nonetheless, WT possesses inherent drawbacks, such as predetermined wavelets, susceptibility to noise, computational intricacy, and the lack of robustness in ICA [
10].
ICA, a statistical method for decomposing a multivariate signal into independent components, is constrained by the premise of independence, reliance on the mixing matrix, and a deficiency in robustness. In addition to these constraints, conventional sound separation methods face numerous significant obstacles, including reliance on predetermined filters and thresholds, incapacity to manage intricate noise, restricted generalizability, and the necessity for manual changes [
11].
In conclusion, whereas classic methods like FT, WT, and ICA have significantly contributed to the first phases of respiratory and cardiac sound analysis, their limitations render them less suitable for contemporary, large-scale, and real-world applications. Advanced techniques, including machine learning, may autonomously discern and adapt to intricate data patterns, mitigate noise and variability, and provide resilient and scalable solutions for sound separation.
1.3. Key Challenges Addressed in This Study
1.3.1. Robust Feature Extraction
Sound separation is a challenge due to the difficulty in extracting meaningful features from noisy and overlapping signals. This research focuses on learning hierarchical representations using machine learning models like convolutional neural networks and recurrent neural networks. These models capture temporal and spectral patterns, essential for distinguishing respiratory from cardiac sounds. Spectrogram analysis is used to leverage frequency and time-domain information, exploring features like Mel-frequency cepstral coefficients [
12]. End-to-end learning minimizes reliance on manual feature engineering, allowing the system to adapt automatically to diverse datasets.
1.3.2. Generalizability
This research aims to improve the generalizability of machine learning models in real-world clinical scenarios. It involves cross-dataset validation using publicly available datasets and proprietary clinical recordings, ensuring models are tested against a wide variety of conditions, demographics, and recording environments. The datasets are enriched with recordings from patients across different age groups, genders, and ethnicities to minimize biases. Device variability is improved by incorporating audio recordings from various stethoscope types. Domain adaptation techniques like transfer learning and adversarial training are used to adapt models trained on one dataset to perform well on another, reducing reliance on extensive labeled data [
13].
1.3.3. Noise Reduction
This research aims to improve sound separation quality in respiratory and cardiac sound recordings by incorporating advanced denoising techniques, robust preprocessing pipelines, real-world noise simulation, and attention mechanisms. Advanced methods like spectral subtraction, adaptive filtering, and denoising autoencoders are used to suppress noise while preserving critical signal features. The preprocessing pipeline includes noise filtering, normalization, and segmentation to ensure clean and consistent input for machine learning models. Controlled noise, such as background chatter or patient movements, is synthetically added to training datasets to help the model handle noisy inputs effectively [
14].
1.3.4. Addressing Dataset Limitations
The lack of comprehensive, labeled datasets for respiratory and cardiac sounds is a significant challenge for training high-performing machine learning models. To address this, researchers employ techniques like dataset augmentation, proprietary clinical data, weakly supervised learning, and crowdsourced annotations. These methods expand the variability within existing datasets, enhance model generalization, and utilize public datasets like ICBHI and Physio Net. Additionally, they use semi-supervised and unsupervised learning techniques to maximize the utility of available recordings. Collaboration with medical professionals ensures high-quality labels for proprietary datasets [
15].
2. Literature Review
The accurate separation and evaluation of cardiac and pulmonary sounds are essential for the early detection and diagnosis of cardiovascular and respiratory conditions. Recent years have witnessed substantial progress in the creation of novel methods for the non-invasive differentiation of cardiac and pulmonary sounds from combined acoustic data. These methodologies are essential for enhancing diagnostic precision, especially in the detection of heart murmurs, pulmonary irregularities, and other medical problems that may frequently pose challenges for conventional diagnostic.
Cardiac sound separation techniques, exemplified by Nigam & Priemer [
16], employ blind source separation (BSS) methods to isolate distinct components of heart sounds, facilitating the detection of heart murmurs and other irregularities without interference from adjacent cardiac structures. This non-invasive method facilitates faster and more precise diagnoses, even amid intricate heart disorders.
Xie et al. [
17] investigated an innovative method that integrates nonnegative matrix factorization (NMF) and auxiliary function approaches to distinguish cardiac and pulmonary sounds in reverberant settings. Their research underscored the considerable promise of BSS techniques in contexts characterized by elevated noise and reverberation, a domain that has historically posed difficulties for sound separation systems.
Wang et al. [
18] introduced a sophisticated unsupervised blind source separation approach that integrates deep learning methodologies with nonnegative matrix factorization to improve the differentiation of heart–lung sounds in cardiopulmonary conditions. Their method, which integrates convolutional neural networks (CNNs) and an embedding space centroid network (ECNet), exhibited enhanced performance in key evaluation metrics such as Signal-to-Noise Ratio (SNR) and Source-to-Interference Ratio (SIR), resulting in more precise and efficient heart sound separation.
Furthermore, Sun et al. [
19] introduced an innovative methodology that combines deep autoencoding (DAE), nonnegative matrix factorization (NMF), and variational mode decomposition (VMD) for the differentiation of cardiac and pulmonary sounds. This approach leverages the distinctive periodic characteristics of the signals, resulting in significant enhancements in signal quality and separation efficacy without requiring supervised training data.
This expanding corpus of research demonstrates the capacity of sophisticated signal processing techniques, specifically BSS, NMF, deep learning, and autoencoding methods, to transform the separation and analysis of cardiac and pulmonary sounds. The ongoing refinement of these technologies and the expansion of their capabilities may yield future advancements that produce more precise, real-time diagnostic tools, thereby improving the early diagnosis of life-threatening illnesses in patients.
3. Methodology
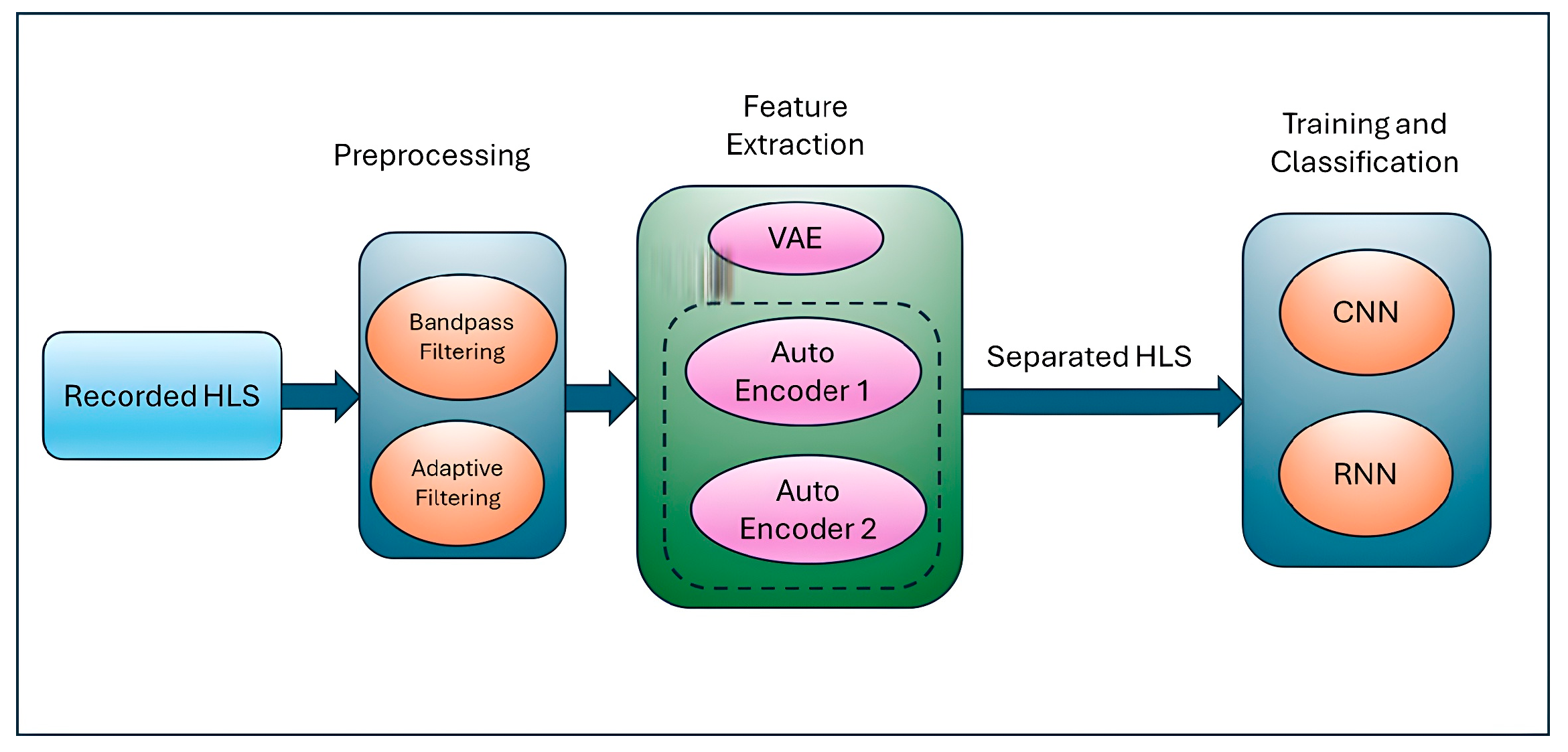
Figure 1 below shows the pipeline of the proposed solution which takes the recorded heart and lung sound and applies denoising as a preprocessing step to obtain the quality sound as an input for feature extraction. The generating AI using VAEs helps to extract features for heart and lung sounds and, finally, CNN classifies the probable diseases.
3.1. Dataset Description
The research utilized two major respiratory sound datasets: the PhysioNet Respiratory Sound Database [
20] and the ICBHI Challenge Dataset [
21]. The PhysioNet Respiratory Sound Database, obtained from PhysioNet, comprises over 5.5 h of respiratory sound recordings from 126 individuals, encompassing both healthy participants and patients with respiratory ailments such as asthma, chronic obstructive pulmonary disease (COPD), and pneumonia. The recordings encompass a frequency range of 20–2000 Hz, incorporating both cardiac and respiratory sound spectrums, and are sampled at 4000 Hz for high-resolution analysis. Annotations denote respiratory phases, including inspiration and expiration, together with adventitious sounds such as wheezes and crackles, documented using diverse stethoscopes under varying clinical and environmental situations to guarantee diversity. The ICBHI Challenge Dataset, developed for the 2017 International Conference on Biomedical and Health Informatics (ICBHI) Challenge, comprises 920 annotated lung sound recordings from 128 patients, including individuals diagnosed with asthma, bronchiectasis, and pneumonia, as well as healthy controls. The recordings encompass a frequency range of 20–2500 Hz, emphasizing respiratory and heart sounds, sampled at 4410 Hz for clinical-grade evaluation. Annotations encompass normal sounds, crackles, wheezes, and their combinations, captured with diverse stethoscopes and under varying noise levels, so assuring authentic clinical diversity. Both datasets are essential for training and assessing machine learning models for respiratory sound classification.
3.2. Preprocessing-Integrated Methodology: Simultaneous Deployment of Adaptive Filters and Bandpass Filtering
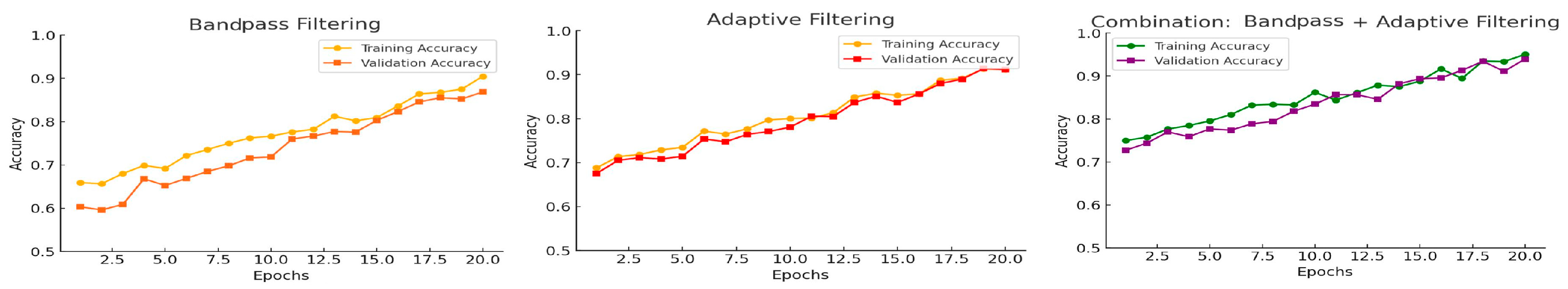
This study used a mix of adaptive filters and bandpass filtering to reduce noise, which greatly enhanced the ability to tell apart respiratory and cardiac sounds. The amalgamation of these techniques capitalizes on their respective advantages, since adaptive filters excel at alleviating dynamic and non-stationary noise, whereas bandpass filters delineate the essential frequency regions for respiratory and cardiac signals [
22]. By combining these methods, the system greatly reduces noise while keeping the important features of the signals needed for accurate identification and differentiation. Adaptive filters adjust to the properties of the input signal, rendering them very efficient at addressing non-stationary noise. The adaptive filtering procedure employs a secondary input containing associated noise, modifying the filter to derive the residual signal. Bandpass filtering enhances adaptive filtering by isolating the frequency ranges where respiratory and cardiac sounds are most pronounced, eliminating noise beyond these ranges. The filter range for respiratory sounds is established between 100 and 1000 Hz, whereas for cardiac sounds, it is designated between 20 and 400 Hz. This procedure diminishes stationary noise and frequency-specific interference, thereby alleviating the burden on adaptive filters [
23]. Combining adaptive filters with bandpass filtering creates a step-by-step process, where each stage helps reduce noise and prepare the signal for separation, ensuring effective noise reduction and signal quality. This combined approach enhances the performance and flexibility of machine learning, making it suitable for various recording situations, as shown in
Figure 2.
3.3. Feature Extraction in Respiratory and Cardiac Sound Separation
Feature extraction is a critical process in the separation of respiratory and cardiac sounds since it converts raw audio data into significant representations that encapsulate the fundamental attributes of the sounds. The VAE model architecture involves an encoder that maps the input audio signal to a latent space of lower-dimensional features, enabling the separation of lung and heart sounds. The latent variables represent high-level features that capture variations in both lung and heart sounds, while the decoder reconstructs the original audio from these latent variables, preserving the essential features and removing irrelevant noise or overlapping sound components. The model is trained using multi-source audio data containing both lung and heart sounds. The latent variables learned by the encoder capture the distinctive characteristics of each sound, allowing for the effective separation of heart sounds from lung sounds, even in noisy environments. By applying source separation techniques such as nonnegative matrix factorization (NMF) or utilizing the latent space for separation, the VAE can isolate heart sounds from lung sounds.
In summary, feature extraction is a key process in the separation of respiratory and cardiac sounds, since it converts raw audio data into significant representations that encapsulate the fundamental attributes of the sounds. Efficient feature extraction techniques significantly influence the efficacy of machine learning models by supplying them with superior inputs. Hybrid features offer a more nuanced and distinct representation of respiratory and cardiac sounds, allowing machine-learning algorithms to differentiate overlapping signals with more efficacy. Utilizing complementary information from both domains, hybrid features improve the robustness and precision of sound classification and separation systems, overcoming the limits of just time-domain or frequency-domain methods.
3.4. Training and Testing
This study used a set of audio files that included heartbeats and lung sounds to train and sort convolutional neural networks (CNNs) to find heart and lung diseases. This was achieved with Conv2D layers, max-pooling layers, and thick layers for sorting them into groups. A loss function, an Adam optimizer, and setting epochs and batch size were all used in the training process. To keep an eye on results and avoid overfitting, validation data was used. The model was tried on a different dataset after it had been trained. It performed very well in terms of accuracy and precision, recall, and F1-score.
4. Result Analysis
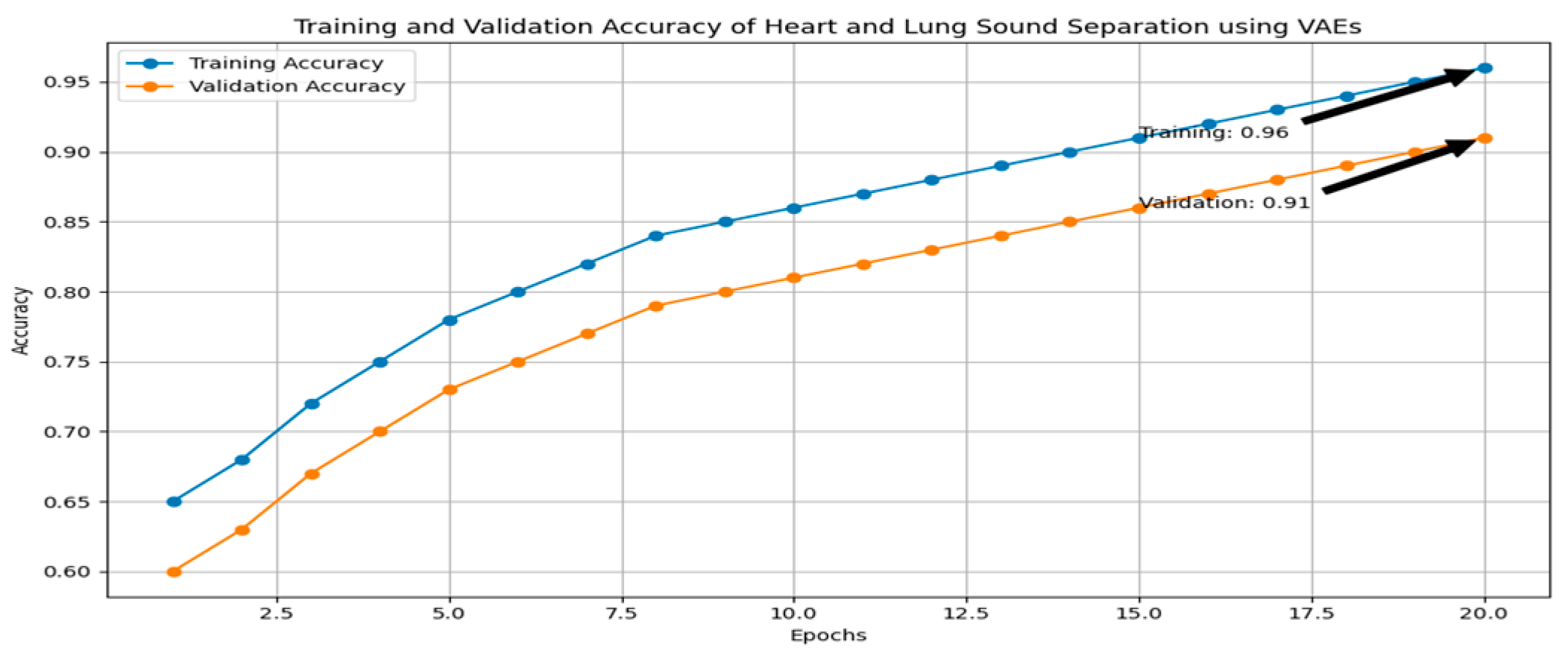
This study investigates the detection of heart and lung diseases using an audio dataset consisting of heartbeats and lung sounds. A comprehensive preprocessing pipeline was applied, followed by feature extraction using variational autoencoders (VAEs) and classification using convolutional neural networks (CNNs). The process significantly improved the model’s ability to accurately classify heart and lung diseases. The first step in the preprocessing phase was to clean the audio signals by reducing noise, which could interfere with the analysis of heart and lung sound patterns. A double denoising strategy was employed to ensure optimal signal quality: adaptive filters and bandpass filtering. These filters dynamically adjust to the changing characteristics of the noise, allowing for the effective removal of background noise that could otherwise distort the heart and lung sounds. Variational autoencoders (VAEs) were used to extract key features from the audio signals, transforming the raw audio data into a more manageable and informative feature space. The extracted features preserved the key characteristics of the heart and lung sounds while reducing noise and irrelevant information, making them more suitable for input into the classification model.
The features extracted through the VAEs were then used to train a CNN model, which consisted of multiple Conv2D layers to capture spatial patterns and temporal correlations within the audio signals. To reduce dimensionality and computational complexity, max-pooling layers were incorporated, followed by dense layers for final classification. The model was trained using a binary cross-entropy loss function and an Adam optimizer to optimize the model during training. After training, the CNN model was evaluated on a separate test dataset, showing high accuracy in classifying heart and lung diseases. The model correctly classified a significant percentage of the heart and lung sound samples, showed strong precision, high recall, and a balanced F1-score, which is crucial in medical diagnoses.
In conclusion, the combination of double denoising techniques, feature extraction using variational autoencoders, and CNN-based classification proved to be an effective approach for heart and lung disease detection from audio signals, as shown in
Figure 3.
5. Conclusions
This study demonstrates the effectiveness of combining advanced preprocessing techniques, feature extraction methods, and deep learning models for heart and lung disease detection using audio signals. By employing double denoising techniques, such as adaptive filtering and bandpass filtering, the quality of the audio data was significantly enhanced, removing noise and isolating the relevant sound frequencies. Feature extraction using variational autoencoders (VAEs) further reduced dimensionality while retaining important characteristics of the heart and lung sounds, making them more suitable for classification. The convolutional neural network (CNN) trained on these refined features successfully classified heart and lung diseases, achieving high accuracy, strong precision, and high recall, which are critical for medical applications. The balanced F1-score further underscores the model’s ability to detect diseases reliably without favoring false positives or negatives. Overall, the combination of preprocessing, feature extraction, and CNN-based classification provides a robust framework for heart and lung disease detection from audio signals. The results of this study confirm that this approach holds significant potential for early and accurate disease detection, contributing to the advancement of non-invasive diagnostic tools in healthcare.
Author Contributions
Methodology, A.J.; supervision, R.K.R. and J.A.; writing—original draft, A.J. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
This study does not include human participants or any experimental procedures involving personal data. It is a review that uses publicly available data from peer-reviewed research that has already been published. No sensitive data, private information, or personally identifiable information was collected or analyzed. Since this study complies with ethical guidelines for non-intervention studies, it does not require formal ethics approval. The analyzed research cited in this article was carried out in accordance with the ethical guidelines established by the corresponding ethics committees or institutional review boards (IRBs). This ensures that the original research adhered to the principles of informed consent, data privacy, and participant protection.
Informed Consent Statement
Not applicable.
Data Availability Statement
This study does not create ethical issues about data security, privacy, or confidentiality because of the nature of the review and the lack of human participants in the research process.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Chaddad, A.; Wu, Y.; Kateb, R.; Bouridane, A. Electroencephalography Signal Processing: A Comprehensive Review and Analysis of Methods and Techniques. Sensors 2023, 23, 6434. [Google Scholar] [CrossRef]
- Cammarasana, S.; Nicolardi, P.; Patanè, G. A Universal Deep Learning Framework for Real-Time Denoising of Ultrasound Images. arXiv 2021, arXiv:2101.09122. [Google Scholar] [CrossRef]
- Altan, G.; Kutlu, Y.; Garbi, Y.; Pekmezci, A.Ö.; Nural, S. Multimedia Respiratory Database (RespiratoryDatabase@TR): Auscultation Sounds and Chest X-rays. arXiv 2021, arXiv:2101.10946. [Google Scholar] [CrossRef]
- Sushkova, O.S.; Morozov, A.A.; Gabova, A.V.; Sarkisova, K.Y. The Diagnostic Value of EEG Wave Trains for Distinguishing Immature Absence Seizures and Sleep Spindles: Evidence from the WAG/Rij Rat Model. Diagnostics 2025, 15, 983. [Google Scholar] [CrossRef]
- Xue, C.; Kowshik, S.S.; Lteif, D.; Puducheri, S.; Jasodanand, V.H.; Zhou, O.T.; Walia, A.S.; Guney, O.B.; Zhang, J.D.; Poésy, S.; et al. AI-based differential diagnosis of dementia etiologies on multimodal data. Nat. Med. 2024, 30, 2977. [Google Scholar] [CrossRef]
- Huang, D.-M.; Huang, J.; Qiao, K.; Zhong, N.; Lu, H.; Wang, W. Deep learning-based lung sound analysis for intelligent stethoscope. Mil. Med. Res. 2023, 10, 44. [Google Scholar] [CrossRef]
- Rocha, B.; Pessoa, D.; Marques, A.; Carvalho, P.; Paiva, R.P. Automatic Classification of Adventitious Respiratory Sounds: A (Un)Solved Problem? Sensors 2020, 21, 57. [Google Scholar] [CrossRef]
- Poh, Y.Y.; Grooby, E.; Tan, K.; Zhou, L.; King, A.; Ramanathan, A.; Malhotra, A.; Harandi, M.; Marzbanrad, F. Real-time Neonatal Chest Sound Separation using Deep Learning. arXiv 2023, arXiv:2310.17116. [Google Scholar] [CrossRef]
- Peng, J.; Li, Y.; LIu, Y.; Wang, M.; Li, H. Adaptive Convolutional Filter for Seismic Noise Attenuation. arXiv 2024, arXiv:2410.18896. [Google Scholar] [CrossRef]
- Kong, Q.; Chen, K.; Liu, H.; Du, X.; Berg-Kirkpatrick, T.; Dubnov, S.; Plumbley, M.D. Universal Source Separation with Weakly Labelled Data. arXiv 2023, arXiv:2305.07447. [Google Scholar] [CrossRef]
- Braiek, H.B.; Khomh, F. Machine Learning Robustness: A Primer. arXiv 2024, arXiv:2404.00897. [Google Scholar] [CrossRef]
- Han, J.; Shaout, A. ENACT-Heart—ENsemble-based Assessment Using CNN and Transformer on Heart Sounds. arXiv 2025, arXiv:2502.16914. [Google Scholar] [CrossRef]
- Jeong, S.G.; Nam, S.; Jung, S.K.; Kim, S. iMedic: Towards Smartphone-based Self-Auscultation Tool for AI-Powered Pediatric Respiratory Assessment. arXiv 2025, arXiv:2504.15743. [Google Scholar] [CrossRef]
- Sharou, K.A.; Li, Z.; Specia, L. Towards a Better Understanding of Noise in Natural Language Processing. In Proceedings of the International Conference on Recent Advances in Natural Language Processing (RANLP 2021); INCOMA Ltd.: Moscow, Russia, 2021; p. 53. [Google Scholar] [CrossRef]
- Wan, Z.; Yu, Q.; Mao, J.; Duan, W.; Ding, C. OpenECG: Benchmarking ECG Foundation Models with Public 1.2 Million Records. arXiv 2025, arXiv:2503.00711. [Google Scholar] [CrossRef]
- Nigam, V.; Priemer, R. Cardiac sound separation. In Proceedings of the Computers in Cardiology, Chicago, IL, USA, 19–22 September 2004; pp. 497–500. [Google Scholar] [CrossRef]
- Xie, Y.; Xie, K.; Yang, Q.; Xie, S. Reverberant blind separation of heart and lung sounds using nonnegative matrix factorization and auxiliary function technique. Biomed. Signal Process. Control 2021, 69, 102899. [Google Scholar] [CrossRef]
- Wang, W.; Wang, S.; Qin, D.; Fang, Y.; Zheng, Y. Heart-lung sound separation by nonnegative matrix factorization and Deep Learning. Biomed. Signal Process. Control 2023, 79, 104180. [Google Scholar] [CrossRef]
- Sun, W.; Zhang, Y.; Chen, F. Research on heart and lung sound separation method based on Dae–NMF–VMD. EURASIP J. Adv. Signal Process. 2024, 2024, 59. [Google Scholar] [CrossRef]
- Zulfiqar, R.; Majeed, F.; Irfan, R.; Rauf, H.T.; Benkhelifa, E.; Belkacem, A.N. Abnormal Respiratory Sounds Classification Using Deep CNN Through Artificial Noise Addition. Front. Med. 2021, 8, 714811. [Google Scholar] [CrossRef]
- Wang, Z.; Sun, Z. Performance evaluation of lung sounds classification using deep learning under variable parameters. EURASIP J. Adv. Signal Process. 2024, 2024, 1–21. [Google Scholar] [CrossRef]
- Han, C.; Kaya, E.M.; Hoefer, K.; Slaney, M.; Carlile, S. Multi-Channel Speech Denoising for Machine Ears. arXiv 2022, arXiv:2202.08793. [Google Scholar] [CrossRef]
- Nossier, S.A.; Wall, J.; Moniri, M.; Glackin, C.; Cannings, N. An Experimental Analysis of Deep Learning Architectures for Supervised Speech Enhancement. Electronics 2020, 10, 17. [Google Scholar] [CrossRef]
| Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).