Infectious Bronchitis Virus (Gammacoronavirus) in Poultry: Genomic Architecture, Post-Translational Modifications, and Structural Motifs
Abstract
1. Introduction
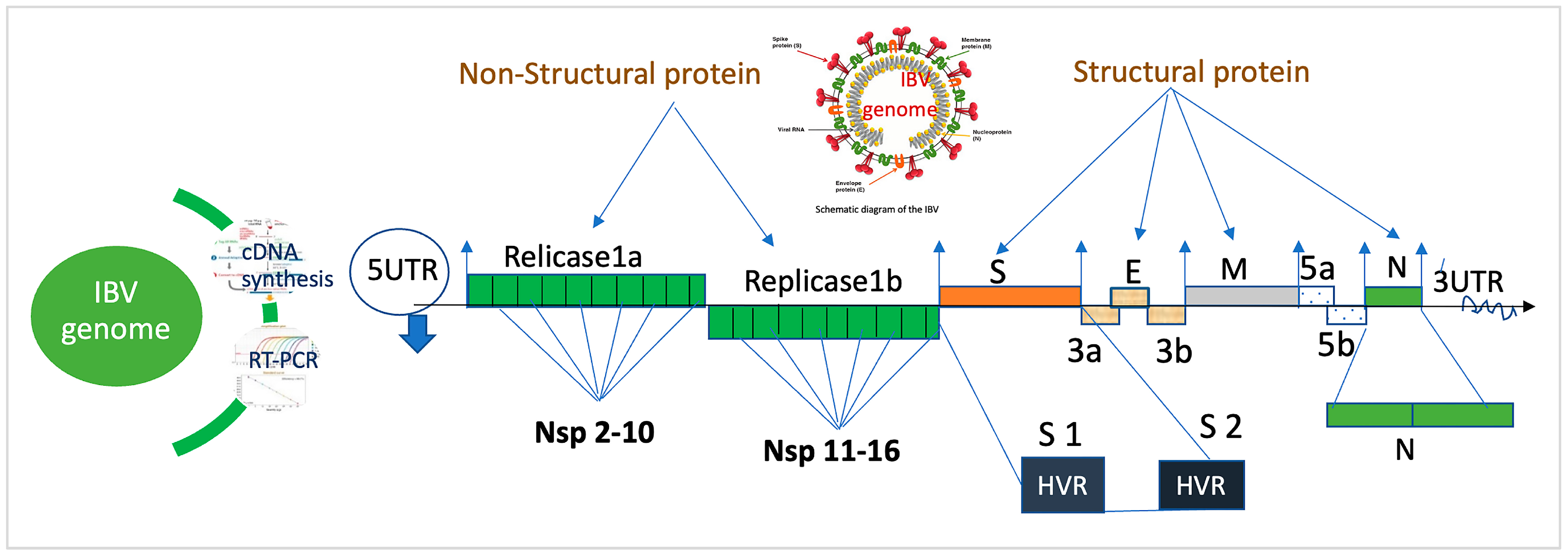
2. Genomic Structure of IBV
3. Diversity of IBV
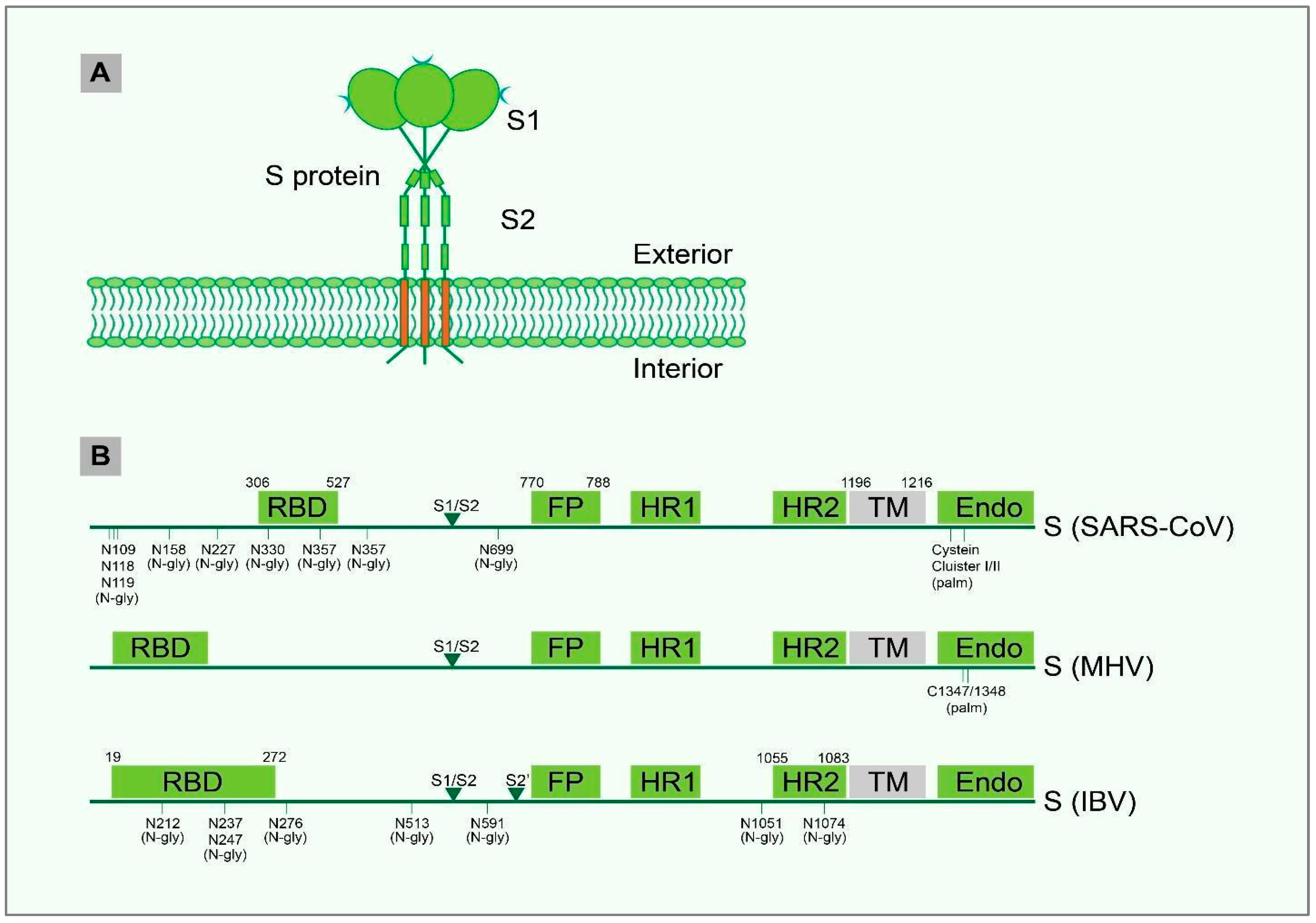
4. PTM and Structural Motif of IBV
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Miłek, J.; Blicharz-Domańska, K. Coronaviruses in Avian Species—Review with Focus on Epidemiology and Diagnosis in Wild Birds. J. Vet. Res. 2018, 62, 249–255. [Google Scholar] [CrossRef] [PubMed]
- De Wit, J.J.; Cook, J.K.A. Spotlight on avian pathology. Avian Pathol. 2020, 49, 4. [Google Scholar] [CrossRef] [PubMed]
- Cavanagh, D.; Gelb, J. Infectious bronchitis. In Diseases of Poultry, 12th ed.; Saif, Y.M., Fadly, A.M., Glisson, J.R., McDougald, L.R., Nolan, L.K., Swayne, D.E., Eds.; Blackwell Publishing: Ames, IA, USA, 2008; pp. 117–135. [Google Scholar]
- Benyeda, Z.; Mato, T.; Suveges, T.; Szabo, E.; Kardi, V.; Abonyi-Toth, Z.; Rusvai, M.; Palya, V. Comparison of the pathogenicity of QX-like, M41 and 793/B infectious bronchitis strains from different pathological conditions. Avian Pathol. 2009, 38, 449–456. [Google Scholar] [CrossRef]
- Ganapathy, K.; Wilkins, M.; Forrester, A.; Lemiere, S.; Cserep, T.; Mcmullin, P.; Jones, R.C. QX-like Infectious Bronchitis Virus isolated from cases of proventriculitis in commercial broilers in England. Vet. Rec. 2012, 171, 597. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Genovese, K.J.; Swaggerty, C.L.; MacKinnon, K.M.; Kogut, M.H. Costimulation with TLR3 and TLR21 ligands synergistically up-regulates Th1-cytokine IFN-gamma and regulatory cytokine IL-10 expression in chicken monocytes. Dev. Comp. Immunol. 2012, 36, 756–760. [Google Scholar] [CrossRef]
- Bhuiyan, Z.A.; Ali, M.Z.; Moula, M.M.; Giasuddin, M.; Khan, Z.U.M. Prevalence and molecular characterization of infectious bronchitis virus isolated from chicken in Bangladesh. Vet. World 2019, 12, 909. [Google Scholar] [CrossRef]
- Xu, G.; Liu, X.Y.; Zhao, Y.; Chen, Y.; Zhao, J.; Zhang, G.Z. Characterization and analysis of an infectious bronchitis virus strain isolated from southern China in 2013. Virol. J. 2016, 13, 40. [Google Scholar] [CrossRef]
- Bande, F.; Arshad, S.S.; Omar, A.R.; Hair-Bejo, M.; Mahmuda, A.; Nair, V. Global distributions and strain diversity of avian infectious bronchitis virus: A review. Anim. Health Res. Rev. 2017, 18, 70–83. [Google Scholar] [CrossRef]
- Khataby, K.; Fellahi, S.; Loutfi, C.; Ennaji, M.M. Assessment of pathogenicity and tissue distribution of infectious bronchitis virus strains (Italy 02 genotype) isolated from Moroccan broiler chickens. BMC Vet. Res. 2016, 12, 94. [Google Scholar] [CrossRef]
- Valastro, V.; Holmes, E.C.; Britton, P.; Fusaro, A.; Jackwood, M.W.; Cattoli, G.; Monne, I. S1 gene-based phylogeny of infectious bronchitis virus: An attempt to harmonize virus classification. Infect. Genet. Evol. 2016, 39, 349–364. [Google Scholar] [CrossRef]
- Legnardi, M.; Tucciarone, C.M.; Franzo, G.; Cecchinato, M. Infectious Bronchitis Virus Evolution, Diagnosis and Control. Vet. Sci. 2020, 7, 79. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Zhao, W.; Han, Z.; Chen, Y.; Zhao, Y.; Sun, J.; Liu, S. Genome characterization, antigenicity and pathogenicity of a novel infectious bronchitis virus type isolated from south China. Infect. Genet. Evol. 2017, 54, 437–446. [Google Scholar] [CrossRef] [PubMed]
- Laconi, A.; Listorti, V.; Franzo, G.; Cecchinato, M.; Naylor, C.; Lupini, C.; Catelli, E. Molecular characterization of whole genome sequence of infectious bronchitis virus 624I genotype confirms the close relationship with Q1 genotype. Transbound. Emerg. Dis. 2019, 66, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Franzo, G.; Listorti, V.; Naylor, C.J.; Lupini, C.; Laconi, A.; Felice, V.; Drigo, M.; Catelli, E.; Cecchinato, M. Molecular investigation of a full-length genome of a Q1-like IBV strain isolated in Italy in 2013. Virus Res. 2015, 210, 77–80. [Google Scholar] [CrossRef]
- Lin, S.Y.; Chen, H.W. Infectious Bronchitis Virus Variants: Molecular Analysis and Pathogenicity Investigation. Int. J. Mol. Sci. 2017, 18, 2030. [Google Scholar] [CrossRef]
- Dhama, K.; Singh, S.D.; Barathidasan, R.; Desingu, P.A.; Chakraborty, S.; Tiwari, R.; Kumar, M.A. Emergence of avian infectious bronchitis virus and its variants need better diagnosis, prevention and control strategies: A global perspective. Pak. J. Biol. Sci. 2014, 17, 751–767. [Google Scholar] [CrossRef]
- Keep, S.; Sives, S.; Stevenson-Leggett, P.; Britton, P.; Vervelde, L.; Bickerton, E. Limited Cross-Protection against Infectious Bronchitis Provided by Recombinant Infectious Bronchitis Viruses Expressing Heterologous Spike Glycoproteins. Vaccines 2020, 8, 330. [Google Scholar] [CrossRef]
- Rohaim, M.A.; El Naggar, R.F.; Abdelsabour, M.A.; Mohamed, M.H.A.; El-Sabagh, I.M.; Munir, M. Evolutionary Analysis of Infectious Bronchitis Virus Reveals Marked Genetic Diversity and Recombination Events. Genes 2020, 11, 605. [Google Scholar] [CrossRef]
- Quinteros, J.A.; Lee, S.W.; Markham, P.F.; Noormohammadi, A.H.; Hartley, C.A.; Legione, A.R.; Browning, G.F. Full genome analysis of Australian infectious bronchitis viruses suggests frequent recombination events between vaccine strains and multiple phylogenetically distant avian coronaviruses of unknown origin. Vet. Microbiol. 2016, 197, 27–38. [Google Scholar] [CrossRef]
- De Wit, J.J.; Cook, J.K.A.; der Heijden, H.M.J.F. Infectious bronchitis virus variants: A review of the history, current situation and control measures. Avian Pathol. 2011, 40, 223–235. [Google Scholar] [CrossRef]
- Cifuentes-Rincón, A.; Lopes, P.D.; Sanmiguel, P.R.A. Genotyping of news variants of the avian infectious bronchitis virus from Tolima department, Colombia. Rev. MVZ Córdoba 2016, 21, 5500–5510. [Google Scholar] [CrossRef][Green Version]
- Sultan, H.A.; Ali, A.; El Feil, W.K.; Bazid, A.H.I.; Zain El-Abideen, M.A.; Kilany, W.H. Protective efficacy of different live attenuated infectious bronchitis virus vaccination regimes against challenge With IBV variant-2 circulating in the Middle East. Front. Vet. Sci. 2019, 6, 341. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-C.; Jeong, S.; Cho, A.Y.; Kim, K.-J.; Kim, J.-Y.; Park, D.-H.; Kim, H.-J.; Kwon, J.-H.; Song, C.-S. Genomic Analysis of Avian Infectious Bronchitis Viruses Recently Isolated in South Korea Reveals Multiple Introductions of GI-19 Lineage (QX Genotype). Viruses 2021, 13, 1045. [Google Scholar] [CrossRef]
- Papineau, A.; Berhane, Y.; Wylie, T.N.; Lung, O. Genome Organization of Canada Goose Coronavirus, A Novel Species Identified in a Mass Die-off of Canada Geese. Sci. Rep. 2019, 9, 5954. [Google Scholar] [CrossRef] [PubMed]
- Decaro, N.; Lorusso, A. Novel human coronavirus (SARS-CoV-2): A lesson from animal coronaviruses. Vet. Microbiol. 2020, 104, 252–253. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Ali, A.; Siddique, R.; Nabi, G. Novel coronavirus is putting the whole world on alert. J. Hosp. Infect. 2020, 104, 252–253. [Google Scholar] [CrossRef]
- Siddell, S.; Snijder, E.J. An introduction to Nidoviruses. In Nidoviruses; Perlman, S., Gallagher, T., Snijder, E., Eds.; ASM Press: Washington, DC, USA, 2008; pp. 1–14. [Google Scholar]
- Zappulli, V.; Ferro, S.; Bonsembiante, F.; Brocca, G.; Calore, A.; Cavicchioli, L.; Centelleghe, C.; Corazzola, G.; De Vreese, S.; Gelain, M.E.; et al. Pathology of Coronavirus Infections: A Review of Lesions in Animals in the One-Health Perspective. Animals 2020, 10, 2377. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.Y.; Zhao, R.; Gao, L.J.; Gao, X.F.; Wang, D.P.; Cao, J.M. SARS-CoV-2: Structure, biology, and structure-based therapeutics development. Front. Cell. Infect. 2020, 10, 587269. [Google Scholar]
- Abro, S.H.; Renström, L.H.M.; Ullman, K.; Isaksson, M.; Zohari, S.; Jansson, D.S.; Belák, S.; Baule, C. Emergence of novel strains of avian infectious bronchitis virus in Sweden. Vet. Microbiol. 2012, 155, 237–246. [Google Scholar] [CrossRef]
- Dent, S.D.; Xia, D.; Wastling, J.M.; Neuman, B.W.; Britton, P.; Maier, H.J. The proteome of the infectious bronchitis virus Beau-R virion. J. Gen. Virol. 2015, 96, 3499. [Google Scholar] [CrossRef]
- Niu, X.; Kong, F.; Hou, Y.J.; Wang, Q. Crucial mutation in the exoribonuclease domain of nsp14 of PEDV leads to high genetic instability during viral replication. Cell Biosci. 2021, 11, 106. [Google Scholar] [CrossRef]
- Agranovsky, A.A. Structure and Expression of Large (+) RNA Genomes of Viruses of Higher Eukaryotes. Biochemistry 2021, 86, 248–256. [Google Scholar] [CrossRef] [PubMed]
- Pasternak, A.O.; Spaan, W.J.M.; Snijder, E.J. Nidovirus transcription: How to make sense? J. Gen. Virol. 2006, 87, 1403–1421. [Google Scholar] [CrossRef]
- Li, T.T.; Li, J.Y.; Huang, T.; Ge, X.Y. Complete genome sequence of a novel strain of infectious bronchitis virus, isolated from chickens in China in 2016. Genome Announc. 2017, 5, e01277–e01317. [Google Scholar] [CrossRef] [PubMed]
- Bhuiyan, M.S.A.; Amin, Z.; Bakar, A.M.S.A.; Saallah, S.; Yusuf, N.H.M.; Shaarani, S.M.; Siddiquee, S. Factor Influences for Diagnosis and Vaccination of Avian Infectious Bronchitis Virus (Gammacoronavirus) in Chickens. Vet. Sci. 2021, 8, 47. [Google Scholar] [CrossRef]
- Imbert, I.; Snijder, E.J.; Dimitrova, M.; Guillemot, J.C.; Lécine, P.; Canard, B. The SARS-Coronavirus PLnc domain of nsp3 as a replication/transcription scaffolding protein. Virus Res. 2008, 133, 136–148. [Google Scholar] [CrossRef]
- Quinteros, J.A.; Noormohammadi, A.H.; Lee, S.W.; Browning, G.F.; Diaz-Méndez, A. Genomics and pathogenesis of the avian coronavirus infectious bronchitis virus. Aust. Vet. J. 2022, 100, 496–512. [Google Scholar] [CrossRef]
- Van Hemert, M.J.; van den Worm, S.H.; Knoops, K.; Mommaas, A.M.; Gorbalenya, A.E.; Snijder, E.J. SARS-Coronavirus Replication/Transcription Complexes Are Membrane-Protected and Need a Host Factor for Activity In Vitro. PLoS Pathog. 2008, 4, e1000054. [Google Scholar] [CrossRef] [PubMed]
- Yoshimoto, F.K. The Proteins of Severe Acute Respiratory Syndrome Coronavirus-2 (SARS CoV-2 or n-COV19), the Cause of COVID-19. Protein J. 2020, 39, 198–216. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.L.; Su, J.L.; Zhao, J.X.; Zhang, G.Z. Complete genome sequence analysis of a predominant infectious bronchitis virus (IBV) strain in China. Virus Genes 2009, 38, 56–65. [Google Scholar] [CrossRef]
- Ziebuhr, J.; Schelle, B.; Karl, N.; Minskaia, E.; Bayer, S.; Siddell, S.G.; Gorbalenya, A.E.; Thiel, V. Human Coronavirus 229E Papain-Like Proteases Have Overlapping Specificities but Distinct Functions in Viral Replication. J. Virol. 2007, 81, 3922–3932. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Oostra, M.; Hagemeijer, M.C.; van Gent, M.; Bekker, C.P.; te Lintelo, E.G.; Rottier, P.J.; de Haan, C.A. Topology and Membrane Anchoring of the Coronavirus Replication Complex: Not All Hydrophobic Domains of nsp3 and nsp6 are Membrane Spanning. J. Virol. 2008, 82, 12392–12405. [Google Scholar] [CrossRef] [PubMed]
- Matthes, N.; Mesters, J.R.; Coutard, B.; Canard, B.; Snijder, E.J.; Moll, R.; Hilgenfeld, R. Non-structural protein Nsp10 of mouse hepatitis virus binds zinc ions and nucleic acids. FEBS Lett. 2006, 580, 4143–4149. [Google Scholar] [CrossRef] [PubMed]
- Cheng, A.; Zhang, W.; Xie, Y.; Jiang, W.; Arnold, E.; Sarafianos, S.G.; Ding, J. Expression, purification, and characterization of SARS coronavirus RNA polymerase. Virology 2005, 335, 165–176. [Google Scholar] [CrossRef] [PubMed]
- Seybert, A.; Hegyi, A.; Siddell, S.G.; Ziebuhr, J. The human coronavirus 229E superfamily 1 helicase has RNA and DNA duplex-unwinding activities with 5′-to-3′ polarity. RNA 2000, 6, 1056–1068. [Google Scholar] [CrossRef]
- Bhardwaj, K.; Guarino, L.; Kao, C.C. The severe acute respiratory syndrome coronavirus Nsp15 protein is an endoribonuclease that prefers manganese as a cofactor. J. Virol. 2004, 78, 12218–12224. [Google Scholar] [CrossRef]
- Snijder, E.J.; Bredenbeek, B.P.; Dobbe, J.C.; Thiel, V.; Ziebuhr, J.; Poon, L.L.; Guan, Y.; Rozanov, M.; Spaan, W.J.; Gorbalenya, A.E. Unique and conserved features of genome and proteome of SARS coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003, 331, 991–1004. [Google Scholar] [CrossRef] [PubMed]
- Abolnik, C. Genomic and single nucleotide polymorphism analysis of infectious bronchitis coronavirus. Infect. Genet. Evol. 2015, 32, 416–424. [Google Scholar] [CrossRef]
- Yamada, Y.; Liu, D.X. Proteolytic activation of the spike protein at a novel RRRR/S motif is implicated in furin-dependent entry, syncytium formation, and infectivity of coronavirus infectious bronchitis virus in cultured cells. J. Virol. 2009, 83, 8744–8758. [Google Scholar] [CrossRef]
- Cavanagh, D.; Davis, P.J. Coronavirus IBV: Removal of spike glycopolypeptide S1 by urea abolishes infectivity and haemagglutination but not attachment to cells. J. Gen. Virol. 1986, 67, 1443–1448. [Google Scholar] [CrossRef]
- Ujike, M.; Taguchi, F. Incorporation of spike and membrane glycoproteins into coronavirus virions. Viruses 2015, 7, 1700–1725. [Google Scholar] [CrossRef] [PubMed]
- Thomas, S. The Structure of the Membrane Protein of SARS-CoV-2 Resembles the Sugar Transporter SemiSWEET. Pathog. Immun. 2020, 5, 342–363. [Google Scholar] [CrossRef] [PubMed]
- Li, F. Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 2016, 3, 237–261. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Liu, X.; Xu, Z.; Wang, J.; Li, Y.; Shen, J.; Lan, Y.; Cheng, C. Degradation and mineralization of ciprofloxacin by gas–liquid discharge non-thermal plasma. Plasma Sci. Technol. 2018, 21, 015501. [Google Scholar] [CrossRef]
- Kim, C.H. SARS-CoV-2 Evolutionary Adaptation toward Host Entry and Recognition of Receptor O-Acetyl Sialylation in Virus-Host Interaction. Int. J. Mol. Sci. 2020, 21, 4549. [Google Scholar] [CrossRef]
- Li, W.; Moore, M.J.; Vasllieva, N.; Sui, J.; Wong, S.K.; Berne, M.A.; Somasundaran, M.; Sullivan, J.L.; Luzuriaga, K.; Greeneugh, T.C.; et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 2003, 426, 450–454. [Google Scholar] [CrossRef]
- Andersen, K.G.; Rambaut, A.; Lipkin, W.I.; Holmes, E.C.; Garry, R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020, 26, 450–452. [Google Scholar] [CrossRef]
- Schwegmann-Wessels, C.; Bauer, S.; Winter, C.; Enjuanes, L.; Laude, H.; Herrler, G. The sialic acid binding activity of the S protein facilitates infection by porcine transmissible gastroenteritis coronavirus. Virol. J. 2011, 8, 435. [Google Scholar] [CrossRef]
- Johnson, M.A.; Pooley, C.; Ignjatovic, J.; Tyack, S.G. A recombinant fowl adenovirus expressing the S1 gene of infectious bronchitis virus protects against challenge with infectious bronchitis virus. Vaccine 2003, 21, 2730–2736. [Google Scholar] [CrossRef]
- Huang, M.; Zou, C.; Liu, Y.; Han, Z.; Xue, C.; Cao, Y. A novel low virulent respiratory infectious bronchitis virus originating from the recombination of QX, TW and 4/91 genotype strains in China. Vet. Microbiol. 2020, 242, 108579. [Google Scholar] [CrossRef]
- Guo, Y.; Tisoncik, J.; McReynolds, S.; Farzan, M.; Prabhakar, B.S.; Gallagher, T.; Rong, L.; Caffrey, M. Identification of a new region of SARS-CoV S protein critical for viral entry. J. Mol. Biol. 2009, 394, 600–605. [Google Scholar] [CrossRef]
- Ng, K.T.; Mohd-Ismail, N.K.; Tan, Y.J. Spike S2 subunit: The dark horse in the race for prophylactic and therapeutic interventions against SARS-CoV-2. Vaccines 2021, 9, 178. [Google Scholar] [CrossRef]
- Kariithi, H.M.; Volkening, J.D.; Goraichuk, I.V.; Ateya, L.O.; Williams-Coplin, D.; Olivier, T.L.; Binepal, Y.S.; Afonso, C.L.; Suarez, D.L. Unique Variants of Avian Coronaviruses from Indigenous Chickens in Kenya. Viruses 2023, 15, 264. [Google Scholar] [CrossRef]
- Bande, F.; Arshad, S.S.; Bejo, M.H.; Moeini, H.; Omar, A.R. Progress and challenges toward the development of vaccines against avian infectious bronchitis. J. Immunol. Res. 2015, 2015, 424860. [Google Scholar] [CrossRef] [PubMed]
- Mo, M.; Huang, B.; Wei, P.; Wei, T.; Chen, Q.; Wang, X.; Li, M.; Fan, W. Complete genome sequences of two Chinese virulent avian coronavirus infectious bronchitis virus variants. J. Virol. 2012, 86, 10903–10904. [Google Scholar] [CrossRef]
- Corse, E.; Machamer, C.E. Infectious bronchitis virus E protein is targeted to the Golgi complex and directs release of virus-like particles. J. Virol. 2000, 74, 4319–4326. [Google Scholar] [CrossRef]
- Raamsman, M.J.B.; Locker, J.K.; de Hooge, A.; de Vries, A.A.F.; Griffiths, G.; Vennema, H.; Rottier, P.J.M. Characterization of Coronavirus mouse hepatitis virus strain A59 small membrane protein E. J. Virol. 2000, 74, 2333–2342. [Google Scholar] [CrossRef] [PubMed]
- Schoeman, D.; Fielding, B.C. Coronavirus envelope protein: Current knowledge. Virol. J. 2019, 16, 69. [Google Scholar] [CrossRef] [PubMed]
- Maeda, J.; Maeda, A.; and Makino, S. Release of Coronavirus E protein in membrane vesicles from virus-infected cells and E protein-expressing cells. Virology 1999, 263, 265–272. [Google Scholar] [CrossRef][Green Version]
- Almazán, F.; Sola, I.; Zuñiga, S.; Marquez-Jurado, S.; Morales, L.; Becares, M.; Enjuanes, L. Coronavirus reverse genetic systems: Infectious clones and replicons. Virus Res. 2014, 189, 262–270. [Google Scholar] [CrossRef]
- Lee, S.; Channappanavar, R.; Kanneganti, T.D. Coronaviruses: Innate immunity, inflammasome activation, inflammatory cell death, and cytokines. Trends Immunol. 2020, 41, 1083–1099. [Google Scholar] [CrossRef] [PubMed]
- Westerbeck, J.W.; Machamer, C.E. The Infectious Bronchitis Coronavirus Envelope Protein Alters Golgi pH to Protect Spike Protein and Promote Release of Infectious Virus. J. Virol. 2019, 93, e00015-19. [Google Scholar] [CrossRef] [PubMed]
- Escors, D.; Ortego, J.; Laude, H.; Enjuanes, L. The membrane M protein carboxy terminus binds to transmissible gastroenteritis coronavirus core and contributes to core stability. J. Virol. 2001, 75, 1312–1324. [Google Scholar] [CrossRef]
- Hurst, K.R.; Koetzner, C.A.; Masters, P.S. Identification of in vivo-interacting domains of the murine coronavirus nucleocapsid protein. J. Virol. 2009, 83, 7221–7234. [Google Scholar] [CrossRef]
- Wong, N.A.; Saier, M.H., Jr. The SARS-Coronavirus Infection Cycle: A Survey of Viral Membrane Proteins, Their Functional Interactions and Pathogenesis. Int. J. Mol. Sci. 2021, 22, 1308. [Google Scholar] [CrossRef]
- Neuman, B.W.; Kiss, G.; Kunding, A.H.; Bhella, D.; Baksh, M.F.; Connelly, S.; Droese, B.; Klaus, J.P.; Makino, S.; Sawicki, S.G.; et al. A structural analysis of M protein in coronavirus assembly and morphology. J. Struct. Biol. 2011, 174, 11–22. [Google Scholar] [CrossRef]
- Fung, T.S.; Liu, D.X. Post-translational modifications of coronavirus proteins: Roles and function. Future Virol. 2018, 13, 405–430. [Google Scholar] [CrossRef]
- De Haan, C.A.; Li, Z.; te Lintelo, E.; Bosch, B.J.; Haijema, B.J.; Rottier, P.J. Murine coronavirus with an extended host range uses heparan sulfate as an entry receptor. J. Virol. 2005, 79, 14451–14456. [Google Scholar] [CrossRef]
- Fehr, A.R.; Perlman, S. Coronaviruses: An overview of their replication and pathogenesis. In Coronaviruses; Springer: Berlin/Heidelberg, Germany, 2015; pp. 1–23. [Google Scholar]
- Luo, H.; Wu, D.; Shen, C.; Chen, K.; Shen, X.; Jiang, H. Severe acute respiratory syndrome coronavirus membrane protein interacts with nucleocapsid protein mostly through their carboxyl termini by electrostatic attraction. Int. J. Biochem. Cell Biol. 2006, 38, 589–599. [Google Scholar] [CrossRef] [PubMed]
- McBride, C.E.; Machamer, C.E. A single tyrosine in the severe acute respiratory syndrome coronavirus membrane protein cytoplasmic tail is important for efficient interaction with spike protein. J. Virol. 2010, 84, 1891–1901. [Google Scholar] [CrossRef]
- Boopathi, S.; Poma, A.B.; Kolandaivel, P. Novel 2019 coronavirus structure, mechanism of action, antiviral drug promises and rule out against its treatment. J. Biomol. Struct. Dyn. 2021, 39, 3409–3418. [Google Scholar] [CrossRef] [PubMed]
- Casais, R.; Davies, M.; Cavanagh, D.; Britton, P. Gene 5 of the avian coronavirus infectious bronchitis virus is not essential for replication. J. Virol. 2005, 79, 8065–8078. [Google Scholar] [CrossRef] [PubMed]
- Zolfaghari Emameh, R.; Eftekhari, M.; Nosrati, H.; Heshmatnia, J.; Falak, R. Identification and characterization of a silent mutation in RNA binding domain of N protein coding gene from SARS-CoV-2. BMC Res. Notes 2021, 14, 10. [Google Scholar] [CrossRef] [PubMed]
- Biacchesi, S. The reverse genetics applied to fish RNA viruses. Vet. Res. 2011, 42, 12. [Google Scholar] [CrossRef]
- Yang, Z.; Wu, N.; Zu, Y.; Fu, Y. Comparative anti-infectious bronchitis virus (IBV) activity of (−)-pinene: Effect on nucleocapsid (N) protein. Molecules 2011, 16, 1044–1054. [Google Scholar] [CrossRef]
- Chang, C.K.; Hou, M.H.; Chang, C.F.; Hsiao, C.D.; Huang, T.H. The SARS coronavirus nucleocapsid protein-forms and functions. Antivir. Res. 2014, 103, 39–50. [Google Scholar] [CrossRef]
- Hiscox, T.; Wurm, L.; Wilson, P.; Britton, D.; Cavanagh, G.; Brooks, G. The coronavirus infectious bronchitis virus nucleoprotein localizes to the nucleolus. J. Virol. 2001, 75, 506–512. [Google Scholar] [CrossRef]
- You, J.H.; Reed, M.L.; Hiscox, J.A. Trafficking motifs in the SARScoronavirus nucleocapsid protein. Biochem. Biophys. Res. Commun. 2007, 358, 1015–1020. [Google Scholar] [CrossRef]
- Caredda, A.; Marongiu, B.; Porcedda, S.; Soro, C. Supereritical Carbon Dioxide Extraction and Characterization of Laurus nobilis Essential Oil. J. Agric. Food Chem. 2002, 50, 1492–1496. [Google Scholar] [CrossRef]
- Meharenna, Y.T.; Li, H.; Hawkes, D.B.; Pearson, A.G.; De Voss, T.L.J. Poulos Crystal Structure of P450cin in a Complex with Its Substrate, 1, 8-Cineole, a Close Structural Homologue to d-Camphor, the Substrate for P450cam. Biochemistry 2004, 43, 9487–9494. [Google Scholar] [CrossRef]
- Thura, M.; Sng, J.X.E.; Ang, K.H.; Li, J.; Gupta, A.; Hong, J.M.; Zeng, Q. Targeting intra-viral conserved nucleocapsid (N) proteins as novel vaccines against SARS-CoVs. Biosci. Rep. 2021, 41, BSR20211491. [Google Scholar] [CrossRef]
- Tan, Y.W.; Fung, T.S.; Shen, H.; Huang, M.; Liu, D.X. Coronavirus infectious bronchitis virus non-structural proteins 8 and 12 form stable complex independent of the non-translated regions of viral RNA and other viral proteins. Virology 2018, 513, 75–84. [Google Scholar] [CrossRef]
- Fang, S.G.; Shen, H.; Wang, J.; Tay, F.P.L.; Liu, D.X. Proteolytic processing of polyproteins 1a and 1ab between non-structural proteins 10 and 11/12 of Coronavirus infectious bronchitis virus is dispensable for viral replication in cultured cells. Virology 2008, 379, 175–180. [Google Scholar] [CrossRef]
- Chen, Y.; Cai, H.; Pan, J.A.; Xiang, N.; Tien, P.; Ahola, T.; Guo, D. Functional screen reveals SARS coronavirus non-structural protein nsp14 as a novel cap N7 methyltransferase. Proc. Natl. Acad. Sci. USA 2009, 106, 3484–3489. [Google Scholar] [CrossRef]
- Narayanan, K.; Ramirez, S.I.; Lokugamage, K.G.; Makino, S. Coronavirus nonstructural protein 1: Common and distinct functions in the regulation of host and viral gene expression. Virus Res. 2015, 202, 89–100. [Google Scholar] [CrossRef]
- Jackwood, M.W.; Hall, D.; Handel, A. Molecular evolution and emergence of avian gammacoronaviruses. Infect. Genet. Evol. 2012, 12, 1305–1311. [Google Scholar] [CrossRef]
- Zhong, Y.; Tan, Y.W.; Liu, D.X. Recent Progress in Studies of Arterivirus- and Coronavirus-Host Interactions. Viruses 2012, 4, 980–1010. [Google Scholar] [CrossRef] [PubMed]
- Neuman, B.W.; Joseph, J.S.; Saikatendu, K.S.; Serrano, P.; Chatterjee, A.; Johnson, M.A.; Liao, L.; Klaus, J.P.; Yates, J.R., 3rd; Wuthrich, K.; et al. Proteomics analysis unravels the functional repertoire of coronavirus nonstructural protein 3. J. Virol. 2008, 82, 5279–5294. [Google Scholar] [CrossRef] [PubMed]
- Graham, R.L.; Sparks, J.S.; Eckerle, L.D.; Sims, A.C.; Denison, M.R. SARS coronavirus replicase proteins in pathogenesis. Virus Res. 2008, 133, 88–100. [Google Scholar] [CrossRef]
- Ziebuhr, J. The coronavirus replicase: Insights into a sophisticated enzyme machinery. Adv. Exp. Med. Biol. 2006, 581, 3–11. [Google Scholar] [PubMed]
- Johnson, M.A.; Jaudzems, K.; Wuthrich, K. NMR structure of the SARS-CoV nonstructural protein 7 in solution at pH 6.5. J. Mol. Biol. 2010, 402, 619–628. [Google Scholar] [CrossRef]
- Subissi, L.; Posthuma, C.C.; Collet, A.; Zevenhoven-Dobbe, J.C.; Gorbalenya, A.E.; Decroly, E.; Snijder, E.J.; Canard, B.; Imbert, I. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl. Acad. Sci. USA 2014, 111, E3900–E3909. [Google Scholar] [CrossRef] [PubMed]
- Hillen, H.S.; Kokic, G.; Farnung, L.; Dienemann, C.; Tegunov, D.; Cramer, P. Structure of replicating SARS-CoV-2 polymerase. Nature 2020, 584, 154–156. [Google Scholar] [CrossRef] [PubMed]
- Shu, T.; Huang, M.; Wu, D.; Ren, Y.; Zhang, X.; Han, Y.; Mu, J.; Wang, R.; Qiu, Y.; Zhang, D.Y.; et al. SARS-Coronavirus-2 Nsp13 Possesses NTPase and RNA Helicase Activities That Can Be Inhibited by Bismuth Salts. Virol. Sin. 2020, 35, 321–329. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Chen, H.; Chen, Z.; Yang, F.; Ye, F.; Zheng, Y.; Yang, J.; Lin, X.; Sun, H.; Wang, L.; et al. Crystal structure of SARS-CoV-2 nsp10 bound to nsp14-ExoN domain reveals an exoribonuclease with both structural and functional integrity. Nucleic Acids Res. 2021, 49, 5382–5392. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, K.; Liu, P.; Leibowitz, J.L.; Kao, C.C. The coronavirus endoribonuclease Nsp15 interacts with retinoblastoma tumor suppressor protein. J. Virol. 2012, 86, 4294–4304. [Google Scholar] [CrossRef]
- Krafcikova, P.; Silhan, J.; Nencka, R.; Boura, E. Structural analysis of the SARS-CoV-2 methyltransferase complex involved in RNA cap creation bound to sinefungin. Nat. Commun. 2020, 11, 3717. [Google Scholar] [CrossRef]
- Ivanov, K.A.; Thiel, V.; Dobbe, J.C.; van der Meer, Y.; Snijder, E.J.; Ziebuhr, J. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J. Virol. 2004, 78, 5619–5632. [Google Scholar] [CrossRef] [PubMed]
- Payne, S. Virus Evolution and Genetics. Viruses 2017, 81–86. [Google Scholar]
- Wang, Y.; Grunewald, M.; Perlman, S. Coronaviruses: An Updated Overview of Their Replication and Pathogenesis. Methods Mol. Biol. 2020, 1, 29. [Google Scholar]
- Chen, Y.; Liu, Q.; Guo, D. Emerging coronaviruses: Genome structure, replication, and pathogenesis. J. Med. Virol. 2020, 92, 418–423. [Google Scholar] [CrossRef]
- Ennaji, Y.; Khataby, K.; Ennaji, M.M. Infectious bronchitis virus in poultry: Molecular epidemiology and factors leading to the emergence and reemergence of novel strains of infectious bronchitis virus. In Emerging and Reemerging Viral Pathogens; Academic Press: Cambridge, MA, USA, 2020; pp. 31–44. [Google Scholar]
- Wang, S.; Sotcheff, S.L.; Gallardo, C.M.; Jaworski, E.; Torbett, B.E.; Routh, A.L. Covariation of viral recombination with single nucleotide variants during virus evolution revealed by CoVaMa. Nucleic Acids Res. 2022, 50, e41. [Google Scholar] [CrossRef] [PubMed]
- Santacroce, L.; Charitos, I.A.; Carretta, D.M.; De Nitto, E.; Lovero, R. The human coronaviruses (HCoVs) and the molecular mechanisms of SARS-CoV-2 infection. J. Mol. Med. 2021, 99, 93–106. [Google Scholar] [CrossRef]
- LKhanh, N.P.; Tan, S.W.; Yeap, S.K.; Satharasinghe, D.A.; Hair-Bejo, M.; Bich, T.N.; Omar, A.R. Molecular characterization of QX-like and variant infectious bronchitis virus strains in Malaysia based on partial genomic sequences comprising the S-3a/3b-EM-intergenic region-5a/5b-N gene order. Avian Dis. 2017, 61, 442–452. [Google Scholar]
- Yan, W.; Qiu, R.; Wang, F.; Fu, X.; Li, H.; Cui, P.; Yang, X. Genetic and pathogenic characterization of a novel recombinant avian infectious bronchitis virus derived from GI-1, GI-13, GI-28, and GI-19 strains in Southwestern China. Poult. Sci. 2021, 100, 101210. [Google Scholar] [CrossRef]
- Jia, W.; Mondal, S.P.; Naqi, S.A. Genetic and antigenic diversity in avian infectious bronchitis virus isolates of the 1940s. Avian Dis. 2002, 46, 437–441. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Parvin, R.; Begum, J.A.; Nooruzzaman, M.; Kabiraj, C.K.; Chowdhury, E.H. Circulation of three genotypes and identification of unique mutations in neutralizing epitopes of infectious bronchitis virus in chickens in Bangladesh. Arch. Virol. 2021, 166, 3093–3103. [Google Scholar] [CrossRef]
- Houta, M.H.; Hassan, K.E.; El-Sawah, A.A.; Elkady, M.F.; Kilany, W.H.; Ali, A.; Abdel-Moneim, A.S. The emergence, evolution and spread of infectious bronchitis virus genotype GI-23. Arch. Virol. 2021, 166, 9–26. [Google Scholar] [CrossRef]
- Leghari, R.A.; Fan, B.; Wang, H.; Bai, J.; Zhang, L.; Abro, S.H.; Jiang, P. Full-length genome sequencing analysis of avian infectious bronchitis virus isolate associated with nephropathogenic infection. Poult. Sci. 2016, 95, 2921–2929. [Google Scholar] [CrossRef]
- Zhang, T.; Han, Z.; Xu, Q.; Wang, Q.; Gao, M.; Wu, W.; Shao, Y.; Li, H.; Kong, X.; Liu, S. Serotype shift of a 793/B genotype infectious bronchitis coronavirus by natural recombination. Infect. Genet. Evol. 2015, 32, 377–387. [Google Scholar] [CrossRef]
- Mendoza-González, L.; Marandino, A.; Panzera, Y.; Tomás, G.; Williman, J.; Techera, C.; Gayosso-Vázquez, A.; Ramírez-Andoney, V.; Alonso-Morales, R.; Realpe-Quintero, M.; et al. Research Note: High genetic diversity of infectious bronchitis virus from Mexico. Poult. Sci. 2022, 101, 102076. [Google Scholar] [CrossRef]
- Rohaim, M.A.; El Naggar, R.F.; Helal, A.M.; Bayoumi, M.M.; El-Saied, M.A.; Ahmed, K.A.; Shabbir, M.Z.; Munir, M. Genetic Diversity and Phylodynamics of Avian Coronaviruses in Egyptian Wild Birds. Viruses 2019, 11, 57. [Google Scholar] [CrossRef]
- Zhang, C.Y.; Wei, J.F.; He, S.H. Adaptive evolution of the spike gene of SARS coronavirus: Changes in positively selected sites in different epidemic groups. BMC Microbiol. 2006, 6, 88. [Google Scholar] [CrossRef]
- Yuan, Y.; He, J.; Gong, L.; Li, W.; Jiang, L.; Liu, J.; Chen, Q.; Yu, J.; Hou, S.; Shi, Y.; et al. Molecular epidemiology of SARS-CoV-2 clusters caused by asymptomatic cases in Anhui Province, China. BMC Infect. Dis. 2020, 20, 930. [Google Scholar] [CrossRef]
- Velazquez-Salinas, L.; Zarate, S.; Eberl, S.; Gladue, D.P.; Novella, I.; Borca, M.V. Positive selection of ORF1ab, ORF3a, and ORF8 genes drives the early evolutionary trends of SARS-CoV-2 during the 2020 COVID-19 pandemic. Front. Microbiol. 2020, 11, 550674. [Google Scholar] [CrossRef]
- Perez, L.; Arce, H.; Perera, C.; Rosell, R.; Frias, M.; Percedo, M.; Tarradas, J.; Doinguez, P.; Nuenez, J.; Ganges, L. Positive selection pressure on the B/C domains of the E2-gene of classical swine fever virus in endemic areas under C-strain vaccination. Infect. Genet. Evol. 2012, 12, 1405–1412. [Google Scholar] [CrossRef]
- Chen, Y.H.; Chen, Y.F. Evidence of selection pressures of neuraminidase gene (NA) of influenza A virus subtype H5N1 on different hosts in Guangxi Province of China. Saudi J. Biol. Sci. 2014, 21, 179–183. [Google Scholar] [CrossRef][Green Version]
- Read, A.F.; Baigent, S.J.; Powers, C.; Kgosana, L.B.; Blackwell, L.; Smith, L.P.; Kennedy, D.A.; Walkden-Brown, S.W.; Nair, V.K. Imperfect vaccination can enhance the transmission of highly virulent pathogens. PLoS Biol. 2015, 13, e1002198. [Google Scholar] [CrossRef]
- Domingo, E. Molecular Basis of Genetic Variation of Viruses: Error-Prone Replication. Virus Popul. 2016, 35–71. [Google Scholar]
- Pérez-Losada, M.; Arenas, M.; Galán, J.C.; Palero, F.; González-Candelas, F. Recombination in viruses: Mechanisms, methods of study, and evolutionary consequences. Infect. Genet. Evol. 2015, 30, 296–307. [Google Scholar] [CrossRef]
- Xu, Y. Genetic diversity and potential recombination between ferret coronaviruses from European and American lineages. J. Infect. 2020, 80, 350–371. [Google Scholar] [CrossRef]
- Lee, C.W.; Jackwood, M.W. Evidence of genetic diversity generated by recombination among avian coronavirus IBV. Arch. Virol. 2000, 145, 2135–2148. [Google Scholar] [CrossRef]
- Pohuang, T.; Chansiripornchai, N.; Tawatsin, A.; Sasipreeyajan, J. Sequence analysis of S1 genes of infectious bronchitis virus isolated in Thailand during 2008–2009: Identification of natural recombination in the field isolates. Virus Genes 2011, 43, 254–260. [Google Scholar] [CrossRef]
- Lowry, K.; Woodman, A.; Cook, J.; Evans, D.J. Evans. Recombination in enteroviruses is a biphasic replicative process involving the generation of greater-than genome length ‘imprecise’ intermediates. PLoS Pathog. 2014, 10, e1004191. [Google Scholar] [CrossRef]
- Gioti, K.; Kottaridi, C.; Voyiatzaki, C.; Chaniotis, D.; Rampias, T.; Beloukas, A. Animal Coronaviruses Induced Apoptosis. Life 2021, 11, 185. [Google Scholar] [CrossRef]
- Almazan, F.; Galan, C.; Enjuanes, L. The nucleoprotein is required for efficient coronavirus genome replication. J. Virol. 2004, 78, 12683–12688. [Google Scholar] [CrossRef]
- Makarenkov, V.; Mazoure, B.; Rabusseau, G.; Legendre, P. Horizontal gene transfer and recombination analysis of SARS-CoV-2 genes helps discover its close relatives and shed light on its origin. BMC Ecol. Evol. 2021, 21, 5. [Google Scholar] [CrossRef]
- Shukla, A.; Hilgenfeld, R. Acquisition of new protein domains by coronaviruses: Analysis of overlapping genes coding for proteins N and 9b in SARS coronavirus. Virus Genes 2015, 50, 29–38. [Google Scholar] [CrossRef]
- Winter, C.; Schwegmann-Wessels, C.; Neumann, U.; Herrler, G. The spike protein of infectious bronchitis virus is retained intracellularly by a tyrosine motif. J. Virol. 2008, 82, 2765–2771. [Google Scholar] [CrossRef]
- Coutard, B.; Valle, C.; de Lamballerie, X.; Canard, B.; Seidah, N.G.; Decroly, E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antivir. Res. 2020, 176, 104742. [Google Scholar] [CrossRef]
- Bosch, B.; Rottier, P. Nidovirus entry into cells. In Nidoviruses; ASM Press: Washington, DC, USA, 2008; pp. 157–178. [Google Scholar]
- Papa, G.; Mallery, D.L.; Albecka, A.; Welch, L.G.; Cattin-Ortolá, J.; Luptak, J.; James, L.C. Furin cleavage of SARS-CoV-2 Spike promotes but is not essential for infection and cell-cell fusion. PLoS Pathog. 2021, 17, e1009246. [Google Scholar] [CrossRef] [PubMed]
- Jackwood, M.W.; Hilt, D.A.; Callison, S.A.; Lee, C.W.; Plaza, H.; Wade, E. Spike glycoprotein cleavage recognition site analysis of infectious bronchitis virus. Avian Dis. 2001, 45, 366–372. [Google Scholar] [CrossRef]
- Simmons, G.; Gosalia, D.N.; Rennekamp, A.J.; Reeves, J.D.; Diamond, S.L.; Bates, P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. USA 2005, 102, 11876–11881. [Google Scholar] [CrossRef] [PubMed]
- Qasim, M.A.; Song, J.; Markley, J.L.; Laskowski, M., Jr. Cleavage of peptide bonds bearing ionizable amino acids at P(1) by serine proteases with hydrophobic S(1) pocket. Biochem. Biophys. Res. Commun. 2010, 400, 507–510. [Google Scholar] [CrossRef][Green Version]
- Goettig, P. Reversed Proteolysis—Proteases as Peptide Ligases. Catalysts 2020, 11, 33. [Google Scholar] [CrossRef]
- Dumez, M.E.; Teller, N.; Mercier, F.; Tanaka, T.; Vandenberghe, I.; Vandenbranden, M.; Devreese, B.; Luxen, A.; Frere, J.M.; Matagne, A.; et al. Activation mechanism of recombinant Der p 3 allergen zymogen: Contribution of cysteine protease Der p 1 and effect of propeptide glycosylation. J. Biol. Chem. 2008, 283, 30606–30617. [Google Scholar] [CrossRef]
- Herman, R.A.; Song, P.; Mirsky, H.P. Trypsin cleavage sites are highly unlikely to occur in celiac-causing restricted epitopes. GM Crops Food 2019, 11, 67–69. [Google Scholar] [CrossRef]
- Kumar, R.; Mehta, D.; Mishra, N.; Nayak, D.; Sunil, S. Role of Host-Mediated Post-Translational Modifications (PTMs) in RNA Virus Pathogenesis. Int. J. Mol. Sci. 2020, 22, 323. [Google Scholar] [CrossRef]
- Masters, P.S. The molecular biology of coronaviruses. Adv. Virus Res. 2006, 66, 193–292. [Google Scholar]
- Cheng, Y.; He, B.; Yang, J.; Ye, F.; Lin, S.; Yang, F.; Chen, Z.; Chen, Z.; Cao, Y.; Lu, G. Crystal structure of the S1 subunit N-terminal domain from DcCoV UAE-HKU23 spike protein. Virology 2019, 535, 74–82. [Google Scholar] [CrossRef]
- Xia, S.; Duan, K.; Zhang, Y.; Zhao, D.; Zhang, H.; Xie, Z.; Li, X.; Peng, C.; Zhang, Y.; Zhang, W.; et al. Effect of an inactivated vaccine against SARS-CoV-2 on safety and immunogenicity outcomes: Interim analysis of 2 randomized clinical trials. JAMA 2020, 324, 951–960. [Google Scholar] [CrossRef]
- Zhang, L.; Jackson, C.B.; Mou, H.; Ojha, A.; Peng, H.; Quinlan, B.D.; Rangarajan, E.S.; Pan, A.; Vanderheiden, A.; Suthar, M.S.; et al. SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat. Commun. 2020, 11, 6013. [Google Scholar] [CrossRef] [PubMed]
- Lontok, E.; Corse, E.; Machamer, C.E. Intracellular targeting signals contribute to localization of coronavirus spike proteins near the virus assembly site. J. Virol. 2004, 78, 5913–5922. [Google Scholar] [CrossRef] [PubMed]
- Schaecher, S.R.; Diamond, M.S.; Pekosz, A. The transmembrane domain of the severe acute respiratory syndrome coronavirus ORF7b protein is necessary and sufficient for its retention in the Golgi complex. Virolo. J. 2008, 82, 9477–9491. [Google Scholar] [CrossRef]
- Winter, C.; Schwegmann-Weßels, C.; Cavanagh, D.; Neumann, U.; Herrler, G. Sialic acid is a receptor determinant for infection of cells by avian Infectious bronchitis virus. J. Gen. Virol. 2006, 87, 1209–1216. [Google Scholar] [CrossRef]
- Wang, J.; Fang, S.; Xiao, H.; Chen, B.; Tam, J.P.; Liu, D.X. Interaction of the coronavirus infectious bronchitis virus membrane protein with beta-actin and its implication in virion assembly and budding. PLoS ONE 2009, 4, e4908. [Google Scholar]
- Nieto-Torres, J.L.; DeDiego, M.L.; Álvarez, E.; Jiménez-Guardeño, J.M.; Regla-Nava, J.A.; Llorente, M.; Kremer, L.; Shuo, S.; Enjuanes, L. Subcellular location and topology of severe acute respiratory syndrome coronavirus envelope protein. Virology 2011, 415, 69–82. [Google Scholar] [CrossRef]
- Schwegmann-Wessels, C.; Al-Falah, M.; Escors, D.; Wang, Z.; Zimmer, G.; Deng, H.; Enjuanes, L.; Naim, H.Y.; Herrler, G.A. novel sorting signal for intracellular localization is present in the S protein of a porcine coronavirus but absent from severe acute respiratory syndrome-associated coronavirus. J. Biol. Chem. 2004, 279, 43661–43666. [Google Scholar] [CrossRef] [PubMed]
- Sobocińska, J.; Roszczenko-Jasińska, P.; Ciesielska, A.; Kwiatkowska, K. Protein Palmitoylation and Its Role in Bacterial and Viral Infections. Front. Immunol. 2018, 8, 2003. [Google Scholar] [CrossRef]
- Linder, M.E. Reversible modification of proteins with thioester-linked fatty acids. In Protein Lipidation; Tamanoi, M., Sigman, D.S., Eds.; Academic Press: San Diego, CA, USA, 2000; pp. 215–240. [Google Scholar]
- Shen, L.F.; Chen, Y.J.; Liu, K.M.; Haddad, A.N.S.; Song, I.W.; Roan, H.Y.; Chen, L.Y.; Yen, J.J.; Chen, Y.J.; Wu, J.Y.; et al. Role of S-Palmitoylation by ZDHHC13 in Mitochondrial function and Metabolism in Liver. Sci. Rep. 2017, 7, 2182. [Google Scholar] [CrossRef]
- Tabaczar, S.; Czogalla, A.; Podkalicka, J.; Biernatowska, A.; Sikorski, A.F. Protein palmitoylation: Palmitoyltransferases and their specificity. Exp. Biol. Med. 2017, 24, 1150–1157. [Google Scholar] [CrossRef] [PubMed]
- Petit, C.M.; Chouljenko, V.N.; Iyer, A.; Colgrove, R.; Farzan, M.; Knipe, D.M.; Kousoulas, K.G. Palmitoylation of the cysteine-rich endodomain of the SARS-coronavirus spike glycoprotein is important for spikemediated cell fusion. Virology 2007, 360, 264–274. [Google Scholar] [CrossRef] [PubMed]
- Tharappel, A.M.; Samrat, S.K.; Li, Z.; Li, H. Targeting crucial host factors of SARS-CoV-2. ACS Infect. Dis. 2020, 6, 2844–2865. [Google Scholar] [CrossRef]
- Thorp, E.B.; Boscarino, J.A.; Logan, H.L.; Goletz, J.T.; Gallagher, T.M. Palmitoylations on murine coronavirus spike proteins are essential for virion assembly and infectivity. J. Virol. 2006, 80, 1280–1289. [Google Scholar] [CrossRef]
- Li, Q.; Liu, Y.; Zhang, L. Cytoplasmic tail determines the membrane trafficking and localization of SARS-CoV-2 spike protein. Fron. Mol. Biosci. 2022, 9, 1004036. [Google Scholar] [CrossRef]
- Sun, M.A.; Wang, Y.; Zhang, Q.; Xia, Y.; Ge, W.; Guo, D. Prediction of reversible disulfide based on features from local structural signatures. BMC Genom. 2017, 18, 279. [Google Scholar] [CrossRef] [PubMed]
- Lopez, L.A.; Riffle, A.J.; Pike, S.L.; Gardner, D.; Hogue, B.G. Importance of conserved cysteine residues in the coronavirus envelope protein. J. Virol. 2008, 82, 3000–3010. [Google Scholar] [CrossRef]
- Nugent, T.; Jones, D.T. Transmembrane protein topology prediction using support vector machines. BMC Bioinform. 2009, 10, 159–169. [Google Scholar] [CrossRef]
- He, M.; Jenkins, P.; Bennett, V. Cysteine 70 of ankyrin-G is S-palmitoylated and is required for function of ankyrin-G in membrane domain assembly. J. Biol. Chem. 2012, 287, 43995–44005. [Google Scholar] [CrossRef]
- Santopolo, S.; Riccio, A.; Santoro, M.G. The biogenesis of SARS-CoV-2 spike glycoprotein: Multiple targets for host-directed antiviral therapy. Biochem. Biophys. Res. Commun. 2021, 538, 80–87. [Google Scholar] [CrossRef]
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292. [Google Scholar] [CrossRef]
- Wissink, E.H.; Kroese, M.V.; Maneschijn-Bonsing, J.G.; Meulenberg, J.J.; van Rijn, P.A.; Rijsewijk, F.A.; Rottier, P.J. Significance of the oligosaccharides of the porcine reproductive and respiratory syndrome virus glycoproteins GP2a and GP5 for infectious virus production. J. Gen. Virol. 2004, 85, 3715–3723. [Google Scholar] [CrossRef] [PubMed]
- Nal, B.; Chan, C.; Kien, F.; Siu, L.; Tse, J.; Chu, K.; Altmeyer, R. Differential maturation and subcellular localization of severe acute respiratory syndrome coronavirus surface proteins S, M and E. J. Gen. Virol. 2005, 86, 1423–1434. [Google Scholar] [CrossRef] [PubMed]
- Thompson, N.; Wakarchuk, W. O-glycosylation and its role in therapeutic proteins. Biosci. Rep. 2022, 42, BSR20220094. [Google Scholar] [CrossRef]
- Zhang, X.Y.; Guo, J.; Wan, X.; Zhou, J.G.; Jin, W.P.; Lu, J.; Wang, W.H.; Yang, A.N.; Liu, D.X.; Shi, Z.L.; et al. Biochemical and antigenic characterization of the structural proteins and their post-translational modifications in purified SARS-CoV-2 virions of an inactivated vaccine candidate. Emerg. Microbes Infect. 2020, 9, 2653–2662. [Google Scholar] [CrossRef] [PubMed]
- Dawood, A.A.; Altobje, M.A. Inhibition of N-linked Glycosylation by Tunicamycin May Contribute to The Treatment of SARS-CoV-2. Microb. Pathog. 2020, 149, 104586. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Schuler, T.; Chen, Z.; Glass, G.E.; Childs, J.E.; Plagemann, P.G. Isolation of lactate dehydrogenase-elevating viruses from wild house mice and their biological and molecular characterization. Virus Res. 2000, 67, 153–162. [Google Scholar] [CrossRef]
- Niemann, H.; Boschek, B.; Evans, D.; Rosing, M.; Tamura, T.; Klenk, H.D. Post-translational glycosylation of coronavirus glycoprotein E1: Inhibition by monensin. EMBO J. 1982, 1, 1499–1504. [Google Scholar] [CrossRef]
- Reis, C.A.; Tauber, R.; Blanchard, V. Glycosylation is a key in SARS-CoV-2 infection. J. Mol. Med. 2021, 99, 1023–1031. [Google Scholar] [CrossRef]
- Cavanagh, D. Coronavirus IBV glycopolypeptides: Size of their polypeptide moieties and nature of their oligosaccharides. J. Gen. Virol. 1983, 64, 1187–1191. [Google Scholar] [CrossRef]
- Pande, S.; Rahardjo, A.; Livingston, B.; Mujacic, M. Monensin, a small molecule ionophore, can be used to increase high mannose levels on monoclonal antibodies generated by Chinese hamster ovary production cell-lines. Biotechnol. Bioeng. 2015, 112, 1383–1394. [Google Scholar] [CrossRef]
- Zheng, J.; Yamada, Y.; Fung, T.S.; Huang, M.; Chia, R.; Liu, D.X. Identification of N-linked glycosylation sites in the spike protein and their functional impact on the replication and infectivity of coronavirus infectious bronchitis virus in cell culture. Virology 2018, 513, 65–74. [Google Scholar] [CrossRef]
- Ingrell, C.R.; Miller, M.L.; Jensen, O.N.; Blom, N. NetPhosYeast: Prediction of protein phosphorylation sites in yeast. Bioinformatics 2007, 23, 895–897. [Google Scholar] [CrossRef] [PubMed]
- Emmott, E.; Munday, D.; Bickerton, E.; Britton, P.; Rodgers, M.A.; Whitehouse, A.; Hiscox, J.A. The cellular interactome of the coronavirus infectious bronchitis virus nucleocapsid protein and functional implications for virus biology. J. Virol. 2013, 87, 9486–9500. [Google Scholar] [CrossRef] [PubMed]
- Spencer, K.A.; Dee, M.; Britton, P.; Hiscox, J.A. Role of phosphorylation clusters in the biology of the coronavirus infectious bronchitis virus nucleocapsid protein. Virology 2008, 370, 373–381. [Google Scholar] [CrossRef]
- Wang, X.; Liao, Y.; Yap, P.L.; Png, K.J.; Tam, J.P.; Liu, D.X. Inhibition of protein kinase R activation and upregulation of GADD34 expression play a synergistic role in facilitating coronavirus replication by maintaining de novo protein synthesis in virus-infected cells. J. Virol. 2009, 83, 12462–12472. [Google Scholar] [CrossRef]
- Reed, M.L.; Howell, G.; Harrison, S.M.; Spencer, K.A.; Hiscox, J.A. Characterization of the nuclear export signal in the coronavirus infectious bronchitis virus nucleocapsid protein. J. Virol. 2007, 81, 4298–4304. [Google Scholar] [CrossRef][Green Version]
- Huang, S.; Yuan, S.; Guo, L.; Yu, Y.; Li, J.; Wu, T.; Liu, T.; Yang, M.; Wu, K.; Liu, H.; et al. Genomic analysis of the immune gene repertoire of amphioxus reveals extraordinary innate complexity and diversity. Genome Res. 2008, 18, 1112–1126. [Google Scholar] [CrossRef] [PubMed]
- Kobe, B.; Kajava, A.V. The leucine-rich repeat as a protein recognition motif. Curr. Opin. Struct. Biol. 2001, 11, 725–732. [Google Scholar] [CrossRef]
- Wei, T.; Gong, J.; Jamitzky, F.; Heckl, W.M.; Stark, R.W.; Rossle, S.C. LRRML: A conformational database and an XML description of leucinerich repeats (LRRs). BMC Struct. Biol. 2008, 8, 47. [Google Scholar] [CrossRef]
- Álvarez, E.; DeDiego, M.L.; Nieto-Torres, J.L.; Jiménez-Guardeño, J.M.; Marcos-Villar, L.; Enjuanes, L. The envelope protein of severe acute respiratory syndrome coronavirus interacts with the non-structural protein 3 and is ubiquitinated. Virology 2010, 402, 281–291. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Zhang, X.; Wu, T.; Wang, Y.; Meng, J.; Liu, Q.; Niu, X.; Wu, Y. The papain-like protease of avian infectious bronchitis virus has deubiquitinating activity. Arch. Virol. 2017, 162, 1943–1950. [Google Scholar] [CrossRef] [PubMed]
- Choi, A.G.; Wong, J.; Marchant, D.; Luo, H. The ubiquitin-proteasome system in positive-strand RNA virus infection. Rev. Med. Virol. 2013, 23, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Luo, H. Interplay between the virus and the ubiquitin-proteasome system: Molecular mechanism of viral pathogenesis. Curr. Opin. Virol. 2016, 17, 1–10. [Google Scholar] [CrossRef]
- Proulx, J.; Borgmann, K.; Park, I.-W. Role of Virally-Encoded Deubiquitinating Enzymes in Regulation of the Virus Life Cycle. Int. J. Mol. Sci. 2021, 22, 4438. [Google Scholar] [CrossRef]
- Lim, K.P.; Ng, L.F.; Liu, D.X. Identification of a novel cleavage activity of the first papain-like proteinase domain encoded by open reading frame 1a of the coronavirus Avian infectious bronchitis virus and characterization of the cleavage products. J. Virol. 2000, 74, 1674–1685. [Google Scholar] [CrossRef]
- Chen, Y.; Savinov, S.N.; Mielech, A.M.; Cao, T.; Baker, S.C.; Mesecar, A.D. X-ray structural and functional studies of the three tandemly linked domains of non-structural protein 3 (nsp3) from murine hepatitis virus reveal conserved functions. J. Biol. Chem. 2015, 290, 25293–25306. [Google Scholar] [CrossRef]

| Protein | Function | References |
|---|---|---|
| Nsp1 | Amino-terminal protein lacking in IBV but existing in other CoVs function is the inhibition of cellular mechanisms, including translation and IFN signaling | [99] |
| Nsp2 | Plays a vital role in assisting IBV protein synthesis by blocking protein kinase phosphorylation of eukaryotic initiation factor 2 (eIF-2alpha), which shuts down protein synthesis | [100] |
| Nsp3 | Involves several domains comprising an acidic domain, an ADP-ribose 1 phosphatase, the PLP PLP1, and the TM domain; nevertheless, SARS-CoV is orthologous to PLP2 of other CoVs | [101] |
| Nsp4 | A membrane-spanning protein, along with Nsp2 and Nsp6 is assumed to anchor the viral replication complex in double-membrane vesicles at the Golgi apparatus | [102] |
| Nsp5 | An enclosing Mpro (cysteine protease) with a Cys–His catalytic dyad and responsible for cleaving the Nsps 4–16 | [103] |
| Nsp6–10 | Membrane-localized proteins that form a complex exhibiting replicase activity in the existence of an RNA primer | [104,105] |
| Nsp11–12 | RdRp for viral RNA replication | [106] |
| Nsp13 | RNA helicase with a function in unwinding or annealing RNA molecules | [107] |
| Nsp14 | ExoN domain that provides support to RNA synthesis, proofreading, and repair | [108] |
| Nsp15 | Endoribonuclease domain stimulated by retinoblastoma protein (pRb) in vitro | [109] |
| Nsp16 | Methyltransferase and RNA cap formation | [110,111] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bhuiyan, M.S.A.; Sarker, S.; Amin, Z.; Rodrigues, K.F.; Saallah, S.; Shaarani, S.M.; Siddiquee, S. Infectious Bronchitis Virus (Gammacoronavirus) in Poultry: Genomic Architecture, Post-Translational Modifications, and Structural Motifs. Poultry 2023, 2, 363-382. https://doi.org/10.3390/poultry2030027
Bhuiyan MSA, Sarker S, Amin Z, Rodrigues KF, Saallah S, Shaarani SM, Siddiquee S. Infectious Bronchitis Virus (Gammacoronavirus) in Poultry: Genomic Architecture, Post-Translational Modifications, and Structural Motifs. Poultry. 2023; 2(3):363-382. https://doi.org/10.3390/poultry2030027
Chicago/Turabian StyleBhuiyan, Md. Safiul Alam, Subir Sarker, Zarina Amin, Kenneth Francis Rodrigues, Suryani Saallah, Sharifudin Md. Shaarani, and Shafiquzzaman Siddiquee. 2023. "Infectious Bronchitis Virus (Gammacoronavirus) in Poultry: Genomic Architecture, Post-Translational Modifications, and Structural Motifs" Poultry 2, no. 3: 363-382. https://doi.org/10.3390/poultry2030027
APA StyleBhuiyan, M. S. A., Sarker, S., Amin, Z., Rodrigues, K. F., Saallah, S., Shaarani, S. M., & Siddiquee, S. (2023). Infectious Bronchitis Virus (Gammacoronavirus) in Poultry: Genomic Architecture, Post-Translational Modifications, and Structural Motifs. Poultry, 2(3), 363-382. https://doi.org/10.3390/poultry2030027








