Integrating Reverse Vaccinology with Immunoinformatics for Rational Vaccine Target Discovery in Mycoplasma genitalium
Abstract
1. Introduction
2. Materials and Methods
2.1. Sequence Retrieval
2.2. Sequence Homology Search
2.3. Sub-Cellular Localization Prediction
2.4. Transmembrane Helices Prediction
2.5. Physico-Chemical Properties Computation
2.6. Virulence Factors Prediction
2.7. Functional Annotation and Domain Prediction
2.8. Signal Peptide Prediction
2.9. Prediction of Antigenicity, Allergenicity, and Toxicity
2.9.1. Antigenicity Prediction
2.9.2. Allergenicity Prediction
2.9.3. Toxicity Prediction
2.10. Homology Modeling of Two Candidate Proteins
2.11. B-Cell and T-Cell Epitope Prediction
2.11.1. B-Cell Epitope Prediction
2.11.2. T-Cell Epitope Prediction
2.12. Molecular Docking
2.13. Molecular Dynamics (MD) Simulations
2.14. Principal Component Analysis (PCA)
2.15. Binding Free Energy Calculations (MM-GBSA)
3. Results
3.1. Identification and Characterization of Hypothetical Proteins
3.2. Physicochemical Properties
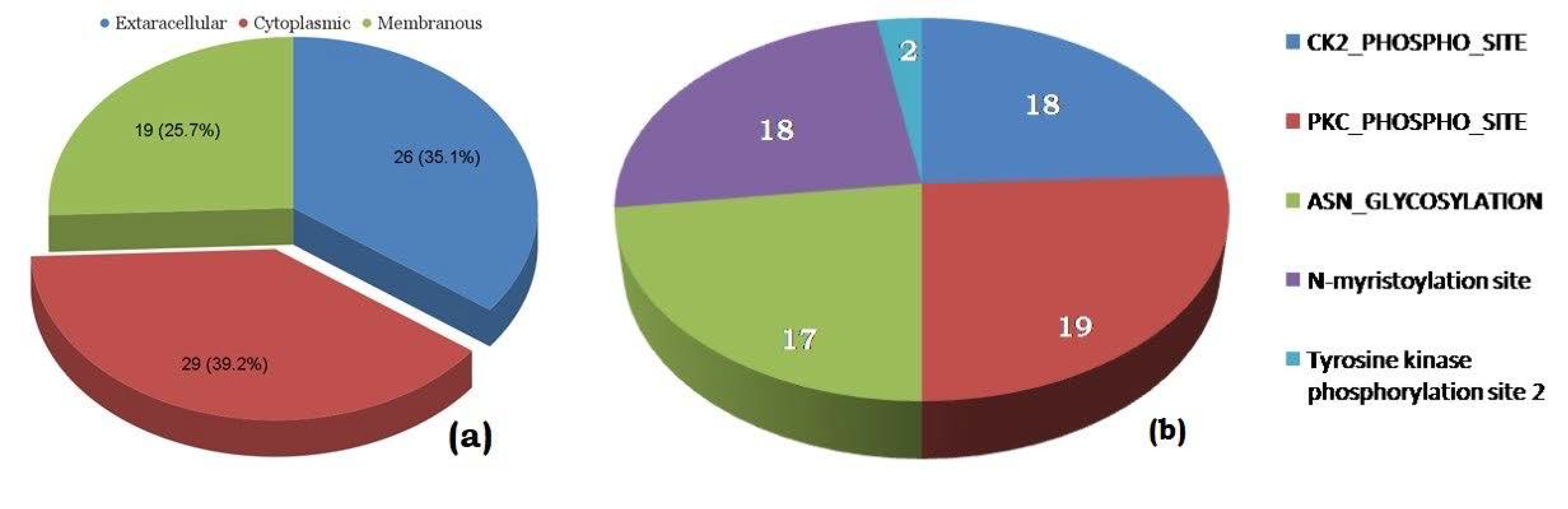
3.3. Sub-Cellular Localization
3.4. Antigenicity and Virulence Assessment
3.5. Functional Annotation
3.6. Prioritization of Vaccine Candidates
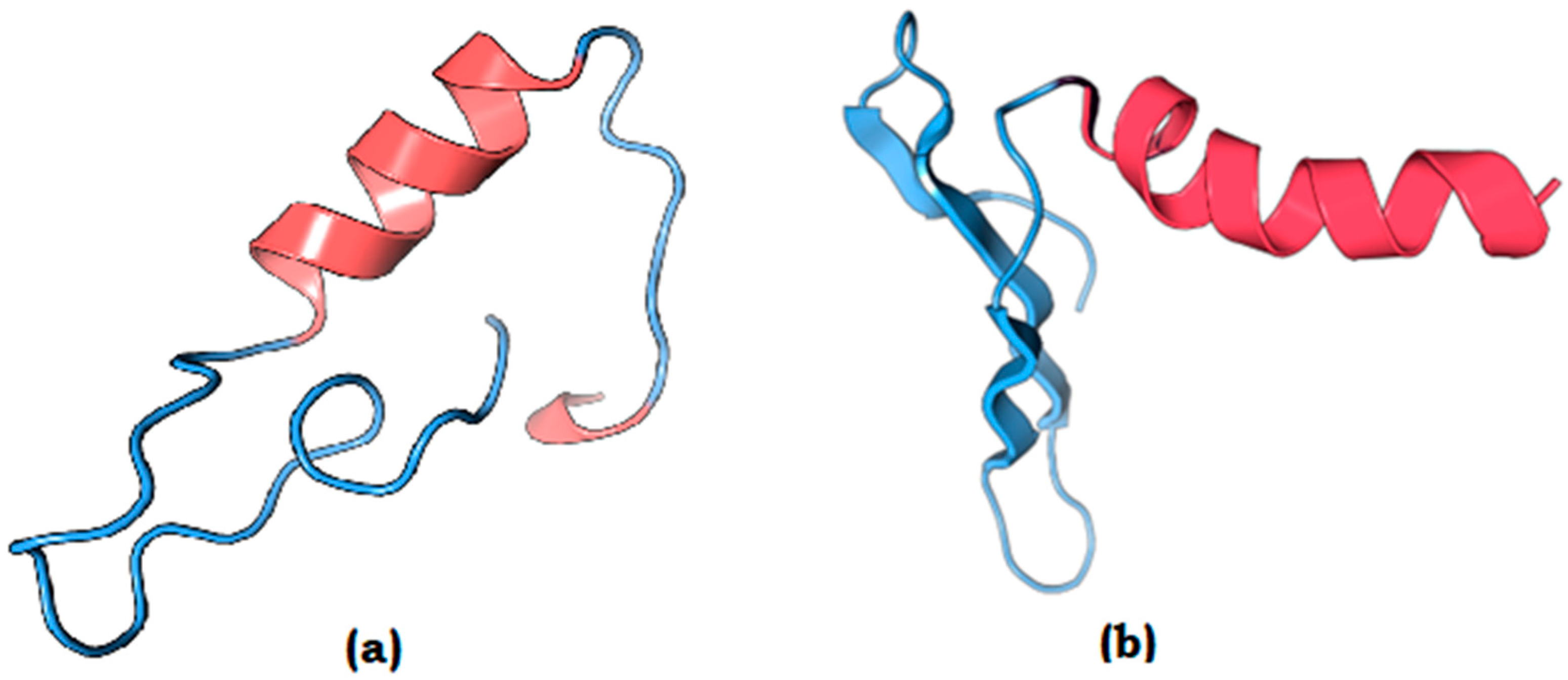
3.7. Homology Modeling of the Potential Vaccine Candidates
3.8. Epitope Prediction
3.8.1. B-Cell Epitope Mapping
3.8.2. T-Cell Epitope Mapping
3.8.3. Epitope Conservancy Across M. genitalium Strains
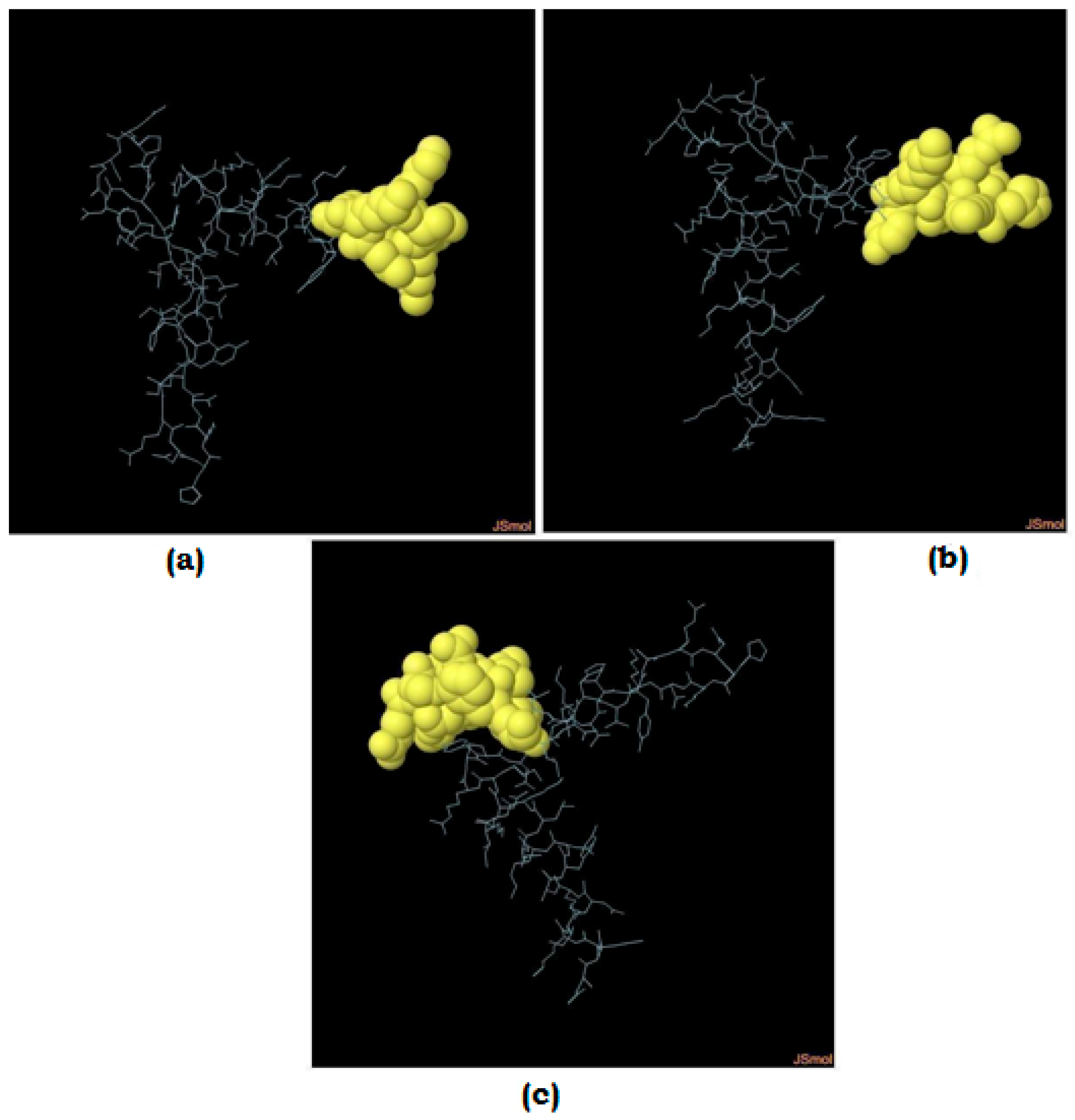
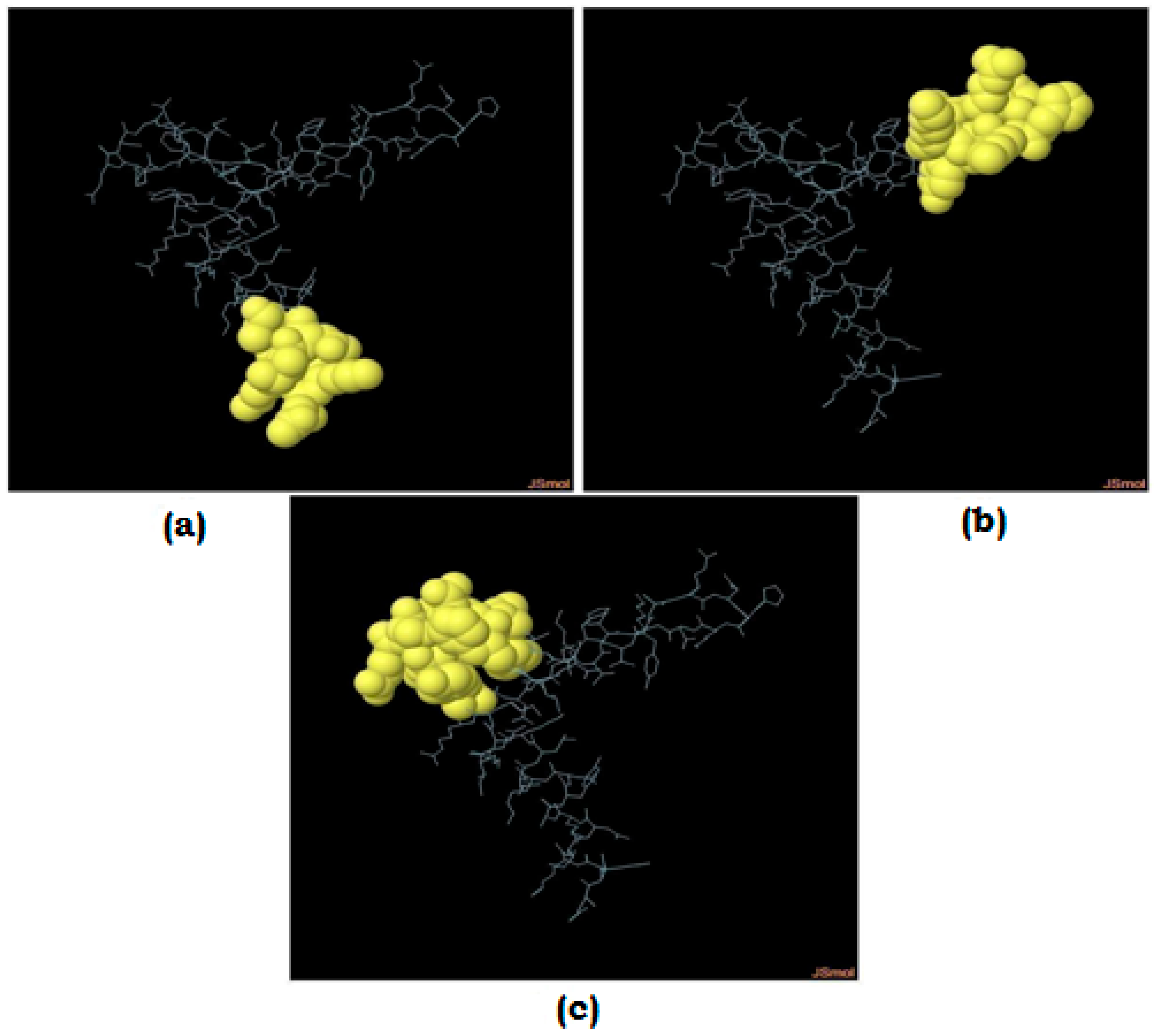
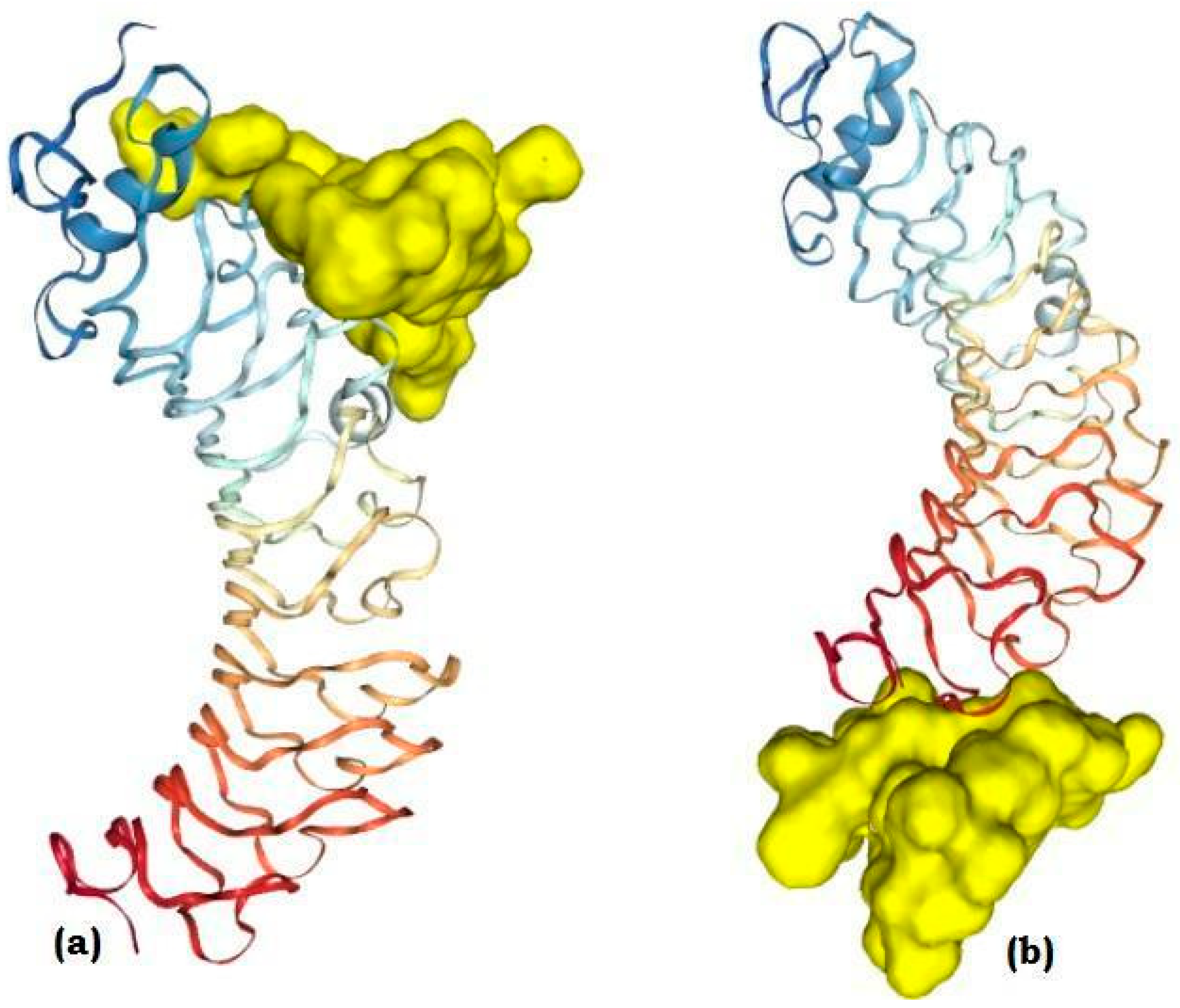
3.9. Molecular Docking
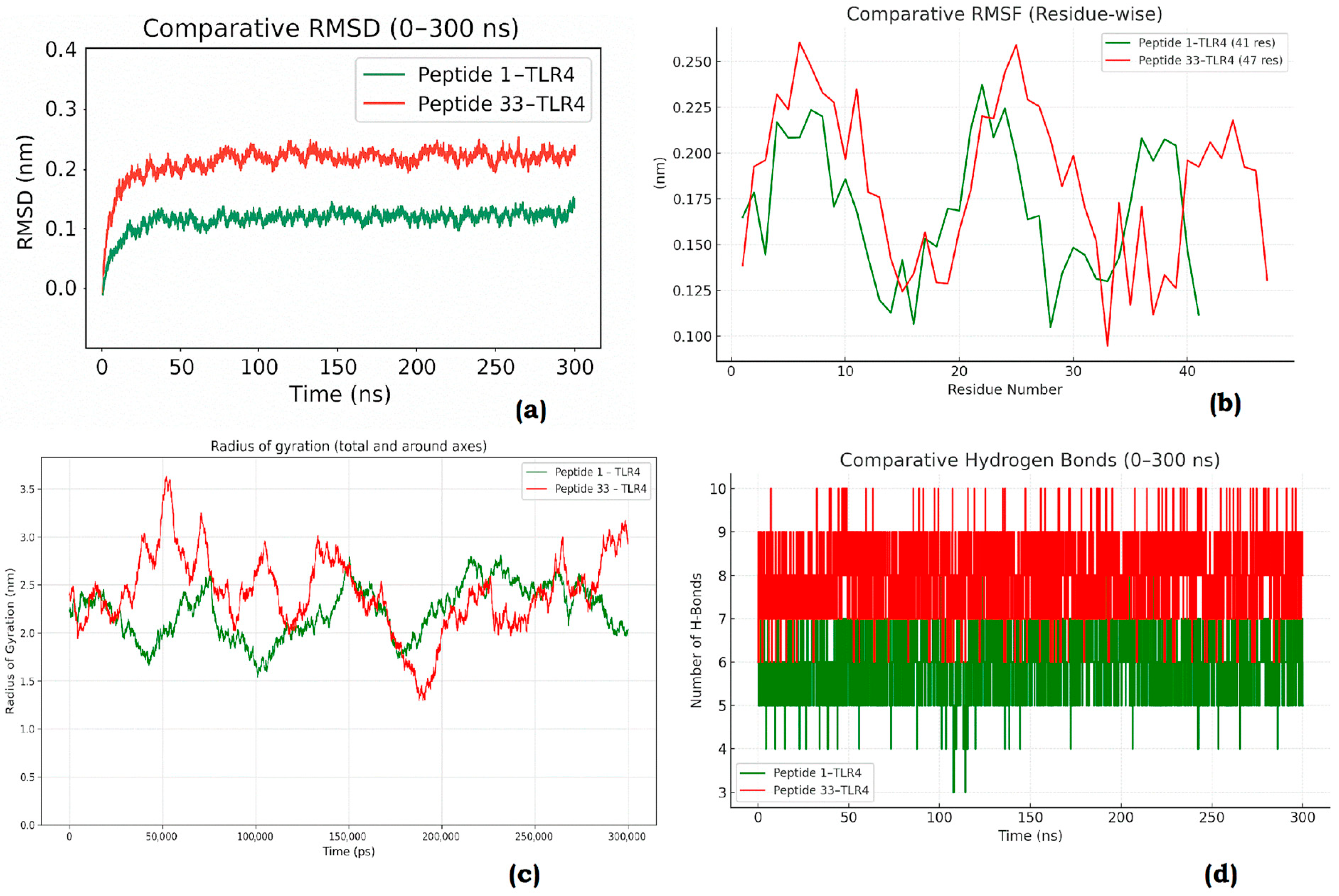
3.10. Molecular Dynamics (MD) Simulations
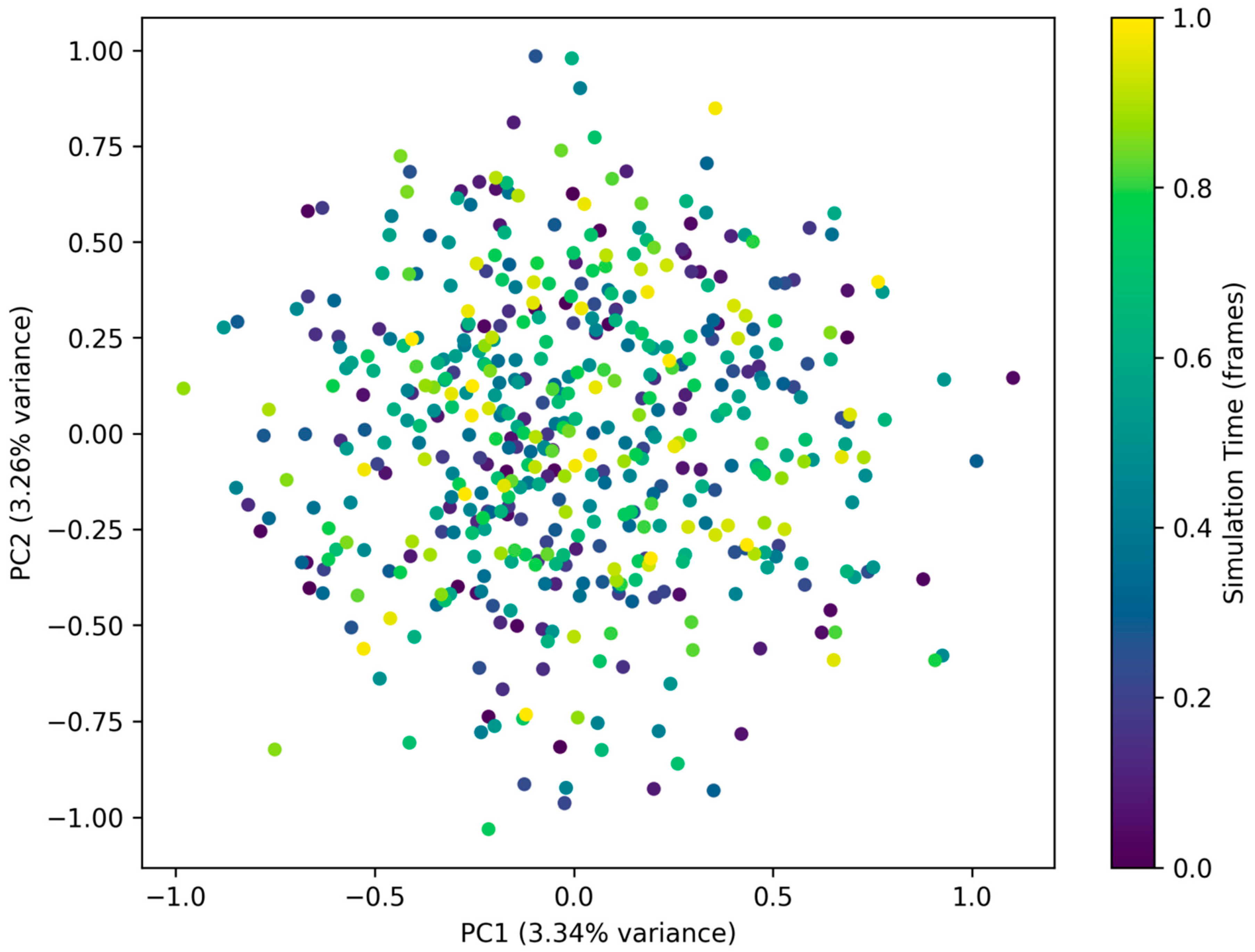
3.11. Principal Component Analysis (PCA)
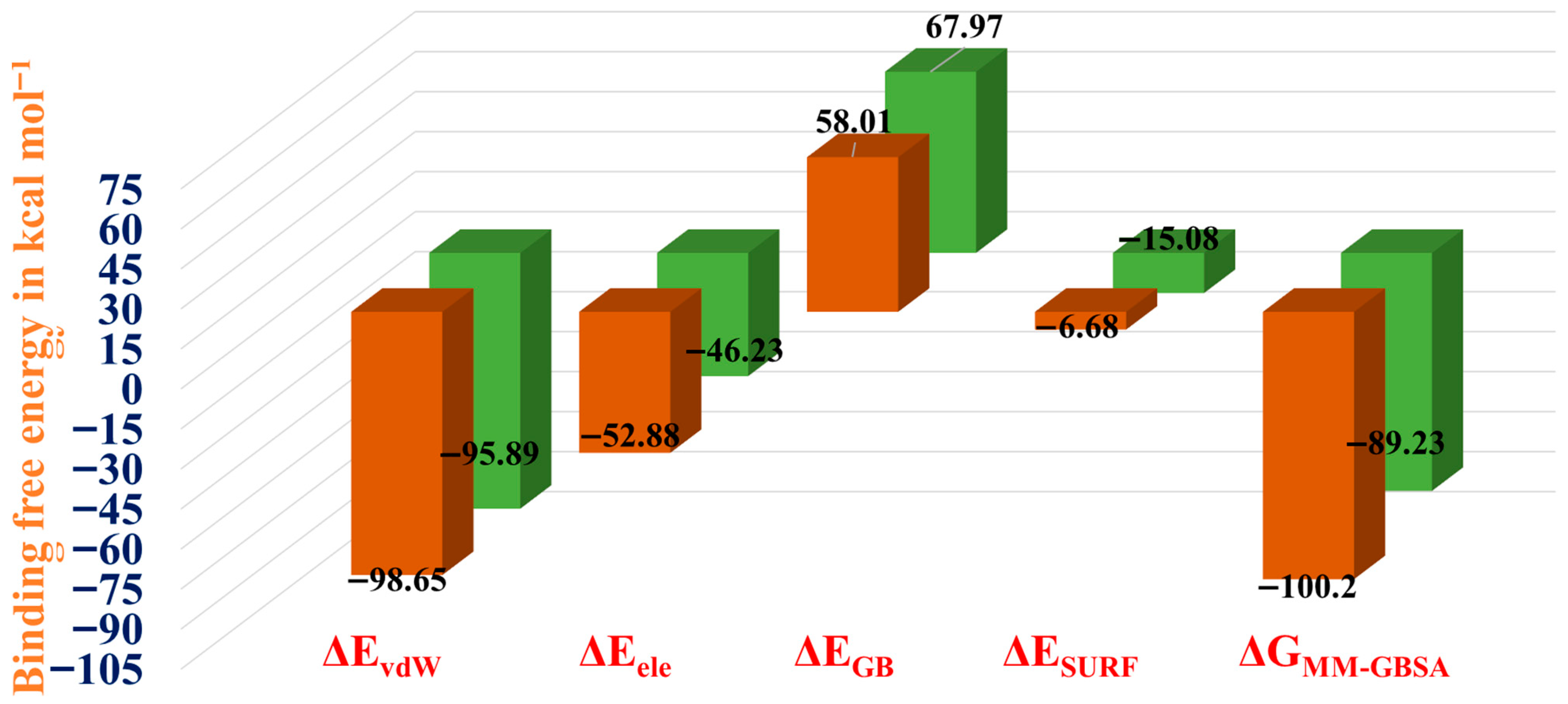
3.12. BindingFree Energy Calculations (MMGBSA)
4. Discussion
5. Conclusions
Limitations of Study
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sethi, S.; Singh, G.; Samanta, P.; Sharma, M. Mycoplasma genitalium: An emerging sexually transmitted pathogen. Indian. J. Med. Res. 2012, 136, 942–955. [Google Scholar]
- Ona, S.; Molina, R.L.; Diouf, K. Mycoplasma genitalium: An Overlooked Sexually Transmitted Pathogen in Women? Infect. Dis. Obstet. Gynecol. 2016, 2016, 4513089. [Google Scholar] [CrossRef]
- Doelman, T.A.; Adriaens, N.; Westerhuis, B.M.; Bruisten, S.M.; Vergunst, C.E.; Bouwman, F.M.; van Dam, A.P. Phenotypic antibiotic resistance of Mycoplasma genitalium and its variation between different macrolide resistance-associated mutations. J. Antimicrob. Chemother. 2025, 80, 465–471. [Google Scholar] [CrossRef] [PubMed]
- Khalid, K.; Hussain, T.; Jamil, Z.; Alrokayan, K.S.; Ahmad, B.; Waheed, Y. Vaccinomics-Aided Development of a Next-Generation Chimeric Vaccine against an Emerging Threat: Mycoplasma genitalium. Vaccines 2022, 10, 1720. [Google Scholar] [CrossRef] [PubMed]
- Razin, S.; Yogev, D.; Naot, Y. Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev. 1998, 62, 1094–1156. [Google Scholar] [CrossRef] [PubMed]
- Kant, R.; Khan, M.S.; Chopra, M.; Saluja, D. Artificial intelligence-driven reverse vaccinology for Neisseria gonorrhoeae vaccine: Prioritizing epitope-based candidates. Front. Mol. Biosci. 2024, 11, 1442158. [Google Scholar] [CrossRef]
- Yueyue, W.; Feichen, X.; Yixuan, X.; Lu, L.; Yiwen, C.; Xiaoxing, Y. Pathogenicity and virulence of Mycoplasma genitalium: Unraveling Ariadne’s Thread. Virulence 2022, 13, 1161–1183. [Google Scholar] [CrossRef]
- Rappuoli, R. Reverse vaccinology, a genome-based approach to vaccine development. Vaccine 2001, 19, 2688–2691. [Google Scholar] [CrossRef]
- Gloanec, N.; Guyard-Nicodème, M.; Chemaly, M.; Dory, D. Reverse vaccinology: A strategy also used for identifying potential vaccine antigens in poultry. Vaccine 2025, 48, 126756. [Google Scholar] [CrossRef]
- Chen, L.; Li, Q.; Nasif, K.F.A.; Xie, Y.; Deng, B.; Niu, S.; Pouriyeh, S.; Dai, Z.; Chen, J.; Xie, C.Y. AI-Driven Deep Learning Techniques in Protein Structure Prediction. Int. J. Mol. Sci. 2024, 25, 8426. [Google Scholar] [CrossRef]
- Olawade, D.B.; Teke, J.; Fapohunda, O.; Weerasinghe, K.; Usman, S.O.; Ige, A.O.; David-Olawade, A.C. Leveraging artificial intelligence in vaccine development: A narrative review. J. Microbiol. Methods 2024, 224, 106998. [Google Scholar] [CrossRef]
- Fatoba, A.J.; Okpeku, M.; Adeleke, M.A. Subtractive Genomics Approach for Identification of Novel Therapeutic Drug Targets in Mycoplasma genitalium. Pathogens 2021, 10, 921. [Google Scholar] [CrossRef] [PubMed]
- Nogueira, W.G.; Jaiswal, A.K.; Tiwari, S.; Ramos, R.T.; Ghosh, P.; Barh, D.; Azevedo, V.; Soares, S.C. Computational identification of putative common genomic drug and vaccine targets in Mycoplasma genitalium. Genomics 2021, 113, 2730–2743. [Google Scholar] [CrossRef] [PubMed]
- Jansen, K.U.; Gruber, W.C.; Simon, R.; Wassil, J.; Anderson, A.S. The impact of human vaccines on bacterial antimicrobial resistance. A review. Environ. Chem. Lett. 2021, 19, 4031–4062. [Google Scholar] [CrossRef] [PubMed]
- Expasy—SIB Swiss Institute of Bioinformatics. Available online: https://www.expasy.org/ (accessed on 5 July 2025).
- GRAVY Calculator. Available online: https://www.gravy-calculator.de/ (accessed on 5 July 2025).
- BLAST: Basic Local Alignment Search Tool. Available online: https://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 5 July 2025).
- Yu, C.S.; Cheng, C.W.; Su, W.C.; Chang, K.C.; Huang, S.W.; Hwang, J.K.; Lu, C.H. CELLO V2.5CELLO V2.5 V2.52GO: A web server for protein sub-cellular Localization prediction with functional gene ontology annotation. PLoS ONE 2014, 9, e99368. [Google Scholar] [CrossRef]
- Yu, N.Y.; Wagner, J.R.; Laird, M.R.; Melli, G.; Rey, S.; Lo, R.; Dao, P.; Sahinalp, S.C.; Ester, M.; Foster, L.J.; et al. PSORTb 3.0: Improved protein sub-cellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 2010, 26, 1608–1615. [Google Scholar] [CrossRef]
- TMHMM 2.0—DTU Health Tech—Bioinformatic Services. Available online: https://services.healthtech.dtu.dk/services/TMHMM-2.0/ (accessed on 5 July 2025).
- Teufel, F.; Almagro Armenteros, J.J.; Johansen, A.R.; Gíslason, M.H.; Pihl, S.I.; Tsirigos, K.D.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 2022, 40, 1023–1025. [Google Scholar] [CrossRef]
- Tusnády, G.E.; Simon, I. The HMMTOP transmembrane topology prediction server. Bioinformatics 2001, 17, 849–850. [Google Scholar] [CrossRef]
- Imai, K.; Asakawa, N.; Tsuji, T.; Akazawa, F.; Ino, A.; Sonoyama, M.; Mitaku, S. SOSUI-GramN: High performance prediction for sub-cellular localization of proteins in gram-negative bacteria. Bioinformation 2008, 2, 417–421. [Google Scholar] [CrossRef]
- Hallgren, J.; Tsirigos, K.D.; Pedersen, M.D.; Almagro Armenteros, J.J.; Marcatili, P.; Nielsen, H.; Krogh, A.; Winther, O. DeepTMHMM predicts alpha and beta transmembrane proteins using deep neural networks. bioRxiv 2022. [Google Scholar] [CrossRef]
- Sharma, A.; Garg, A.; Ramana, J.; Gupta, D. VirulentPred 2.0: An improved method for prediction of virulent proteins in bacterial pathogens. Protein Sci. 2023, 32, e4808. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Raghava, G.P.S. VICMpred: An SVM-based method for the prediction of functional proteins of Gram-negative bacteria using amino acid patterns and composition. Genom. Proteom. Bioinform. 2006, 4, 42–47. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.; Tosatto, S.C.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam v35.0Pfam v35.0 v35.0: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef]
- Conserved Domains Database (CDD) and Resources. Available online: https://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml (accessed on 5 July 2025).
- De Castro, E.; Sigrist, C.J.; Gattiker, A.; Bulliard, V.; Langendijk-Genevaux, P.S.; Gasteiger, E.; Bairoch, A.; Hulo, N. ScanProsite: Detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 2006, 34, W362–W365. [Google Scholar] [CrossRef]
- Doytchinova, I.A.; Flower, D.R. VaxiJen v2.0VaxiJen v2.0 v2.0: A server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinform. 2007, 8, 4. [Google Scholar] [CrossRef]
- Dimitrov, I.; Bangov, I.; Flower, D.R.; Doytchinova, I. AllerTOP v.2—A server for in silico prediction of allergens. J. Mol. Model. 2014, 20, 2278. [Google Scholar] [CrossRef]
- Rathore, A.S.; Choudhury, S.; Arora, A.; Tijare, P.; Raghava, G.P.S. ToxinPred 3.0: An improved method for predicting the toxicity of peptides. Comput. Biol. Med. 2024, 179, 108926. [Google Scholar] [CrossRef]
- Pan, X.; Zuallaert, J.; Wang, X.; Shen, H.B.; Campos, E.P.; Marushchak, D.O.; De Neve, W. ToxDL: Deep learning using primary structure and domain embeddings for assessing protein toxicity. Bioinformatics 2021, 36, 5159–5168. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Yuan, S.; Chan, H.S.; Hu, Z. Using PyMOL as a Platform for Computational Drug Design. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2017, 7, e1298. [Google Scholar] [CrossRef]
- PyMOL|Pymol.org. Available online: https://www.pymol.org/ (accessed on 5 July 2025).
- Jespersen, M.C.; Peters, B.; Nielsen, M.; Marcatili, P. BepiPred-2.0: Improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res. 2017, 45, W24–W29. [Google Scholar] [CrossRef]
- Immune Epitope Database and Analysis Resource (IEDB)|NIAID: National Institute of Allergy and Infectious Diseases. Available online: https://www.niaid.nih.gov/research/immune-epitope-database (accessed on 5 July 2025).
- Vita, R.; Blazeska, N.; Marrama, D.; IEDB Curation Team Members; Duesing, S.; Bennett, J.; Greenbaum, J.; De Almeida Mendes, M.; Mahita, J.; Wheeler, D.K.; et al. The Immune Epitope Database (IEDB): 2024 update. Nucleic Acids Res. 2025, 53, D436–D443. [Google Scholar] [CrossRef]
- Hoof, I.; Peters, B.; Sidney, J.; Pedersen, L.E.; Sette, A.; Lund, O.; Buus, S.; Nielsen, M. NetMHCpan, a method for MHC class I binding prediction beyond humans. Immunogenetics 2009, 61, 1–13. [Google Scholar] [CrossRef]
- Han, J.; Kim, H.J.; Lee, S.C.; Hong, S.; Park, K.; Jeon, Y.H.; Kim, D.; Cheong, H.K.; Kim, H.S. Structure-Based Rational Design of a Toll-like Receptor 4 (TLR4) Decoy Receptor with High Binding Affinity for a Target Protein. PLoS ONE 2012, 7, e30929. [Google Scholar] [CrossRef] [PubMed]
- Lamiable, A.; Thévenet, P.; Rey, J.; Vavrusa, M.; Derreumaux, P.; Tufféry, P. PEP-FOLD3: Faster de novo structure prediction for linear peptides in solution and in complex. Nucleic Acids Res. 2016, 44, W449–W454. [Google Scholar] [CrossRef]
- Yan, Y.; Tao, H.; He, J.; Huang, S.-Y. The HDOCK server for integrated protein–protein docking. Nat. Protoc. 2020, 15, 1829–1852. [Google Scholar] [CrossRef] [PubMed]
- Rashidieh, B.; Valizadeh, M.; Assadollahi, V.; Ranjbar, M.M. Molecular dynamics simulation on the low sensitivity of mutants of NEDD-8 activating enzyme for MLN4924 inhibitor as a cancer drug. Am. J. Cancer Res. 2015, 5, 3400–3406. [Google Scholar]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- David, C.C.; Jacobs, D.J. Principal Component Analysis: A Method for Determining the Essential Dynamics of Proteins. Methods Mol Biol. 2014, 1084, 193–226. [Google Scholar]
- Jaadi, Z. Principal Component Analysis (PCA): Explained Step-by-Step. Built In, 1 April 2021. Available online: https://builtin.com/data-science/step-step-explanation-principal-component-analysis (accessed on 29 August 2025).
- Adelusi, T.I.; Bolaji, O.Q.; Ojo, T.O.; Adegun, I.P.; Adebodun, S. Molecular Mechanics with Generalized Born Surface Area (MMGBSA) Calculations and Docking Studies Unravel some Antimalarial Compounds Using Heme O Synthase as Therapeutic Target. ChemistrySelect 2023, 8, e202303686. [Google Scholar] [CrossRef]
- Sobolev, O.V.; Afonine, P.V.; Moriarty, N.W.; Hekkelman, M.L.; Joosten, R.P.; Perrakis, A.; Adams, P.D. A global Ramachandran score identifies protein structures with unlikely stereochemistry. Structure 2020, 28, 1249–1258.e2. [Google Scholar] [CrossRef] [PubMed]
- Wiederstein, M.; Sippl, M.J. ProSA-Web: Interactive Web Service for the Recognition of Errors in Three-Dimensional Structures of Proteins. Nucleic Acids Res. 2007, 35, W407–W410. [Google Scholar] [CrossRef] [PubMed]
- Colovos, C.; Yeates, T.O. Verification of protein structures: Patterns of nonbonded atomic interactions. Protein Sci. 1993, 2, 1511–1519. [Google Scholar] [CrossRef] [PubMed]
- Haggerty, C.L.; Taylor, B.D. Mycoplasma genitalium: An emerging cause of pelvic inflammatory disease. Infect. Dis. Obstet. Gynecol. 2011, 2011, 959816. [Google Scholar] [CrossRef]
- Barik, K.; Arya, P.K.; Singh, A.K.; Kumar, A. Potential therapeutic targets for combating Mycoplasma genitalium. 3 Biotech 2023, 13, 9. [Google Scholar] [CrossRef]
- Duan, Y.; Hao, Y.; Feng, H.; Shu, J.; He, Y. Research progress on Haemophilus parasuis vaccines. Front. Vet. Sci. 2025, 12, 1492144. [Google Scholar] [CrossRef]
- Mukhopadhyay, H.; Bairagi, A.; Mukherjee, A.; Prasad, A.K.; Roy, A.D.; Nayak, A. Multidrug Resistant Acinetobacter Baumannii: A Study on Its Pathogenesis and Therapeutics. Curr. Res. Microb. Sci. 2025, 8, 100331. [Google Scholar] [CrossRef]
- Calderwood, S.K.; Gong, J.; Murshid, A. Extracellular HSPs: The Complicated Roles of Extracellular HSPs in Immunity. Front. Immunol. 2016, 7, 159. [Google Scholar] [CrossRef]
- Sen, T.; Verma, N.K. Functional Annotation and Curation of Hypothetical Proteins Present in A Newly Emerged Serotype 1c of Shigella flexneri: Emphasis on Selecting Targets for Virulence and Vaccine Design Studies. Genes 2020, 11, 340. [Google Scholar] [CrossRef] [PubMed]
- Lazar, V.; Oprea, E.; Ditu, L.-M. Resistance, Tolerance, Virulence and Bacterial Pathogen Fitness-Current State and Envisioned Solutions for the Near Future. Pathogens 2023, 12, 746. [Google Scholar] [CrossRef] [PubMed]
- Guarra, F.; Colombo, G. Computational Methods in Immunology and Vaccinology: Design and Development of Antibodies and Immunogens. J. Chem. Theory Comput. 2023, 19, 5315–5333. [Google Scholar] [CrossRef] [PubMed]
- Pranavathiyani, G.; Prava, J.; Rajeev, A.C.; Pan, A. Novel Target Exploration from Hypothetical Proteins of Klebsiella pneumoniae MGH 78578 Reveals a Protein Involved in Host-Pathogen Interaction. Front. Cell. Infect. Microbiol. 2020, 10, 109. [Google Scholar] [CrossRef]
- Dowling, A.J. Novel gain of function approaches for vaccine candidate identification in Burkholderia pseudomallei. Front. Cell. Infect. Microbiol. 2012, 2, 139. [Google Scholar] [CrossRef]
| S. No. | Seq.ID | No. of Amino Acid Residues | Mol. Wt. | Theoretical pI | Total Number of Negatively Charged Residues | Total Number of Positively Charged Residues | Extinction Coefficients | Instability Index | Aliphatic Index | Grand Average of Hydropathicity (GRAVY) |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fig|2097.70.peg.1 | 193 | 20,901.76 | 7.02 | 16 | 16 | 26,930 | 20.04 | 58.08 | −0.628 |
| 2 | fig|2097.70.peg.2 | 87 | 10,066.52 | 5.36 | 11 | 8 | 5960 | 21.22 | 106.32 | −0.272 |
| 3 | fig|2097.70.peg.3 | 286 | 33,582.61 | 5.98 | 50 | 49 | 32,430 | 37.88 | 69.97 | −1.049 |
| 4 | fig|2097.70.peg.12 | 129 | 15,317.56 | 5.09 | 18 | 14 | 7450 | 26.78 | 87.67 | −0.195 |
| 5 | fig|2097.70.peg.28 | 835 | 96,004.28 | 4.15 | 168 | 69 | 67,285 | 51.44 | 67.05 | −0.918 |
| 6 | fig|2097.70.peg.32 | 320 | 37,185.56 | 9.89 | 8 | 27 | 59,485 | 26.17 | 129.75 | 0.736 |
| 7 | fig|2097.70.peg.33 | 599 | 68,777.93 | 7.31 | 74 | 74 | 55,240 | 55.84 | 71.8 | −0.907 |
| 8 | fig|2097.70.peg.34 | 218 | 24,708.47 | 9.81 | 15 | 20 | 6990 | 53.28 | 88.53 | −0.417 |
| 9 | fig|2097.70.peg.35 | 178 | 20,555.91 | 9.83 | 14 | 24 | 11,920 | 49.76 | 89.33 | −0.043 |
| 10 | fig|2097.70.peg.36 | 286 | 32,450.21 | 9.8 | 13 | 21 | 47,900 | 31.56 | 126.47 | 0.735 |
| 11 | fig|2097.70.peg.40 | 59 | 7252.32 | 10.71 | 1 | 9 | 23,950 | 23.22 | 59.66 | −0.605 |
| 12 | fig|2097.70.peg.41 | 254 | 29,759.81 | 9.69 | 12 | 24 | 50,880 | 16.96 | 101.65 | 0.426 |
| 13 | fig|2097.70.peg.46 | 756 | 88,407.1 | 4.61 | 164 | 101 | 39,880 | 39.47 | 86.56 | −1.028 |
| 14 | fig|2097.70.peg.49 | 212 | 25,044.97 | 9.49 | 22 | 31 | 30,940 | 36.27 | 91.04 | −0.338 |
| 15 | fig|2097.70.peg.60 | 336 | 37,119.09 | 9.16 | 27 | 33 | 63,370 | 27.92 | 64.14 | −0.678 |
| 16 | fig|2097.71.peg.1 | 409 | 45,698.09 | 9.06 | 33 | 39 | 73,340 | 26.13 | 73.67 | −0.535 |
| 17 | fig|2097.71.peg.2 | 1444 | 159,334.7 | 8.56 | 131 | 137 | 215,090 | 30.96 | 74.25 | −0.489 |
| 18 | fig|2097.71.peg.9 | 92 | 10,079.51 | 9.44 | 4 | 8 | 8940 | 18.16 | 75.22 | −0.311 |
| 19 | fig|2097.71.peg.1 | 259 | 28,803.14 | 8.87 | 23 | 27 | 45,045 | 34.68 | 71.08 | −0.727 |
| 20 | fig|2097.71.peg.1 | 319 | 35,001.18 | 9.52 | 28 | 38 | 33,460 | 33.04 | 63.51 | −0.732 |
| 21 | fig|2097.71.peg.48 | 154 | 18,148.6 | 5.38 | 13 | 12 | 24,410 | 23.09 | 129.22 | 0.763 |
| 22 | fig|2097.71.peg.50 | 409 | 48,520.11 | 8.69 | 61 | 67 | 37,275 | 32.36 | 97.02 | −0.59 |
| 23 | fig|2097.71.peg.51 | 375 | 43,187.64 | 9.36 | 21 | 33 | 38,390 | 30.73 | 125.49 | 0.74 |
| 24 | fig|2097.71.peg.54 | 279 | 31,687.37 | 10.15 | 18 | 40 | 40,450 | 36.16 | 104.44 | 0.016 |
| 25 | fig|2097.71.peg.57 | 90 | 10,472.54 | 9.85 | 9 | 18 | 3105 | 17.2 | 120.22 | −0.258 |
| 26 | fig|2097.71.peg.59 | 1113 | 130,580.1 | 6.81 | 146 | 144 | 157,945 | 34.53 | 93.44 | −0.393 |
| 27 | fig|2097.71.peg.60 | 108 | 11,335.68 | 9.8 | 4 | 10 | 5960 | 22.38 | 67.78 | −0.501 |
| 28 | fig|2097.71.peg.61 | 123 | 12,983.36 | 9.57 | 8 | 12 | 15,470 | 43.25 | 64.31 | −0.68 |
| 29 | fig|2097.71.peg.62 | 40 | 4645.41 | 6.04 | 5 | 5 | 5500 | 38.74 | 109.5 | −0.185 |
| 30 | fig|2097.71.peg.63 | 176 | 19,106.87 | 6.31 | 14 | 13 | 26,930 | 16.55 | 59.83 | −0.569 |
| 31 | fig|2097.69.peg.1 | 295 | 32,373.67 | 8.51 | 24 | 26 | 60,390 | 30.86 | 62.85 | −0.633 |
| 32 | fig|2097.69.peg.2 | 67 | 7973.49 | 10.01 | 5 | 11 | 17,990 | 10.21 | 113.28 | −0.021 |
| 33 | fig|2097.69.peg.3 | 196 | 21,380.8 | 9.43 | 14 | 19 | 27,960 | 42.28 | 68.16 | −0.591 |
| 34 | fig|2097.69.peg.4 | 137 | 14,915.57 | 6.15 | 20 | 19 | 6990 | 48.74 | 58.32 | −0.932 |
| 35 | fig|2097.69.peg.6 | 196 | 23,299.79 | 9.52 | 21 | 31 | 35,870 | 26.84 | 89.49 | −0.626 |
| 36 | fig|2097.69.peg.7 | 347 | 40,052.08 | 5.92 | 40 | 36 | 48,820 | 34.76 | 86.8 | −0.467 |
| 37 | fig|2097.69.peg.8 | 113 | 13,267.1 | 8.95 | 13 | 16 | 19,940 | 41.21 | 84.6 | −0.566 |
| 38 | fig|2097.69.peg.10 | 44 | 5011.15 | 12.01 | 1 | 7 | NA * | 17.28 | 152.95 | 0.648 |
| 39 | fig|2097.69.peg.12 | 556 | 62,239.63 | 6.59 | 65 | 64 | 62,690 | 26.98 | 78.2 | −0.474 |
| 40 | fig|2097.69.peg.13 | 262 | 29,206.1 | 6.76 | 28 | 28 | 10,430 | 32.75 | 94.2 | −0.154 |
| 41 | fig|2097.69.peg.14 | 218 | 24,887.78 | 9.43 | 19 | 26 | 14,900 | 26.18 | 105.96 | −0.041 |
| 42 | fig|2097.69.peg.16 | 970 | 108,126.5 | 8.59 | 70 | 76 | 131,015 | 28.48 | 102.71 | 0.166 |
| 43 | fig|2097.69.peg.25 | 340 | 39,661.11 | 8.57 | 52 | 55 | 15,930 | 40.38 | 82.91 | −0.843 |
| 44 | fig|2097.69.peg.25 | 340 | 39,661.11 | 8.57 | 52 | 55 | 15,930 | 40.38 | 82.91 | −0.843 |
| 45 | fig|2097.69.peg.27 | 115 | 13,060.69 | 9.7 | 3 | 7 | 22,460 | 25.83 | 133.13 | 1.027 |
| 46 | fig|2097.69.peg.34 | 76 | 9189.53 | 7.97 | 18 | 19 | NA * | 54.49 | 73.03 | −1.301 |
| 47 | fig|2097.69.peg.35 | 83 | 8385.38 | 10 | 2 | 8 | 2980 | 24.3 | 70.6 | −0.447 |
| 48 | fig|2097.69.peg.36 | 173 | 19,009.5 | 9.82 | 11 | 23 | 39,085 | 39.31 | 69.88 | −0.657 |
| 49 | fig|2097.69.peg.37 | 162 | 18,141.3 | 9.4 | 12 | 16 | 27,960 | 32.91 | 73.4 | −0.591 |
| 50 | fig|2097.69.peg.38 | 135 | 14,805.63 | 7.87 | 21 | 22 | 6990 | 53.49 | 61.41 | −0.897 |
| 51 | fig|2097.69.peg.43 | 256 | 30,481.32 | 9.77 | 17 | 29 | 42,400 | 22.24 | 99.3 | 0.094 |
| 52 | fig|2097.69.peg.44 | 550 | 64,213.61 | 9.1 | 50 | 61 | 67,630 | 30.66 | 100.27 | −0.039 |
| 53 | fig|2097.69.peg.56 | 226 | 26,256.39 | 9.43 | 14 | 22 | 26,720 | 25.91 | 127.26 | 0.617 |
| 54 | fig|2097.69.peg.57 | 630 | 74,234.51 | 6.24 | 83 | 79 | 77,030 | 42.19 | 100.98 | −0.23 |
| 55 | fig|2097.69.peg.58 | 620 | 72,815.2 | 8.96 | 69 | 80 | 67,185 | 29.89 | 96.66 | −0.216 |
| 56 | fig|2097.69.peg.62 | 294 | 34,572.15 | 7.69 | 35 | 36 | 13,535 | 28.33 | 98.78 | −0.235 |
| 57 | fig|2097.69.peg.73 | 85 | 9072.92 | 7.88 | 7 | 8 | 13,980 | 41.4 | 50.47 | −0.831 |
| 58 | fig|2097.69.peg.78 | 411 | 47,823.61 | 9.46 | 37 | 55 | 25,445 | 21.63 | 96.74 | −0.237 |
| 59 | fig|2097.69.peg.81 | 102 | 11,303.66 | 9.3 | 7 | 10 | 18,450 | 35 | 67.94 | −0.31 |
| 60 | fig|2097.69.peg.83 | 345 | 39,454.58 | 5.03 | 53 | 37 | 14,440 | 36.9 | 94.06 | −0.59 |
| 61 | fig|2097.69.peg.84 | 1802 | 215,902.4 | 8.66 | 308 | 321 | 82,060 | 41.81 | 80.1 | −1.164 |
| 62 | fig|2097.69.peg.92 | 147 | 17,470.55 | 4.91 | 32 | 23 | 14,440 | 28.41 | 79.66 | −0.941 |
| 63 | fig|2097.69.peg.94 | 216 | 25,626.25 | 8.77 | 30 | 34 | 51,005 | 33.82 | 74.95 | −0.73 |
| 64 | fig|2097.69.peg.98 | 167 | 19,345.06 | 7.12 | 14 | 14 | 27,055 | 39.95 | 92.22 | −0.421 |
| 65 | fig|2097.69.peg.103 | 122 | 13,513.01 | 5.18 | 18 | 15 | 2980 | 50.58 | 83.11 | −0.676 |
| 66 | fig|2097.69.peg.112 | 42 | 4607.35 | 10 | 1 | 6 | 2980 | 24.91 | 97.38 | −0.388 |
| 67 | fig|2097.69.peg.113 | 167 | 18,121.11 | 7.96 | 15 | 16 | 22,460 | 42.51 | 70.72 | −0.762 |
| 68 | fig|2097.69.peg.114 | 47 | 5275.49 | 9.52 | 2 | 5 | 11,000 | 10.95 | 142.77 | 0.84 |
| 69 | fig|2097.69.peg.115 | 165 | 18,406.57 | 9.16 | 13 | 16 | 27,960 | 34.63 | 69.09 | −0.602 |
| 70 | fig|2097.69.peg.116 | 77 | 8395.48 | 9.35 | 11 | 14 | 5500 | 32.06 | 60.91 | −0.906 |
| 71 | fig|2097.69.peg.117 | 59 | 7015.26 | 10.01 | 5 | 12 | 23,490 | 33.69 | 85.59 | −0.58 |
| 72 | fig|2097.69.peg.118 | 33 | 4217.09 | 10.67 | 0 | 11 | 11,460 | 69.82 | 73.94 | −1.364 |
| 73 | fig|2097.69.peg.119 | 55 | 6996.4 | 10.75 | 4 | 18 | 13,980 | 61.56 | 70.91 | −1.387 |
| 74 | fig|2097.69.peg.120 | 713 | 76,591.36 | 5.56 | 68 | 62 | 68,300 | 27.37 | 77.64 | −0.287 |
| S. No. | Seq ID | Final Location | TM Helix | Signal Peptide | Toxicity | |
|---|---|---|---|---|---|---|
| ToxinPred | ToxiDL | |||||
| 1 | fig|2097.70.peg.1 | Extracellular | No | No | Non-Toxic | Non-Toxic |
| 2 | fig|2097.70.peg.3 | Extracellular | No | No | Non-Toxic | Non-Toxic |
| 3 | fig|2097.70.peg.33 | Extracellular | No | No | Non-Toxic | Non-Toxic |
| 4 | fig|2097.70.peg.60 | Extracellular | No | No | Non-Toxic | Non-Toxic |
| 5 | fig|2097.71.peg.1 | Extracellular | Yes | Yes | Non-Toxic | Non-Toxic |
| 6 | fig|2097.71.peg.9 | Extracellular | No | No | Non-Toxic | Non-Toxic |
| 7 | fig|2097.71.peg.1 | Extracellular | No | No | Non-Toxic | Non-Toxic |
| 8 | fig|2097.71.peg.1 | Extracellular | No | No | Non-Toxic | Non-Toxic |
| 9 | fig|2097.71.peg.61 | Extracellular | No | No | Non-Toxic | Non-Toxic |
| 10 | fig|2097.71.peg.63 | Extracellular | No | No | Non-Toxic | Non-Toxic |
| 11 | fig|2097.69.peg.1 | Extracellular | No | No | Non-Toxic | Non-Toxic |
| 12 | fig|2097.69.peg.3 | Extracellular | No | No | Non-Toxic | Non-Toxic |
| 13 | fig|2097.69.peg.4 | Extracellular | No | No | Non-Toxic | Non-Toxic |
| 14 | fig|2097.69.peg.35 | Extracellular | No | No | Non-Toxic | Non-Toxic |
| 15 | fig|2097.69.peg.36 | Extracellular | No | No | Non-Toxic | Non-Toxic |
| 16 | fig|2097.69.peg.37 | Extracellular | No | No | Non-Toxic | Non-Toxic |
| 17 | fig|2097.69.peg.38 | Extracellular | No | No | Non-Toxic | Non-Toxic |
| 18 | fig|2097.69.peg.73 | Extracellular | No | No | Non-Toxic | Non-Toxic |
| 19 | fig|2097.69.peg.81 | Extracellular | Yes | Yes | Non-Toxic | Non-Toxic |
| 20 | fig|2097.69.peg.113 | Extracellular | No | No | Non-Toxic | Non-Toxic |
| 21 | fig|2097.69.peg.115 | Extracellular | No | No | Non-Toxic | Non-Toxic |
| 22 | fig|2097.69.peg.116 | Extracellular | No | No | Non-Toxic | Non-Toxic |
| 23 | fig|2097.69.peg.117 | Extracellular | No | No | Non-Toxic | Non-Toxic |
| Seq ID | Virulence | Antigenicity | Antigenicity Scores | Allergenicity |
|---|---|---|---|---|
| fig|2097.70.peg.1 | Virulent | Antigenic | 0.6703 | PROBABLE NON-ALLERGEN |
| fig|2097.70.peg.33 | Virulent | Antigenic | 0.6128 | PROBABLE NON-ALLERGEN |
| fig|2097.69.peg.81 | Virulent | Antigenic | 0.4724 | PROBABLE NON-ALLERGEN |
| fig|2097.70.peg.3 | Virulent | Antigenic | 0.4723 | PROBABLE NON-ALLERGEN |
| fig|2097.71.peg.63 | Virulent | Antigenic | 0.472 | PROBABLE NON-ALLERGEN |
| fig|2097.69.peg.36 | Virulent | Antigenic | 0.4696 | PROBABLE NON-ALLERGEN |
| fig|2097.69.peg.113 | Virulent | Antigenic | 0.463 | PROBABLE NON-ALLERGEN |
| fig|2097.70.peg.60 | Virulent | Antigenic | 0.451 | PROBABLE NON-ALLERGEN |
| fig|2097.69.peg.35 | Virulent | Antigenic | 0.4507 | PROBABLE NON-ALLERGEN |
| fig|2097.69.peg.116 | Virulent | Antigenic | 0.4492 | PROBABLE NON-ALLERGEN |
| fig|2097.69.peg.115 | Virulent | Antigenic | 0.4478 | PROBABLE NON-ALLERGEN |
| fig|2097.71.peg.1 | Virulent | Antigenic | 0.4312 | PROBABLE NON-ALLERGEN |
| fig|2097.71.peg.9 | Virulent | Antigenic | 0.4298 | PROBABLE NON-ALLERGEN |
| fig|2097.71.peg.1 | Virulent | Antigenic | 0.4122 | PROBABLE NON-ALLERGEN |
| fig|2097.69.peg.4 | Virulent | Antigenic | 0.3864 | PROBABLE NON-ALLERGEN |
| fig|2097.69.peg.73 | Virulent | Antigenic | 0.386 | PROBABLE NON-ALLERGEN |
| fig|2097.71.peg.1 | Virulent | Antigenic | 0.3701 | PROBABLE NON-ALLERGEN |
| fig|2097.71.peg.61 | Virulent | Antigenic | 0.3603 | PROBABLE NON-ALLERGEN |
| fig|2097.69.peg.38 | Virulent | Antigenic | 0.3585 | PROBABLE NON-ALLERGEN |
| fig|2097.69.peg.117 | Virulent | Antigenic | 0.3464 | PROBABLE NON-ALLERGEN |
| fig|2097.69.peg.3 | Virulent | Antigenic | 0.338 | PROBABLE NON-ALLERGEN |
| fig|2097.69.peg.37 | Virulent | Antigenic | 0.3189 | PROBABLE NON-ALLERGEN |
| fig|2097.69.peg.1 | Virulent | Antigenic | 0.2927 | PROBABLE NON-ALLERGEN |
| fig|2097.71.peg.1 | ||||
| S. No. | Start | End | Predicted Peptide Regions | Length |
| 1 | 5 | 45 | FANTNLDWGENKQKQFVENQLGYKETTSTNSHNFHSKSFTQ | 41 |
| 2 | 68 | 121 | GSVGYDSSSSSSSTKDQALAWSTTTSLDSKTGYRDLVTNDTGLNGPINGSFSIQ | 54 |
| 3 | 131 | 159 | SGNHTNSSGSSGPIKTAYPVKKDQKSTVK | 29 |
| 4 | 161 | 177 | NSLINATPLNSYGDEGI | 17 |
| 5 | 188 | 189 | QG | 2 |
| fig|2097.70.peg.33 | ||||
| No. | Start | End | Predicted Peptide Regions | Length |
| 1 | 5 | 12 | QKAKINKA | 8 |
| 2 | 21 | 22 | NK | 2 |
| 3 | 35 | 81 | HKNKVHALYQDPESGNIFSLKKRKQLASNYPLFELTSDNPISFTNNI | 47 |
| T-Cell Epitope for MHC-I | |||||
| Seq ID | Start | End | Length | Predicted Peptide Regions | Score |
| fig|2097.70.peg.33 | 118 | 129 | 12 | YTDEKKVPLINY | 0.953554 |
| fig|2097.71.peg.1 | 140 | 148 | 9 | SSGPIKTAY | 0.654153 |
| T-Cell Epitope for MHC-II | |||||
| Seq ID | Start | End | Length | Predicted Peptide Regions | Score |
| fig|2097.70.peg.33 | 317 | 331 | 15 | PKPVVDLKPQRIEPR | 0.9528 |
| fig|2097.71.peg.1 | 97 | 111 | 15 | KTGYRDLVTNDTGLN | 0.6913 |
| Peptide-Receptor Complex | ΔEvdW | ΔEele | ΔEGB | ΔESURF | ΔGMM-GBSA |
|---|---|---|---|---|---|
| Complex 1 | −98.65 | −52.88 | 58.01 | −6.68 | −100.2 |
| Complex 2 | −95.89 | −46.23 | 67.97 | −15.08 | −89.23 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Taneja, J.; Kant, R.; Saluja, D. Integrating Reverse Vaccinology with Immunoinformatics for Rational Vaccine Target Discovery in Mycoplasma genitalium. Venereology 2025, 4, 14. https://doi.org/10.3390/venereology4030014
Taneja J, Kant R, Saluja D. Integrating Reverse Vaccinology with Immunoinformatics for Rational Vaccine Target Discovery in Mycoplasma genitalium. Venereology. 2025; 4(3):14. https://doi.org/10.3390/venereology4030014
Chicago/Turabian StyleTaneja, Jyoti, Ravi Kant, and Daman Saluja. 2025. "Integrating Reverse Vaccinology with Immunoinformatics for Rational Vaccine Target Discovery in Mycoplasma genitalium" Venereology 4, no. 3: 14. https://doi.org/10.3390/venereology4030014
APA StyleTaneja, J., Kant, R., & Saluja, D. (2025). Integrating Reverse Vaccinology with Immunoinformatics for Rational Vaccine Target Discovery in Mycoplasma genitalium. Venereology, 4(3), 14. https://doi.org/10.3390/venereology4030014





