Abstract
Serine integrases are emerging as one of the most powerful biological tools for biotechnology. Over the past decade, many research papers have been published on the use of serine integrases in synthetic biology. In this review, we aim to systematically summarize the various studies ranging from structure and the catalytic mechanism to genetic design and interdisciplinary applications. First, we introduce the classification, structure, and catalytic model of serine integrases. Second, we present a timeline with milestones that describes the representative achievements. Then, we summarize the applications of serine integrases in genome engineering, genetic design, and DNA assembly. Finally, we discuss the potential of serine integrases for advancing interdisciplinary research. We anticipate that serine integrases will be further expanded as a versatile genetic toolbox for synthetic biology applications.
1. Introduction
As the genetic information carrier, DNA plays the core role in leading mRNA transcription, directing protein translation, and programming cellular behaviors. The variable change of DNA sequences may reprogram life to confer desired characteristics. Over the past two decades, synthetic biology, which focuses on (re)designing and (re)constructing new biological parts, devices, systems, and organisms, has emerged with intense demands for simple, reliable, and efficient DNA manipulating tools [1]. To meet this demand, synthetic biologists have concentrated on the study of site-specific DNA-modifying enzymes that can catalyze DNA variations with precision, prediction, and high efficiency [2].
Recombinases are DNA-modifying enzymes that recognize specific double strand DNA sequences and catalyze DNA–DNA site-specific recombination. Comparing and aligning the recombinase amino acid sequences indicate two subfamilies of recombinases with distinct catalytic mechanisms: tyrosine recombinases and serine recombinases (also called serine integrases). Tyrosine recombinases cleave single-strand DNA and form covalent 3′-phosphotyrosine bonds [2,3] with the DNA backbone and rejoin DNA strands via a Holliday-Junction-like intermediate state, whereas serine integrases cleave double strand DNA and form covalent 5′-phosphoserine bonds [2,4,5] with a DNA backbone and perform as an “assembly cleavage-rotation-ligation-disassembly” process [2]. In comparison to some tyrosine recombinases with similar DNA recognition sites and reversible reactions (e.g., Cre [6], and FLP [3,7]), serine integrases can recognize and catalyze recombination events between two different and specific DNA sites (approximately 50 bp for each) called attP (attachment site in Phage) and attB (attachment site in Bacteria). Depending on the orientations of attP/attB sites, serine integrases can catalyze DNA sequences as deletion, integration, recombination, and inversion [8]. In general, serine integrase-based DNA recombination is a one-way irreversible reaction; however, this reaction can be reversed when a kind of accessory factor protein (Recombination Directionality Factor, RDF) exists [9].
With the fast development of synthetic biology, serine integrases have been used as one of the powerful genetic tools with their unique features of site-specific, orthogonality, irreversibility, high affinity, and high efficiency [8,10]. Serine integrases are widely used in diverse ways, including genome engineering, biological part and genetic circuit design, and DNA assembly. Moreover, serine integrases also advance multidisciplinary research such as chemical engineering, materials science and engineering, and biomedical engineering.
This review summarizes representative serine integrase-associated synthetic biology research achievements over the past decade. We highlight their innovative designs and potential challenges and envision how serine integrases will accelerate synthetic biology and other interdisciplinary fields in the following decades. We also discuss the engineering framework of the “design-build-test-learn” cycle for creating novel approaches with serine integrases to alter living organisms. Finally, we boldly imagine how serine integrases will extend the synthetic biology field and where serine integrases can be utilized beyond life.
2. Mechanism of Site-Specific Recombination Mediated by Serine Integrases
Serine integrases are usually discovered from bacteriophages for catalyzing their DNA integration into the recipient genome via site-specific recombination events between the attP–attB attachment site pairs [11]. When integration is finished, two new sites are formed: attL (attachment site on the left) and attR (attachment site on the right). When the host (e.g., bacteria) is converted to a lysogenic state, the prophage will evade the bacterial chromosome by expressing serine integrases with RDFs, which leads to a periodic reverse recombination event [8].
The general structural model and catalytic domains/motifs of the serine integrase subfamily have been identified clearly (Figure 1a) [12,13]. The NTD (N-Terminal catalytic Domain) contains highly conserved residues including serine (catalytic residue), tyrosine, and arginine [14]. NTD performs its function by cleavage and ligation during the recombination process (Figure 1b) [14]. The flexible αE domain plays an important role in the DNA-protein binding process [15]. RD (Recombinase Domain) mediates the attachment between attP/attB sites and the serine integrase monomer [15]. ZD (Zinc ribbon Domain) [16,17] leads to the conformationally distinct of integrase–attP and integrase–attB complexes; CC (Coiled-Coil motif) [15,18], which is embedded in the two ZDs, can assemble the two complexes of “attP-serine integrases dimer” and “attB-serine integrases dimer” to form a DNA-protein homologous tetramer [14,19]. A recently proposed structural model showed the recombination event containing six steps: (1) DNA-protein dimerization [19], (2) assembly of tetramer complex [19,20,21], (3) double strand DNA cleavage within 2 bp overhangs [8], (4) complex 180° rotation [20,22,23], (5) DNA re-ligation [20], and (6) disassembly of tetramer complex [13] (Figure 1b).
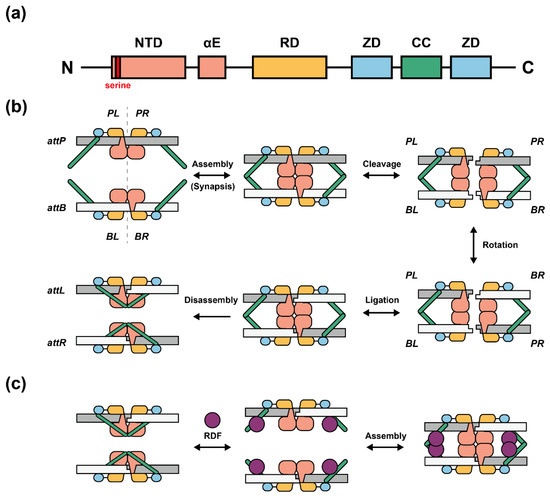
Figure 1.
Domains and proposed recombination model of serine integrases. (a) General structural domains and motifs of serine integrases. NTD: N-Terminal catalytic Domain; αE: flexible linker; RD: Recombinase Domain; ZD: Zinc ribbon Domain; CC: Coiled-Coil motif. ZD is embedded between the two ZDs; CC is divided into two antiparallel helical segments. (b) Proposed model of serine integrase-mediated recombination. First, integrase dimers specifically bind to attP or attB site depending on the ZD binding position. Second, dimer–attP and dimer–attB complexes will be automatically assembled as homologous tetramer by stabilization between CCs interaction. Third, integrase monomers cleave attP and attB sites and form 5′-phosphoserine linkages, DNA half-sites, and 3′ dinucleotide overhangs (2 bp). Then, PL-BL or PR-BR dimeric complexes will rotate 180° along the horizontal axis. After that, PL-BR and BL-PR dimeric complexes will be formed by ligation between DNA strands, called attL and attR. Finally, the CCs will be conformationally changed and form a new internal interaction along with the same DNA double strand rather than two heterologous DNA strands, which leads to disassembly and inhibits reversible exchange. (c) Proposed structural model of serine integrase-mediated reverse recombination. RDFs bind to CCs and alter the integrase–attL and integrase–attR internal interactions. The released CCs may interact with each other located on two heterologous DNA strands and reassemble again.
RDF is a small protein encoded by bacteriophages that can alter the recombination direction [24]. Some serine integrases and their paired RDFs have been characterized such as Bxb1 (RDF: gp47) [25], phiC31 (RDF: gp3) [26,27], phiBT1 (RDF: gp3) [28], A118 (RDF: Gp44) [29], and TP901-1 (RDF: orf7) [30]. When RDF exists, a possible model of reverse recombination indicates that RDF may bind to CCs to prohibit the internal dimeric DNA-protein interaction (Figure 1c) [15].
3. Orientation of att Sites
As the origin of serine integrases, bacteriophages facilitate their invasion via site-specific circular DNA integration and later convert into a lysogenic state to excise (delete) their linearized DNA from the host genome by co-existence of integrase and RDF (Figure 2a) [11]. This process inspires researchers to rearrange the orientation of attP/attB sites to utilize them for other synthetic biology and bioengineering applications. For example, when attP/attB sites are located in two different linearized DNA, the two strands will exchange partial fragments specifically to create two recombined DNA strands (Figure 2b) [31]. This strategy can also be developed as multiple linear DNA assembly in one pot [32]. In addition, when recombination occurs between two circular DNA (e.g., plasmids), these two molecules will be assembled into a merged, large circular DNA [33]. When RDF exists, the merged circular DNA can be disassembled and separated into two independent circular DNA molecules (Figure 2c). Furthermore, when attP/attB sites are oppositely located in the same DNA strand, serine integrases (with or without RDFs) enable the inversion of the internal DNA sequence (Figure 2d) [8]. In summary, depending on the orientation of attP/attB sites, serine integrases can rearrange DNA sequences as integration/deletion, recombination, assembly/disassembly, and inversion.
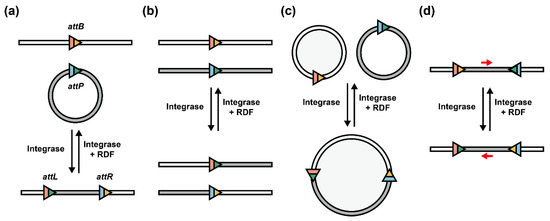
Figure 2.
Serine integrases with different att site orientations. Serine integrases catalyze attP and attB recombination to make attL and attR sites. When integrases and RDFs exist simultaneously, the direction will be reversed. (a) Integration mediated by serine integrases, and deletion when RDFs exist. (b) Recombination between two linear DNA strands. (c) Circular DNA assembly and disassembly. (d) Inversion of internal DNA sequence located in att sites. Red arrow: direction of internal DNA sequence.
4. Recent Achievements of Serine Integrases in Synthetic Biology
In 2000, the landmarks in synthetic biology were the inventions of the genetic toggle switch [34] and repressilator [35]. Despite the traditional applications of serine integrase in genomic engineering, a series of serine integrase-associated synthetic biology research has recently emerged to utilize them for developing new biotechnologies, toolboxes, genetic circuit design, life programming, and other multidisciplinary fields.
In 2012, Bonnet et al. first created a rewriteable recombinase addressable data (RAD) module, which utilized serine integrases and excisionases to invert DNA sequences (e.g., promoter) as reversible inversion [36]. This innovative design opened the next decade of emerging studies of serine integrases in synthetic biology (Figure 3). Next year, two papers reported significant designs, which were inspired by electronics engineering. Siuti et al. [37] and Bonnet et al. [38] brought Boolean logic circuits to serine integrase-based genetic logic circuit design (e.g., AND, OR, and NOT gates). Meanwhile, Bonnet et al. reported a genetic amplifier via the RNA polymerase flow by the programmable serine integrases [38]. After that, a series of serine integrase-based biological parts and genetic circuit designs were established nearly every year, for instance, genetic memory circuits [37,39,40], population-based logic circuits [41,42], state machines [43], synthetic feedback loops [44], comprehensive layered circuit systems [45,46], coding sequence manipulation [47], binary counting module [48], genetic cascades [49], “keys match locks” model [50], and cellular differentiation circuits [41,42,51].
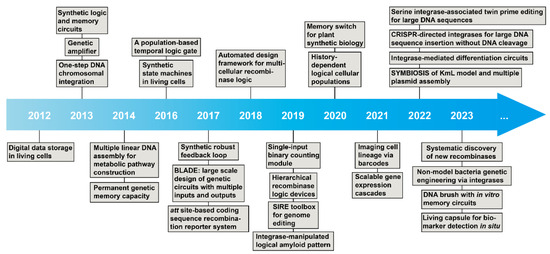
Figure 3.
Timeline of milestones in serine integrase-associated synthetic biology.
Another serine integrase utilization is in vitro linear DNA assembly for defined purposes. In 2014, Colloms et al. first reported a strategy for constructing metabolic pathways via assembling multiple linearized DNA fragments by orthogonal serine integrases [32]. Furthermore, other achievements obtained during the last decade consist of genome engineering (e.g., one-step integration [52], SIRE toolbox [53], CRISPR-Cas plus serine integrase for large DNA genome manipulation [54,55], and non-model bacterial genome engineering [56]), bioinformatics-assisted design (e.g., automatic circuit design [57] and systematic discovery of new recombinases [58]), engineering living materials (e.g., amyloid pattern [59], and DNA brush [60]), plant engineering [61,62], imaging cell lineage [63], and living therapeutics [64,65].
5. Serine Integrase Applications
Depending on the purposes, serine integrases can be utilized as DNA manipulation tools both in vitro and in vivo. First, serine integrases are broadly developed as diverse genome engineering tools in different hosts (e.g., bacteria [66], yeast [67], mammalian cells [68,69,70,71], animals [63,72], and plants [61,62,73,74]), and catalyze various reactions (e.g., integration and deletion) (Figure 4a). Second, serine integrases inspire the creative designs of new biological parts (Figure 4b) and the assembly of comprehensive genetic circuits (Figure 4c). Furthermore, orthogonal serine integrase systems enable in vitro assembly of either linear DNA (Figure 4d) [32] or circular DNA (Figure 4e) [50]. Both can produce large biobricks and assemble several independent modules for the designed applications, for example, the biosynthesis of carotenoid [75], erythromycin [76], and the co-expression of chromoproteins [50].
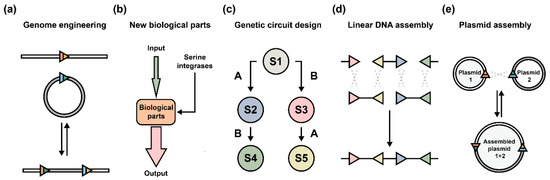
Figure 4.
Application of serine integrases in different ways. (a) The development of serine integrase-based genome engineering strategies includes exogenous DNA integration and genome sequence manipulation (e.g., deletion and inversion). (b) Designs of new biological parts with programmable serine integrases as controllers. (c) Serine integrase-based genetic circuits organize multiple biological parts to achieve more complex functions in living cells. In general, engineered circuits consist of several input signals, multilayered genetic regulators (including logic gates, amplifiers, and memory modules), and diverse output signals. Meanwhile, the host organism can be programmed into predictable states (e.g., S1–S5 means different states: state 1 to state 5). Serine integrases are represented by A and B. (d) Assembly of linear DNA fragments require the matched attP/attB sites localized on fragment ends. Orthogonal serine integrases or orthogonal 2 bp overhangs should be engineered and applied. (e) Assembly of circular DNA (e.g., plasmids) requires orthogonal attP/attB sites located in different DNA molecules. Multiple rounds of assembly can produce predicted and complicated larger plasmids.
5.1. Genome Engineering
Chromosomal DNA carries genetic information for programming organisms’ behaviors. It is important to use precise and efficient methods for modifying and manipulating chromosomes. Currently, several strategies have been used for genome engineering, yet each has its own advantages and disadvantages. First, transposons enable random integration into chromosomes and are suitable for cell library construction [77], but they lack precise and predictable recombination properties. Second, recA-dependent DNA crossover reaction provides a way to recombine DNA strands at any desired locus in the chromosome [78]; however, its efficiency is not good enough. Third, Lambda-Red-mediated homologous recombination, which relies on DNA fragments with short (approximately 50 bp) homology arms, and the Lambda-phage proteins Exo, Bet, and Gam provide an efficient, alternative way to delete or replace native chromosome DNA sequence [79]; however, its ability to recombine more than 2.5 kb DNA fragments is profoundly difficult and also it might be not suitable for most eukaryotic organisms. Finally, the recently developed CRISPR-Cas systems [80,81] and derived DNA editing technologies empowered researchers with more flexible methods, but a versatile protein-RNA/DNA complex might lead to off-target activity, which potentially cause unpredictable harm to the hosts.
Serine integrase-mediated attP–attB site-specific single integration depends on the pre-existed attB site (pseudosite) [82] on the host chromosome (or post-integrated attB site) and attP site on the exogenous DNA sequence. For instance, in 2013, St-Pierre et al. developed a one-step DNA-chromosome integration system: the initial target strain with several engineered orthogonal attB sites could be recombined with integration vectors (consisting of one attP site, one cloning module, and one FLP-excisable integration module) to enable strain carrying expected biobricks (Figure 5a) [52]. Also, with the assistance of CRISPR-Cas technologies, post-integrated attB sites could be generated by twin prime editing [55] or Cas9 nickase-reverse transcriptase fusion protein [54].
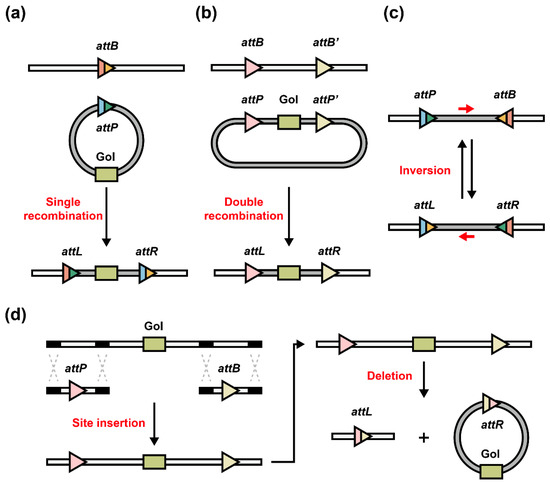
Figure 5.
Serine integrases mediate genome engineering. (a) Single-site integration: pre- or post-existed host chromosomal attB site and exogenous circular DNA (contains attP site and genes of interest (GoI)) can be recombined as genome integration. (b) Double-site recombination and integration: two orthogonal attP/attB pairs (the pairs are from two distinct serine integrases, respectively, or the pairs are derived from one serine integrase with two different 2 bp overhangs with orthogonality) can be both recombined by serine integrases as genome integration. (c) Inversion: DNA sequences can be inverted by serine integrase-mediated attP/attB recombination to produce attL/attR sites, while when RDFs exist, reversible recombination from attL/attR sites occurs and reproduces attP/attB sites. Red arrow: direction of internal DNA sequence. (d) Deletion: the internal DNA sequence localized between two attP/attB sites can be deleted from genome.
Another approach reported by Snoeck et al. presented a methodology called SIRE, which utilized single serine integrase with two orthogonal attP/attB pairs (the 2 bp cleavage overhangs are different) to knock-in target DNA sequences (Figure 5b) and up to 10.3 kb gene cluster could be successfully knocked into the E. coli MG1655 genome. Additionally, a SIRE-based yeast knock-out method has also been successfully developed [53]. To achieve the inversion of genomic DNA, the target sequence should be localized into one pair of attP/attB sites (Figure 5c). Recent studies exhibited the example of inversion (approximately 40 kb sequence) in human cells [55]. Some other studies reported the successful fusion of inactivated Cas9 (dCas9) [83] with integrases to achieve the functions (genomic deletion and integration) with alternative guide RNAs rather than specific att sites, which represented a step toward programmable, scarless, and flexible genome editing strategies [84,85].
5.2. Biological Parts
Biological parts can be defined as specific DNA sequences that encode for unique biological functions. Biological parts can be classified as basic parts and composite parts. A basic part is considered as a minimal element like the promoter, ribosome binding site, coding sequence, terminator, insulator, and attP/attB site. A composite part works as a well-ordered organization with several basic parts [86]. Some representative composite parts are the operator [87], toggle switch [34], repressilator [35], and allocator [88].
Regarding serine integrase-associated biological parts, they are generally organized with irreplaceable attP/attB sites and other alterable basic elements. In 2013, Bonnet et al. proposed a “genetic amplifier” concept organized with serine integrases and orthogonal RNA polymerases (Figure 6a) [38]. A weak input could be amplified within genetic logic gates, producing a stronger output signal by serine integrase-mediated RNA polymerase conversion in the E. coli strain. Moreover, subsequent research by Courbet et al. extended this part’s application for detecting pathological biomarkers in human clinical samples [89]. Then, Zhao et al. reported a serine integrase (with or without RDF)-based binary to decimal converter module that N latches could exponentially count to 2N − 1 (Figure 6b) [48]. This module could be regulated reversibly and efficiently by inducible (pTet and pBAD systems) serine integrases and their RDFs. After multiple repeated operations, the module still worked well with a high recombination efficiency of more than 90%. Then, Bernabé-Orts et al. expanded the toggle switch model into the plant and developed a whole plant memory switch with phiC31 and its RDF (Figure 6c). Such a module was assembled with two coding sequences and one reversible, attP/attB altered terminator-promoter part [62]. This research extended the toolbox for plant synthetic biology [61,90]. Another notable design by Ba et al. proposed a model called “keys match locks” [50]. This biological part design was built up with the ordered composition by attP/attB sites, promoters, and terminators (Figure 6d). The schematic model could be programmed by three orthogonal serine integrases, which acted as “keys”, and the designed parts performed as “locks”. When attP-attB deletion occurs, the inner terminator sequence could be deleted irreversibly. Until all terminators were deleted and promoters were present, the “keys” were matched with “locks” and the downstream output signals could be generated. Like the mentioned “binary to decimal” design, this model was similarly able to utilize limited input signals to generate exponential output signals as N to 2N − 1. Lastly, serine integrase-based biological part regulation was utilized in eukaryotic cells for imaging cell lineage. Chow et al. [63] constructed a system named integrase-editable memory by engineered mutagenesis with optical in situ readout (intMEMOIR), which enabled researchers to reconstruct cell lineage relationships in cultured mouse cell lines and flies (Figure 6e). The serine integrase-mediated barcode design could be changed into up to 59,049 different states, which can be imaged and analyzed with single-cell clonal history, spatial-temporal location, and gene expression.
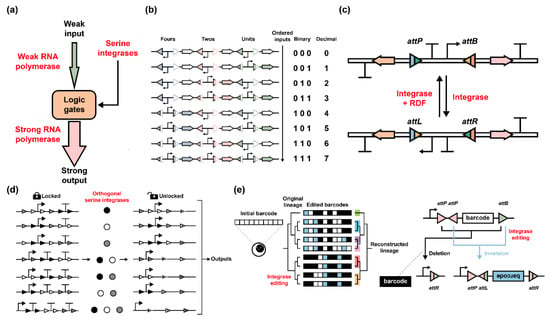
Figure 6.
Engineered biological parts with serine integrases. (a) A weak input signal induces expression of weak RNA polymerase. After serine integrase-programmed regulation, strong RNA polymerase (for example, T7 RNA polymerase) can be expressed and facilitate strong outputs. The “logic gate” part consists of orthogonal attP/attB sites and terminators, which are manipulated by programmed orthogonal serine integrases. (b) A binary-to-decimal converter enables N inputs to be 2N − 1 outputs. The red, green, and blue triangles indicate orthogonal attP/attB sites, and the red, green, and blue arrows indicate RFP, GFP, and CFP, respectively. In this model, Zhao et al. proposed that three orthogonal serine integrases (with or without RDFs) could alter the module into eight different states with the decimal numbers 0 to 7 [48]. (c) Design of genetic memory switch in plants. This module consists of two coding sequences and one reversible part (attP/attB sites, terminator, and promoter). Integrase and its RDF can catalyze reversion to control ON/OFF states. (d) A “keys match locks” model is organized with “locks” modules and “keys” integrases. The matches of orthogonal serine integrases with att sites are supposed to be “keys”, and the ordered composition of attP/attB sites, promoters, and terminators are recognized as distinct “locks”. When supposed terminators are excised and promoters still remain, this situation means “keys” correctly match “locks” and the downstream output signals will be generated. (e) Integrase-editable memory barcodes enable cell lineage reconstruction. In this scheme, the original single cell with “initial barcode” (left) is organized with ten different barcode parts (right). Each part is organized with two inverted attP sites and one attB site. When serine integrases exist, the barcode will be either deleted (black) or inverted (blue), and as a result, one barcode can be converted into three different states and ten barcodes can theoretically generate 310 = 59,049 states. FISH readout methods enable barcode detection and cell lineage reconstruction.
5.3. Genetic Circuit Design
Organisms are nearly all-organized with intelligent genetic codes. Well-regulated genetic circuits enable life to rapidly respond to environmental signals. Synthetic biologists are seeking genetic circuits and trying to engineer them toward minimal leakage, maximal response threshold, simple regulation strategies, and extended circuit classifications [87]. In general, genetic circuits are organized by multiple basic biological parts, and circuit dynamic properties are displayed by the cooperation of multi-layered parts [91]. Most protein-based regulators are proteins that interact with DNA and ligands. For example, lactose-responded lacI repressor can interact with a specific DNA site called lacO [87]. However, these regulatory proteins are unable to modify DNA sequences permanently. Serine integrase-associated genetic circuits have been widely engineered as permanent layers.
In 2013, Siuti et al. proposed synthetic genetic circuits that associated serine integrases with logic gates and memory circuits in living cells [37]. For proof of concept, two orthogonal serine integrases Bxb1 and phiC31 were utilized to build up several Boolean logic gates like AND, OR, and NOT gates (Figure 7a). A simple demonstration was the regulation of a promoter–terminator(s)–output circuit. Serine integrases regulated these circuits via reversion/deletion of basic parts, while the GFP fluorescence signal could be measured as an output signal. This study represents a serine integrase-based genetic circuit design milestone, inspiring others to design many more complex circuits (e.g., multiple layers with diverse inputs and outputs [46]) to control cells’ behaviors. Afterward, Yang et al. expanded the memory circuits by identifying 34 new phage integrases and several engineered memory switches were built up that could record up to 1.375 bytes of information (Figure 7b) [39]. This achievement demonstrated that multiple orthogonal integrases could be programmed, and the genetic information could be permanently recorded in living cells. Then, Hsiao et al. presented a two-input temporal logic gate that could sense and record the order of the inputs, the timing between inputs, and the duration of input pulses (Figure 7c) [42]. The overlaps of two orthogonal integrases’ attP/attB sites enabled E. coli cells to produce distinct output signals referring to altered input times, which provided a time-dependent, integrase-based genetic circuit design. Furthermore, a population-level distribution containing multiple single cells could be utilized to deduce the order, timing, and duration of transient environmental inductions. To meet this demand, Roquet et al. developed the genetic state machines in living cells to regulate complex gene expression, in which three inducible serine integrases could make one strain to be converted into sixteen different states by the variation of orthogonal attP/attB orders (Figure 7d) [43].
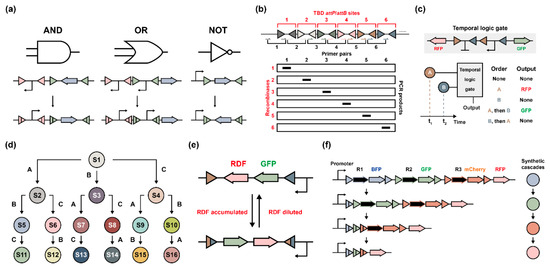
Figure 7.
Serine integrase-based genetic circuit design. (a) Representative Boolean logic gates of AND, OR, and NOT, showing the principle of serine integrase-based genetic circuit design. Two orthogonal attP/attB pairs by serine integrases can alter the orientation of promoters and genes of interest. (b) Design of identification strategy for potential bacteriophage integrase candidates. The possible attP/attB sites are ordered on the same DNA landing pad called memory array. Each primer pair binds to the intermediate between attachment site pairs and can be amplified by PCR if inverted. PCR products indicate the activity and specificity of each serine integrase and its correct attachment site pair. (c) Temporal logic gates can be modified by serine integrases either at the same time or at a distinct time. Only two orthogonal integrases can cause five different temporal events. (d) Three inducible serine integrases (represented as A, B, and C) can result in sixteen different states in one living cell. The induction order determines which state the cell should be in (S1 means state 1, others were in a similar way). (e) An engineered negative feedback loop is constructed with one promoter, one pair attP/attB site, one output signal GFP, and alternative RDFs. The serine integrases are expressed under another inducible system to regulate their relative amount. When the single cell is in the initial state (up), GFP and RDFs are co-expressed and accumulated. When the cell is in the second state (down), RDFs reach a suitable amount and the “serine integrase + RDF” complex will take hold and catalyze a reverse reaction that converts the cell back to its initial state. In general, each cell is independently dynamic alternating; however, the cell population may act as a balanced state. (f) A programmed genetic cascade is ordered with several single layers. Each layer contains one recombinase with its matched attP/attB sites and one reporter gene (fluorescent proteins). Once the cascade is triggered, the first layer will be activated that causes the expression of reporter 1 (BFP) and recombinase 1 (R1). When recombinase 1 is accumulated beyond a threshold, it will delete layer 1 by attP1/attB1 site-specific recombination and leave one gap. Then, the promoter will trigger layer 2 and repeat the same reaction in sequence. After rounds, a genetic cascade will be accomplished and the fluorescence can be measured to determine the cascade process.
Despite the one-way permanent record, more flexible feedback loops should be determined because it is necessary for living organisms to adapt to the environment with reversible responses and intensified signal records. Therefore, both a negative feedback loop and a positive feedback loop (gene expression cascade) were reported by Folliard et al. [44] and Kim et al. [49], respectively. For the negative feedback loop, researchers designed a serine integrase (with RDF)-based negative feedback loop to demonstrate that the capability of reversible genetic circuits could regulate cell population behavior (Figure 7e). In each single cell, the relative ratio between “integrases” and “integrases + RDFs” worked as feedback control factors to alternate this cell’s performance, which was organized as the population with a relative-balanced state [44]. On the contrary, the scalable cascade circuit enabled the sequential expression of multiple genes by one trigger (Figure 7f). The engineered all-in-one circuit could exhibit a fluorescent protein expression cascade, and alternatively trigger a gene-editing cascade [49].
5.4. DNA Assembly
Basic biological parts can be predictably assembled to build up complex genetic circuits for functionalization. In molecular cloning, the general strategy for biological part construction and assembly often uses one of the following methods: restriction enzyme digestion and ligation, TOPO cloning, golden gate assembly, gateway cloning, and gibson assembly [50]. In addition, serine integrases, which are a classic DNA manipulable subfamily, can also be engineered to develop useful molecular cloning methods.
Until now, serine integrase-based molecular cloning strategies have been developed for linear DNA fragment assembly and circular DNA molecule assembly. Colloms et al. reported a rapid procedure that phiC31 integrase could catalyze five linear DNA fragments with overlapped orthogonal attP/attB pairs to successfully assemble and produce an integrated plasmid with an entire metabolic pathway (Figure 4d) [32]. To demonstrate the generalization, the authors chose and characterized three pathways: the carotenoid biosynthetic pathway, the lycopene biosynthetic pathway, and the violacein biosynthetic pathway. In these tests, the number of integrated genes could be easily measured by cell pellet color visualization and restriction enzyme digestion. Moreover, this strategy could also substitute, add, and vary assembled genes. In another study, Wang et al. tested four serine integrases (phiBT1, TG1, phiRv1, and Bxb1) for screening the best one for in vitro DNA assembly, in which Bxb1 displayed the highest efficiency and was utilized to assemble a three-gene lycopene biosynthetic pathway as a proof [92]. Similarly, Gao et al. assembled the erythromycin gene cluster by serine integrases for the production of a secondary metabolite in Streptomyces [76]. Abioye et al. created a high fidelity one-pot DNA assembly strategy for the construction of the β-carotenoid pathway [75]. In summary, serine integrase-based linear DNA fragment assembly provides an alternative strategy for molecular cloning.
All above mentioned approaches typically require the preparation of multiple linear DNA fragments in advance by polymerase chain reaction (PCR), which suggests that there is a lack of a simple strategy for multi-circular DNA molecule assembly. Recently, Ba et al. established a one-pot plasmid assembly approach and demonstrated that orthogonal integrases could facilitate the assembly of engineered “Donor” and “Acceptor” plasmids in vitro to construct composite plasmids (Figure 4e). As an example, the authors assembled distinct chromoprotein genes, generating novel colored E. coli strains [50].
6. Serine Integrases Accelerate the Synthetic Biology Research
In 2000, the innovation of the genetic toggle switch [34] and repressilator [35] represented the beginning of synthetic biology research. After more than 10 years, the first serine integrase-based genetic converter was designed in 2012 [36]. After that, serine integrase accelerated synthetic biology over the next decade with many remarkable milestones (Figure 3). The emerged designs covered multidisciplinary fields. Here, we systematically summarize the published papers and propose a scheme within the “design-build-test-learn” cycle, including three modules as “input-process-output” genetic workflow, and three independent dimensions as host organism, external carrier, and enabling technology (Figure 8).
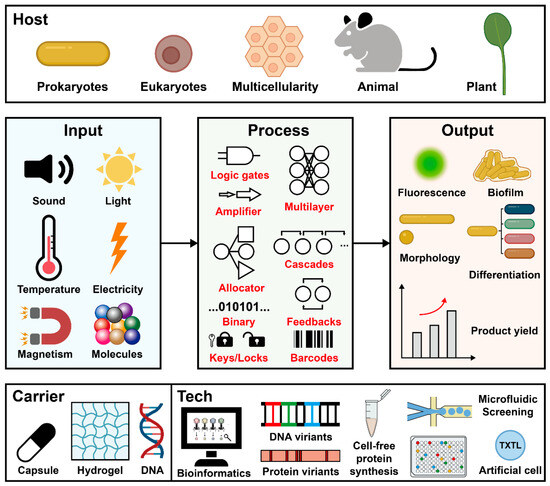
Figure 8.
Schematic overview of serine integrases-based synthetic biology. TXTL: transcription-translation.
Input signals enable cells to sense and respond the external environment for altering their metabolism and behavior (Figure 8, middle left). Up to now, the main inducers for serine integrase expression are chemical molecules [43] such as arabinose, anhydrotetracycline, 2,4-diacetylphloroglucinol (DAPG), and some metal ions like cadmium [93] with strictly regulated inducible circuits to prohibit leakage. In addition, some studies showed other physical factor-induced serine (or tyrosine) integrase-manipulated genetic circuits. For example, two tyrosine recombinases Cre and FLP could be redesigned to respond to light and worked as optogenetic tools [94,95]. Although other physical factors have not been utilized as inducers for regulating integrase, the well-optimized designs that respond to sound [96], temperature [97], electricity [98], and magnetism [99] have been reported and may be potentially utilized.
The signal process occurs when the recipient receives the input signals and converts them into genetic information (Figure 8, middle). Unique properties of serine integrase like site-specific, orthogonality, predictable, and high efficiency, empower it as a genetic information processor. The simple Boolean logic gates could be designed as single-layered circuits (e.g, binary counting module [48], and “keys match locks” module [50]), and also could be assembled as multilayered networks [100] such as amplifier [38], allocator [100], genetic cascades [49], feedback loops [44], and modified genetic barcodes [63].
Depending on genetic information processing, the host can export diverse outputs (Figure 8, middle right). The general output signal is fluorescence, including fluorescent proteins and luciferases. Additionally, engineered microorganisms could produce amyloid fibers [59] and natural products [76,82,101] as output signals. Furthermore, the processed signals enable bacteria to reprogram their phenotypes. An impressive work by Kasari et al. reported the decoupling of E. coli growth and production by removing the chromosomal origin of replication (relying on the phiC31 expression under the control of temperature-sensitive cI857 repressor), which caused the elongation phenotype of E. coli as filaments [102]. E. coli cells could also become spheres by removing the mreB gene [103]. Another phenotype change is cell differentiation. Recently, Zúñiga et al. [41] and Williams et al. [51] engineered two integrase-mediated differentiation circuit series to create multicellular diversities from one E. coli mother cell.
As previously reported, serine integrases were active in a wide range of hosts, including not only prokaryotes (E. coli [66], Pseudomonas [104], Rhodococcus [105], and other non-model bacteria [56]), but also eukaryotes (Saccharomyces cerevisiae [67] and mammalian cell lines [68,69,70,71,106,107]), animals (Drosophila [63] and mouse [72]), and plants (tobacco [62,73] and Arabidopsis [61,74]) (Figure 8, top). However, it is necessary to provide other carriers for microorganisms to resist extreme environments for practical applications such as capsules for engineering living therapeutics [64], hydrogels for bacteria protection [108], and in situ DNA brushes [60] for memory materials design (Figure 8, bottom left).
It is worth noting that the application of serine integrases in synthetic biology could not be achieved without the support of diverse enabling technologies (Figure 8, bottom right). The most important ones are bioinformatics and artificial intelligence, which can provide powerful tools for the new recombinase discovery [39,58,109,110,111,112,113,114], bioinformatic modeling and prediction [115,116,117,118], structure analysis [19,22,119,120], etc. Relying on the supports of computer science, serine integrases and their att sites could be designed, analyzed, and evolved [121] as variants with diverse properties (e.g., affinity [17,106,122], catalytic efficiency [106,123,124,125,126], specificity [12,120,127], orthogonality [124,128], and chimerism [129]). Furthermore, other technologies including cell-free protein synthesis [130], microfluidics [131], and artificial cells [132,133] will support the research of serine integrases (or tyrosine recombinases) in different synthetic biology applications.
7. Conclusions
In this review, we systematically summarized the achievements of serine integrases over the past decade. First, we introduced the structure and catalytic mechanism of serine integrases. Second, we outlined the landmarks within the time frame of the past ten years. Then, we classified the applications of serine integrases such as genome engineering, genetic design, DNA assembly, among others. Finally, a schematic overview of serine integrase-based synthetic biology exhibited the interdisciplinary crosstalk among synthetic biology, materials science and engineering, chemical engineering, biomedical engineering, and bioinformatics.
Looking forward, more research and developments of serine integrases will continue to expand in the next decade due to their close relationship with synthetic biology. Such direction will inspire the researchers to create more intelligent synthetic organisms and living/non-living hybrid systems for more compelling and useful applications.
Author Contributions
Conceptualization, J.L.; writing—original draft preparation, F.B.; writing—review and editing, J.L., F.B., Y.Z., L.W. and W.-Q.L.; supervision, J.L.; funding acquisition, J.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (No. 31971348 and No. 32171427).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Meng, F.; Ellis, T. The second decade of synthetic biology: 2010–2020. Nat. Commun. 2020, 11, 5174. [Google Scholar] [CrossRef]
- Grindley, N.D.F.; Whiteson, K.L.; Rice, P.A. Mechanisms of Site-Specific Recombination. Annu. Rev. Biochem. 2006, 75, 567–605. [Google Scholar] [CrossRef]
- Evans, B.R.; Chen, J.-W.; Parsons, R.L.; Bauer, T.K.; Teplow, D.B.; Jayaram, M. Identification of the Active Site Tyrosine of Flp Recombinase. Possible Relevance of its Location to the Mechanism of Recombination. J. Biol. Chem. 1990, 265, 18504–18510. [Google Scholar] [CrossRef]
- Smith, M.C.A.; Till, R.; Brady, K.; Soultanas, P.; Thorpe, H.; Smith, M.C.M. Synapsis and DNA cleavage in phiC31 integrase-mediated site-specific recombination. Nucleic Acids Res. 2004, 32, 2607–2617. [Google Scholar] [CrossRef]
- Thorpe, H.M.; Smith, M.C.M. In vitro site-specific integration of bacteriophage DNA catalyzed by a recombinase of the resolvase/invertase family. Proc. Natl. Acad. Sci. USA 1998, 95, 5505–5510. [Google Scholar] [CrossRef]
- Abremski, K.; Hoess, R. Bacteriophage P1 Site-specific Recombination. Purification and Properties of the Cre Recombinase Protein. J. Biol. Chem. 1984, 259, 1509–1514. [Google Scholar] [CrossRef]
- Andrews, B.J.; Proteau, G.A.; Beatty, L.G.; Sadowski, P.D. The FLP Recombinase of the 2 micron Circle DNA of Yeast: Interaction with Its Target Sequences. Cell 1985, 40, 795–803. [Google Scholar] [CrossRef]
- Merrick, C.A.; Zhao, J.; Rosser, S.J. Serine Integrases: Advancing Synthetic Biology. ACS Synth. Biol. 2018, 7, 299–310. [Google Scholar] [CrossRef]
- Olorunniji, F.J.; McPherson, A.L.; Rosser, S.J.; Smith, M.C.M.; Colloms, S.D.; Stark, W.M. Control of serine integrase recombination directionality by fusion with the directionality factor. Nucleic Acids Res. 2017, 45, 8635–8645. [Google Scholar] [CrossRef]
- Stark, W.M. Making serine integrases work for us. Curr. Opin. Microbiol. 2017, 38, 130–136. [Google Scholar] [CrossRef]
- Groth, A.C.; Calos, M.P. Phage Integrases: Biology and Applications. J. Mol. Biol. 2004, 335, 667–678. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Sharp, R.; Rutherford, K.; Gupta, K.; Van Duyne, G.D. Serine Integrase attP Binding and Specificity. J. Mol. Biol. 2018, 430, 4401–4418. [Google Scholar] [CrossRef] [PubMed]
- Rutherford, K.; Van Duyne, G.D. The ins and outs of serine integrase site-specific recombination. Curr. Opin. Struct. Biol. 2014, 24, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Yuan, P.; Gupta, K.; Van Duyne, G.D. Tetrameric Structure of a Serine Integrase Catalytic Domain. Structure 2008, 16, 1275–1286. [Google Scholar] [CrossRef]
- Gupta, K.; Sharp, R.; Yuan, J.B.; Li, H.; Van Duyne, G.D. Coiled-coil interactions mediate serine integrase directionality. Nucleic Acids Res. 2017, 45, 7339–7353. [Google Scholar] [CrossRef]
- Prorocic, M.M.; Wenlong, D.; Olorunniji, F.J.; Akopian, A.; Schloetel, J.-G.; Hannigan, A.; McPherson, A.L.; Stark, W.M. Zinc-finger recombinase activities in vitro. Nucleic Acids Res. 2011, 39, 9316–9328. [Google Scholar] [CrossRef]
- McEwan, A.R.; Raab, A.; Kelly, S.M.; Feldmann, J.; Smith, M.C.M. Zinc is essential for high-affinity DNA binding and recombinase activity of phiC31 integrase. Nucleic Acids Res. 2011, 39, 6137–6147. [Google Scholar] [CrossRef]
- McEwan, A.R.; Rowley, P.A.; Smith, M.C.M. DNA binding and synapsis by the large C-terminal domain of phiC31 integrase. Nucleic Acids Res. 2009, 37, 4764–4773. [Google Scholar] [CrossRef]
- Keenholtz, R.A.; Rowland, S.-J.; Boocock, M.R.; Stark, W.M.; Rice, P.A. Structural basis for catalytic activation of a serine recombinase. Structure 2011, 19, 799–809. [Google Scholar] [CrossRef][Green Version]
- Chang, Y.; Johnson, R.C. Controlling tetramer formation, subunit rotation and DNA ligation during Hin-catalyzed DNA inversion. Nucleic Acids Res. 2015, 43, 6459–6472. [Google Scholar] [CrossRef][Green Version]
- Dhar, G.; McLean, M.M.; Heiss, J.K.; Johnson, R.C. The Hin recombinase assembles a tetrameric protein swivel that exchanges DNA strands. Nucleic Acids Res. 2009, 37, 4743–4756. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Trejo, C.S.; Rock, R.S.; Stark, W.M.; Boocock, M.R.; Rice, P.A. Snapshots of a molecular swivel in action. Nucleic Acids Res. 2018, 46, 5286–5296. [Google Scholar] [CrossRef] [PubMed]
- Olorunniji, F.J.; Buck, D.E.; Colloms, S.D.; McEwan, A.R.; Smith, M.C.M.; Stark, W.M.; Rosser, S.J. Gated rotation mechanism of site-specific recombination by phiC31 integrase. Proc. Natl. Acad. Sci. USA 2012, 109, 19661–19666. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, P.; Bibb, L.A.; Hatfull, G.F. Two-step site selection for serine-integrase-mediated excision: DNA-directed integrase conformation and central dinucleotide proofreading. Proc. Natl. Acad. Sci. USA 2008, 105, 3238–3243. [Google Scholar] [CrossRef]
- Ghosh, P.; Wasil, L.R.; Hatfull, G.F. Control of Phage Bxb1 Excision by a Novel Recombination Directionality Factor. PLoS Biol. 2006, 4, e186. [Google Scholar] [CrossRef]
- Fan, H.-F.; Hsieh, T.-S.; Ma, C.-H.; Jayaram, M. Single-molecule analysis of phiC31 integrase-mediated site-specific recombination by tethered particle motion. Nucleic Acids Res. 2016, 44, 10804–10823. [Google Scholar] [CrossRef]
- Khaleel, T.; Younger, E.; McEwan, A.R.; Varghese, A.S.; Smith, M.C.M. A phage protein that binds phiC31 integrase to switch its directionality. Mol. Microbiol. 2011, 80, 1450–1463. [Google Scholar] [CrossRef]
- Zhang, L.; Zhu, B.; Dai, R.; Zhao, G.; Ding, X. Control of Directionality in Streptomyces Phage phiBT1 Integrase-Mediated Site-Specific Recombination. PLoS ONE 2013, 8, e80434. [Google Scholar]
- Mandali, S.; Gupta, K.; Dawson, A.R.; Van Duyne, G.D.; Johnson, R.C. Control of Recombination Directionality by the Listeria Phage A118 Protein Gp44 and the Coiled-Coil Motif of Its Serine Integrase. J. Bacteriol. 2017, 199, e00019-17. [Google Scholar] [CrossRef]
- Breuner, A.; Brondsted, L.; Hammer, K. Novel Organization of Genes Involved in Prophage Excision Identified in the Temperate Lactococcal Bacteriophage TP901-1. J. Bacteriol. 1999, 181, 7291–7297. [Google Scholar] [CrossRef]
- Marken, J.P.; Murray, R.M. Addressable and adaptable intercellular communication via DNA messaging. Nat. Commun. 2023, 14, 2358. [Google Scholar] [CrossRef]
- Colloms, S.D.; Merrick, C.A.; Olorunniji, F.J.; Stark, W.M.; Smith, M.C.M.; Osbourn, A.; Keasling, J.D.; Rosser, S.J. Rapid metabolic pathway assembly and modification using serine integrase site-specific recombination. Nucleic Acids Res. 2014, 42, e23. [Google Scholar] [CrossRef]
- Neil, K.; Allard, N.; Jordan, D.; Rodrigue, S. Assembly of large mobilizable genetic cargo by double recombinase operated insertion of DNA (DROID). Plasmid 2019, 104, 102419. [Google Scholar] [CrossRef]
- Gardner, T.S.; Cantor, C.R.; Collins, J.J. Construction of a genetic toggle switch in Escherichia coli. Nature 2000, 403, 339–342. [Google Scholar] [CrossRef]
- Elowitz, M.B.; Leibler, S. A synthetic oscillatory network of transcriptional regulators. Nature 2000, 403, 335–338. [Google Scholar] [CrossRef]
- Bonnet, J.; Subsoontorn, P.; Endy, D. Rewritable digital data storage in live cells via engineered control of recombination directionality. Proc. Natl. Acad. Sci. USA 2012, 109, 8884–8889. [Google Scholar] [CrossRef]
- Siuti, P.; Yazbek, J.; Lu, T.K. Synthetic circuits integrating logic and memory in living cells. Nat. Biotechnol. 2013, 31, 448–452. [Google Scholar] [CrossRef]
- Bonnet, J.; Yin, P.; Ortiz, M.E.; Subsoontorn, P.; Endy, D. Amplifying Genetic Logic Gates. Science 2013, 340, 599–603. [Google Scholar] [CrossRef]
- Yang, L.; Nielsen, A.A.K.; Fernandez-Rodriguez, J.; McClune, C.J.; Laub, M.T.; Lu, T.K.; Voigt, C.A. Permanent genetic memory with >1-byte capacity. Nat. Methods 2014, 11, 1261–1266. [Google Scholar] [CrossRef]
- Siuti, P.; Yazbek, J.; Lu, T.K. Engineering genetic circuits that compute and remember. Nat. Protoc. 2014, 9, 1292–1300. [Google Scholar] [CrossRef]
- Zúñiga, A.; Guiziou, S.; Mayonove, P.; Meriem, Z.B.; Camacho, M.; Moreau, V.; Ciandrini, L.; Hersen, P.; Bonnet, J. Rational programming of history-dependent logic in cellular populations. Nat. Commun. 2020, 11, 4758. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, V.; Hori, Y.; Rothemund, P.W.; Murray, R.M. A population-based temporal logic gate for timing and recording chemical events. Mol. Syst. Biol. 2016, 12, 869. [Google Scholar] [CrossRef] [PubMed]
- Roquet, N.; Soleimany, A.P.; Ferris, A.C.; Aaronson, S.; Lu, T.K. Synthetic recombinase-based state machines in living cells. Science 2016, 353, aad8559. [Google Scholar] [CrossRef] [PubMed]
- Folliard, T.; Steel, H.; Prescott, T.P.; Wadhams, G.; Rothschild, L.J.; Papachristodoulou, A. A Synthetic Recombinase-Based Feedback Loop Results in Robust Expression. ACS Synth. Biol. 2017, 6, 1663–1671. [Google Scholar] [CrossRef]
- Guiziou, S.; Mayonove, P.; Bonnet, J. Hierarchical composition of reliable recombinase logic devices. Nat. Commun. 2019, 10, 456. [Google Scholar] [CrossRef]
- Weinberg, B.H.; Pham, N.T.H.; Caraballo, L.D.; Lozanoski, T.; Engel, A.; Bhatia, S.; Wong, W.W. Large-scale design of robust genetic circuits with multiple inputs and outputs for mammalian cells. Nat. Biotechnol. 2017, 35, 453–462. [Google Scholar] [CrossRef]
- Bland, M.J.; Ducos-Galand, M.; Val, M.-E.; Mazel, D. An att site-based recombination reporter system for genome engineering and synthetic DNA assembly. BMC Biotechnol. 2017, 17, 62. [Google Scholar] [CrossRef]
- Zhao, J.; Pokhilko, A.; Ebenhöh, O.; Rosser, S.J.; Colloms, S.D. A single-input binary counting module based on serine integrase site-specific recombination. Nucleic Acids Res. 2019, 47, 4896–4909. [Google Scholar] [CrossRef]
- Kim, T.; Weinberg, B.; Wong, W.; Lu, T.K. Scalable recombinase-based gene expression cascades. Nat. Commun. 2021, 12, 2711. [Google Scholar] [CrossRef]
- Ba, F.; Liu, Y.; Liu, W.-Q.; Tian, X.; Li, J. SYMBIOSIS: Synthetic manipulable biobricks via orthogonal serine integrase systems. Nucleic Acids Res. 2022, 50, 2973–2985. [Google Scholar] [CrossRef]
- Williams, R.L.; Murray, R.M. Integrase-mediated differentiation circuits improve evolutionary stability of burdensome and toxic functions in E. coli. Nat. Commun. 2022, 13, 6822. [Google Scholar] [CrossRef]
- St-Pierre, F.; Cui, L.; Priest, D.G.; Endy, D.; Dodd, I.B.; Shearwin, K.E. One-Step Cloning and Chromosomal Integration of DNA. ACS Synth. Biol. 2013, 2, 537–541. [Google Scholar] [CrossRef] [PubMed]
- Snoeck, N.; De Mol, M.L.; Van Herpe, D.; Goormans, A.; Maryns, I.; Coussement, P.; Peters, G.; Beauprez, J.; De Maeseneire, S.L.; Soetaert, W. Serine integrase recombinational engineering (SIRE): A versatile toolbox for genome editing. Biotechnol. Bioeng. 2019, 116, 364–374. [Google Scholar] [CrossRef] [PubMed]
- Yarnall, M.T.N.; Ioannidi, E.I.; Schmitt-Ulms, C.; Krajeski, R.N.; Lim, J.; Villiger, L.; Zhou, W.; Jiang, K.; Garushyants, S.K.; Roberts, N.; et al. Drag-and-drop genome insertion of large sequences without double-strand DNA cleavage using CRISPR-directed integrases. Nat. Biotechnol. 2023, 41, 500–512. [Google Scholar] [CrossRef] [PubMed]
- Anzalone, A.V.; Gao, X.D.; Podracky, C.J.; Nelson, A.T.; Koblan, L.W.; Raguram, A.; Levy, J.M.; Mercer, J.A.M.; Liu, D.R. Programmable deletion, replacement, integration and inversion of large DNA sequences with twin prime editing. Nat. Biotechnol. 2022, 40, 731–740. [Google Scholar] [CrossRef]
- Elmore, J.R.; Dexter, G.N.; Baldino, H.; Huenemann, J.D.; Francis, R.; Peabody, G.L.; Martinez-Baird, J.; Riley, L.A.; Simmons, T.; Coleman-Derr, D.; et al. High-throughput genetic engineering of nonmodel and undomesticated bacteria via iterative site-specific genome integration. Sci. Adv. 2023, 9, eade1285. [Google Scholar] [CrossRef]
- Guiziou, S.; Ulliana, F.; Moreau, V.; Leclere, M.; Bonnet, J. An Automated Design Framework for Multicellular Recombinase Logic. ACS Synth. Biol. 2018, 7, 1406–1412. [Google Scholar] [CrossRef]
- Durrant, M.G.; Fanton, A.; Tycko, J.; Hinks, M.; Chandrasekaran, S.S.; Perry, N.T.; Schaepe, J.; Du, P.P.; Lotfy, P.; Bassik, M.C.; et al. Systematic discovery of recombinases for efficient integration of large DNA sequences into the human genome. Nat. Biotechnol. 2023, 41, 488–499. [Google Scholar] [CrossRef]
- Kalyoncu, E.; Ahan, R.E.; Ozcelik, C.E.; Seker, U.O.S. Genetic Logic Gates Enable Patterning of Amyloid Nanofibers. Adv. Mater. 2019, 31, 1902888. [Google Scholar] [CrossRef]
- Avidan, N.; Levy, M.; Daube, S.S.; Bar-Ziv, R.H. Toward Memory in a DNA Brush: Site-Specific Recombination Responsive to Polymer Density, Orientation, and Conformation. J. Am. Chem. Soc. 2023, 145, 9729–9736. [Google Scholar] [CrossRef]
- Guiziou, S.; Maranas, C.J.; Chu, J.C.; Nemhauser, J.L. An integrase toolbox to record gene-expression during plant development. Nat. Commun. 2023, 14, 1844. [Google Scholar] [CrossRef]
- Bernabé-Orts, J.M.; Quijano-Rubio, A.; Vazquez-Vilar, M.; Mancheño-Bonillo, J.; Moles-Casas, V.; Selma, S.; Gianoglio, S.; Granell, A.; Orzaez, D. A memory switch for plant synthetic biology based on the phage phiC31 integration system. Nucleic Acids Res. 2020, 48, 3379–3394. [Google Scholar] [CrossRef] [PubMed]
- Chow, K.-H.K.; Budde, M.W.; Granados, A.A.; Cabrera, M.; Yoon, S.; Cho, S.; Huang, T.-H.; Koulena, N.; Frieda, K.L.; Cai, L.; et al. Imaging cell lineage with a synthetic digital recording system. Science 2021, 372, eabb3099. [Google Scholar] [CrossRef] [PubMed]
- Inda-Webb, M.E.; Jimenez, M.; Liu, Q.; Phan, N.V.; Ahn, J.; Steiger, C.; Wentworth, A.; Riaz, A.; Zirtiloglu, T.; Wong, K.; et al. Sub-1.4 cm3 capsule for detecting labile inflammatory biomarkers in situ. Nature 2023, 620, 386–392. [Google Scholar] [CrossRef] [PubMed]
- Archer, E.J.; Robinson, A.B.; Süel, G.M. Engineered E. coli that Detect and Respond to Gut Inflammation through Nitric Oxide Sensing. ACS Synth. Biol. 2012, 1, 451–457. [Google Scholar] [CrossRef] [PubMed]
- Riley, L.A.; Payne, I.C.; Tumen-Velasquez, M.; Guss, A.M. Simple and Rapid Site-Specific Integration of Multiple Heterologous DNAs into the Escherichia coli Chromosome. J. Bacteriol. 2023, 205, e0033822. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Brown, W.R.A. Comparison and optimization of ten phage encoded serine integrases for genome engineering in Saccharomyces cerevisiae. BMC Biotechnol. 2016, 16, 13. [Google Scholar] [CrossRef]
- Gomide, M.S.; Sales, T.T.; Barros, L.R.C.; Limia, C.G.; de Oliveira, M.A.; Florentino, L.H.; Barros, L.M.G.; Robledo, M.L.; José, G.P.C.; Almeida, M.S.M.; et al. Genetic switches designed for eukaryotic cells and controlled by serine integrases. Commun. Biol. 2020, 3, 255. [Google Scholar] [CrossRef]
- Pristovšek, N.; Nallapareddy, S.; Grav, L.M.; Hefzi, H.; Lewis, N.E.; Rugbjerg, P.; Hansen, H.G.; Lee, G.M.; Andersen, M.R.; Kildegaard, H.F. Systematic Evaluation of Site-Specific Recombinant Gene Expression for Programmable Mammalian Cell Engineering. ACS Synth. Biol. 2019, 8, 758–774. [Google Scholar] [CrossRef]
- Gaidukov, L.; Wroblewska, L.; Teague, B.; Nelson, T.; Zhang, X.; Liu, Y.; Jagtap, K.; Mamo, S.; Tseng, W.A.; Lowe, A.; et al. A multi-landing pad DNA integration platform for mammalian cell engineering. Nucleic Acids Res. 2018, 46, 4072–4086. [Google Scholar] [CrossRef]
- Stoll, S.M.; Ginsburg, D.S.; Calos, M.P. Phage TP901-1 Site-Specific Integrase Functions in Human Cells. J. Bacteriol. 2002, 184, 3657–3663. [Google Scholar] [CrossRef] [PubMed]
- Low, B.E.; Hosur, V.; Lesbirel, S.; Wiles, M.V. Efficient targeted transgenesis of large donor DNA into multiple mouse genetic backgrounds using bacteriophage Bxb1 integrase. Sci. Rep. 2022, 12, 5424. [Google Scholar] [CrossRef] [PubMed]
- Shao, M.; Kumar, S.; Thomson, J.G. Precise excision of plastid DNA by the large serine recombinase Bxb1. Plant Biotechnol. J. 2014, 12, 322–329. [Google Scholar] [CrossRef] [PubMed]
- Thomson, J.G.; Chan, R.; Smith, J.; Thilmony, R.; Yau, Y.-Y.; Wang, Y.; Ow, D.W. The Bxb1 recombination system demonstrates heritable transmission of site-specific excision in Arabidopsis. BMC Biotechnol. 2012, 12, 9. [Google Scholar] [CrossRef]
- Abioye, J.; Lawson-Williams, M.; Lecanda, A.; Calhoon, B.; McQue, A.L.; Colloms, S.D.; Stark, W.M.; Olorunniji, F.J. High fidelity one-pot DNA assembly using orthogonal serine integrases. Biotechnol. J. 2023, 18, 2200411. [Google Scholar] [CrossRef]
- Gao, H.; Taylor, G.; Evans, S.K.; Fogg, P.C.M.; Smith, M.C.M. Application of serine integrases for secondary metabolite pathway assembly in Streptomyces. Synth. Syst. Biotechnol. 2020, 5, 111–119. [Google Scholar] [CrossRef]
- English, M.A.; Alcantar, M.A.; Collins, J.J. A self-propagating, barcoded transposon system for the dynamic rewiring of genomic networks. Mol. Syst. Biol. 2023, 19, e11398. [Google Scholar] [CrossRef]
- Wright, W.D.; Shah, S.S.; Heyer, W.-D. Homologous recombination and the repair of DNA double-strand breaks. J. Biol. Chem. 2018, 293, 10524–10535. [Google Scholar] [CrossRef]
- Datsenko, K.A.; Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex Genome Engineering Using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Effendi, S.S.W.; Ng, I.-S. Reprogramming T7RNA Polymerase in Escherichia coli Nissle 1917 under Specific Lac Operon for Efficient p-Coumaric Acid Production. ACS Synth. Biol. 2022, 11, 3471–3481. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.S.; Larson, M.H.; Gilbert, L.A.; Doudna, J.A.; Weissman, J.S.; Arkin, A.P.; Lim, W.A. Repurposing CRISPR as an RNA-Guided Platform for Sequence-Specific Control of Gene Expression. Cell 2013, 152, 1173–1183. [Google Scholar] [CrossRef] [PubMed]
- Standage-Beier, K.; Brookhouser, N.; Balachandran, P.; Zhang, Q.; Brafman, D.A.; Wang, X. RNA-Guided Recombinase-Cas9 Fusion Targets Genomic DNA Deletion and Integration. CRISPR J. 2019, 2, 209–222. [Google Scholar] [CrossRef]
- Chaikind, B.; Bessen, J.L.; Thompson, D.B.; Hu, J.H.; Liu, D.R. A programmable Cas9-serine recombinase fusion protein that operates on DNA sequences in mammalian cells. Nucleic Acids Res. 2016, 44, 9758–9770. [Google Scholar] [CrossRef]
- Anderson, J.C.; Dueber, J.E.; Leguia, M.; Wu, G.C.; Goler, J.A.; Arkin, A.P.; Keasling, J.D. BglBricks: A flexible standard for biological part assembly. J. Biol. Eng. 2010, 4, 1. [Google Scholar] [CrossRef]
- English, M.A.; Gayet, R.V.; Collins, J.J. Designing Biological Circuits: Synthetic Biology Within the Operon Model and Beyond. Annu. Rev. Biochem. 2021, 90, 221–244. [Google Scholar] [CrossRef]
- Segall-Shapiro, T.H.; Meyer, A.J.; Ellington, A.D.; Sontag, E.D.; Voigt, C.A. A ‘resource allocator’ for transcription based on a highly fragmented T7 RNA polymerase. Mol. Syst. Biol. 2014, 10, 742. [Google Scholar] [CrossRef]
- Courbet, A.; Endy, D.; Renard, E.; Molina, F.; Bonnet, J. Detection of pathological biomarkers in human clinical samples via amplifying genetic switches and logic gates. Sci. Transl. Med. 2015, 7, 289ra83. [Google Scholar] [CrossRef]
- Lloyd, J.P.B.; Ly, F.; Gong, P.; Pflueger, J.; Swain, T.; Pflueger, C.; Fourie, E.; Khan, M.A.; Kidd, B.N.; Lister, R. Synthetic memory circuits for stable cell reprogramming in plants. Nat. Biotechnol. 2022, 40, 1862–1872. [Google Scholar] [CrossRef]
- Brophy, J.A.N.; Voigt, C.A. Principles of genetic circuit design. Nat. Methods 2014, 11, 508–520. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Tang, B.; Ye, Y.; Mao, Y.; Lei, X.; Zhao, G.; Ding, X. Bxb1 integrase serves as a highly efficient DNA recombinase in rapid metabolite pathway assembly. Acta Biochim. Biophys. Sin. 2017, 49, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Akboğa, D.; Saltepe, B.; Bozkurt, E.U.; Şeker, U.Ö.Ş. A Recombinase-Based Genetic Circuit for Heavy Metal Monitoring. Biosensors 2022, 12, 122. [Google Scholar] [CrossRef]
- Sheets, M.B.; Tague, N.; Dunlop, M.J. An optogenetic toolkit for light-inducible antibiotic resistance. Nat. Commun. 2023, 14, 1034. [Google Scholar] [CrossRef]
- Sheets, M.B.; Wong, W.W.; Dunlop, M.J. Light-Inducible Recombinases for Bacterial Optogenetics. ACS Synth. Biol. 2020, 9, 227–235. [Google Scholar] [CrossRef]
- Yang, Y.; Yang, Y.; Liu, D.; Wang, Y.; Lu, M.; Zhang, Q.; Huang, J.; Li, Y.; Ma, T.; Yan, F.; et al. In-vivo programmable acoustic manipulation of genetically engineered bacteria. Nat. Commun. 2023, 14, 3297. [Google Scholar] [CrossRef]
- Piraner, D.I.; Abedi, M.H.; Moser, B.A.; Lee-Gosselin, A.; Shapiro, M.G. Tunable thermal bioswitches for in vivo control of microbial therapeutics. Nat. Chem. Biol. 2017, 13, 75–80. [Google Scholar] [CrossRef]
- Tschirhart, T.; Kim, E.; McKay, R.; Ueda, H.; Wu, H.-C.; Pottash, A.E.; Zargar, A.; Negrete, A.; Shiloach, J.; Payne, G.F.; et al. Electronic control of gene expression and cell behaviour in Escherichia coli through redox signalling. Nat. Commun. 2017, 8, 14030. [Google Scholar] [CrossRef]
- Carlsen, R.W.; Edwards, M.R.; Zhuang, J.; Pacoret, C.; Sitti, M. Magnetic steering control of multi-cellular bio-hybrid microswimmers. Lab Chip 2014, 14, 3850–3859. [Google Scholar] [CrossRef] [PubMed]
- Müller, M.; Ausländer, S.; Spinnler, A.; Ausländer, D.; Sikorski, J.; Folcher, M.; Fussenegger, M. Designed cell consortia as fragrance-programmable analog-to-digital converters. Nat. Chem. Biol. 2017, 13, 309–316. [Google Scholar] [CrossRef]
- Li, L.; Zheng, G.; Chen, J.; Ge, M.; Jiang, W.; Lu, Y. Multiplexed site-specific genome engineering for overproducing bioactive secondary metabolites in actinomycetes. Metab. Eng. 2017, 40, 80–92. [Google Scholar] [CrossRef] [PubMed]
- Kasari, M.; Kasari, V.; Kärmas, M.; Jõers, A. Decoupling Growth and Production by Removing the Origin of Replication from a Bacterial Chromosome. ACS Synth. Biol. 2022, 11, 2610–2622. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.-N.; Ma, B.-G. OptoCRISPRi-HD: Engineering a Bacterial Green-Light-Activated CRISPRi System with a High Dynamic Range. ACS Synth. Biol. 2023, 12, 1708–1715. [Google Scholar] [CrossRef] [PubMed]
- Khan, N.; Yeung, E.; Farris, Y.; Fansler, S.J.; Bernstein, H.C. A broad-host-range event detector: Expanding and quantifying performance between Escherichia coli and Pseudomonas species. Synth. Biol. 2020, 5, ysaa002. [Google Scholar] [CrossRef]
- Round, J.W.; Robeck, L.D.; Eltis, L.D. An Integrative Toolbox for Synthetic Biology in Rhodococcus. ACS Synth. Biol. 2021, 10, 2383–2395. [Google Scholar] [CrossRef]
- Chao, G.; Travis, C.; Church, G. Measurement of large serine integrase enzymatic characteristics in HEK293 cells reveals variability and influence on downstream reporter expression. FEBS J. 2021, 288, 6410–6427. [Google Scholar] [CrossRef]
- Xu, Z.; Thomas, L.; Davies, B.; Chalmers, R.; Smith, M.; Brown, W. Accuracy and efficiency define Bxb1 integrase as the best of fifteen candidate serine recombinases for the integration of DNA into the human genome. BMC Biotechnol. 2013, 13, 87. [Google Scholar] [CrossRef]
- Peng, R.; Ba, F.; Li, J.; Cao, J.; Zhang, R.; Liu, W.-Q.; Ren, J.; Liu, Y.; Li, J.; Ling, S. Embedding Living Cells with a Mechanically Reinforced and Functionally Programmable Hydrogel Fiber Platform. Adv. Mater. 2023. [Google Scholar] [CrossRef]
- Zhang, G.; Yao, S.; Liu, Y.; Fang, H.; Song, Y.; Wang, C.; Wei, D.; Feng, J. Systematic Discovery of a New Catalogue of Tyrosine-Type Integrases in Bacterial Genomic Islands. Appl. Environ. Microbiol. 2023, 89, e0173822. [Google Scholar] [CrossRef]
- Jelicic, M.; Schmitt, L.T.; Paszkowski-Rogacz, M.; Walder, A.; Schubert, N.; Hoersten, J.; Sürün, D.; Buchholz, F. Discovery and characterization of novel Cre-type tyrosine site-specific recombinases for advanced genome engineering. Nucleic Acids Res. 2023, 51, 5285–5297. [Google Scholar] [CrossRef]
- Askora, A.; Kawasaki, T.; Fujie, M.; Yamada, T. In vitro characterization of the site-specific recombination system based on genus Habenivirus phiRSM small serine integrase. Mol. Genet. Genom. 2021, 296, 551–559. [Google Scholar] [CrossRef] [PubMed]
- Yoon, B.; Kim, I.; Nam, J.-A.; Chang, H.-I.; Ha, C.H. In vivo and in vitro characterization of site-specific recombination of a novel serine integrase from the temperate phage EFC-1. Biochem. Biophys. Res. Commun. 2016, 473, 336–341. [Google Scholar] [CrossRef] [PubMed]
- Miura, T.; Hosaka, Y.; Yang, Y.-Z.; Nishizawa, T.; Asayama, M.; Takahashi, H.; Shirai, M. In vivo and in vitro characterization of site-specific recombination of actinophage R4 integrase. J. Gen. Appl. Microbiol. 2011, 57, 45–57. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Ou, X.; Zhao, G.; Ding, X. Highly Efficient In Vitro Site-Specific Recombination System Based on Streptomyces Phage phiBT1 Integrase. J. Bacteriol. 2008, 190, 6392–6397. [Google Scholar] [CrossRef]
- Landau, J.; Cuba Samaniego, C.; Giordano, G.; Franco, E. Computational characterization of recombinase circuits for periodic behaviors. iScience 2023, 26, 105624. [Google Scholar] [CrossRef]
- Poole, W.; Pandey, A.; Shur, A.; Tuza, Z.A.; Murray, R.M. BioCRNpyler: Compiling chemical reaction networks from biomolecular parts in diverse contexts. PLoS Comput. Biol. 2022, 18, e1009987. [Google Scholar] [CrossRef]
- Bowyer, J.E.; Ding, C.; Weinberg, B.H.; Wong, W.W.; Bates, D.G. A mechanistic model of the BLADE platform predicts performance characteristics of 256 different synthetic DNA recombination circuits. PLoS Comput. Biol. 2020, 16, e1007849. [Google Scholar] [CrossRef]
- Pokhilko, A.; Zhao, J.; Ebenhöh, O.; Smith, M.C.M.; Stark, W.M.; Colloms, S.D. The mechanism of phiC31 integrase directionality: Experimental analysis and computational modelling. Nucleic Acids Res. 2016, 44, 7360–7372. [Google Scholar]
- Abe, K.; Takahashi, T.; Sato, T. Extreme C-terminal element of SprA serine integrase is a potential component of the “molecular toggle switch” which controls the recombination and its directionality. Mol. Microbiol. 2021, 115, 1110–1121. [Google Scholar] [CrossRef]
- Gaj, T.; Mercer, A.C.; Gersbach, C.A.; Gordley, R.M.; Barbas 3rd, C.F. Structure-guided reprogramming of serine recombinase DNA sequence specificity. Proc. Natl. Acad. Sci. USA 2011, 108, 498–503. [Google Scholar] [CrossRef]
- Han, P.; Ma, Y.; Fu, Z.; Guo, Z.; Xie, J.; Wu, Y.; Yuan, Y.-J. A DNA Inversion System in Eukaryotes Established via Laboratory Evolution. ACS Synth. Biol. 2021, 10, 2222–2230. [Google Scholar] [CrossRef] [PubMed]
- Sumikawa, T.; Ohno, S.; Watanabe, T.; Yamamoto, R.; Yamano, M.; Mori, T.; Mori, K.; Tobimatsu, T.; Sera, T. Site-Specific Integration by Recruitment of a Complex of phiC31 Integrase and Donor DNA to a Target Site by Using a Tandem, Artificial Zinc-Finger Protein. Biochemistry 2018, 57, 6868–6877. [Google Scholar] [CrossRef]
- Zhang, Q.; Azarin, S.M.; Sarkar, C.A. Model-guided engineering of DNA sequences with predictable site-specific recombination rates. Nat. Commun. 2022, 13, 4152. [Google Scholar] [CrossRef] [PubMed]
- Jusiak, B.; Jagtap, K.; Gaidukov, L.; Duportet, X.; Bandara, K.; Chu, J.; Zhang, L.; Weiss, R.; Lu, T.K. Comparison of Integrases Identifies Bxb1-GA Mutant as the Most Efficient Site-Specific Integrase System in Mammalian Cells. ACS Synth. Biol. 2019, 8, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Keenholtz, R.A.; Grindley, N.D.F.; Hatfull, G.F.; Marko, J.F. Crossover-site sequence and DNA torsional stress control strand interchanges by the Bxb1 site-specific serine recombinase. Nucleic Acids Res. 2016, 44, 8921–8932. [Google Scholar] [CrossRef][Green Version]
- Gupta, M.; Till, R.; Smith, M.C.M. Sequences in attB that affect the ability of phiC31 integrase to synapse and to activate DNA cleavage. Nucleic Acids Res. 2007, 35, 3407–3419. [Google Scholar] [CrossRef]
- Hoersten, J.; Ruiz-Gómez, G.; Lansing, F.; Rojo-Romanos, T.; Schmitt, L.T.; Sonntag, J.; Pisabarro, M.T.; Buchholz, F. Pairing of single mutations yields obligate Cre-type site-specific recombinases. Nucleic Acids Res. 2022, 50, 1174–1186. [Google Scholar] [CrossRef]
- Singh, S.; Rockenbach, K.; Dedrick, R.M.; VanDemark, A.P.; Hatfull, G.F. Cross-talk between Diverse Serine Integrases. J. Mol. Biol. 2014, 426, 318–331. [Google Scholar] [CrossRef][Green Version]
- Farruggio, A.P.; Calos, M.P. Serine integrase chimeras with activity in E. coli and HeLa cells. Biol. Open 2014, 3, 895–903. [Google Scholar] [CrossRef][Green Version]
- Pandey, A.; Rodriguez, M.L.; Poole, W.; Murray, R.M. Characterization of Integrase and Excisionase Activity in a Cell-Free Protein Expression System Using a Modeling and Analysis Pipeline. ACS Synth. Biol. 2023, 12, 511–523. [Google Scholar] [CrossRef]
- Li, L.; Zhang, R.; Chen, L.; Tian, X.; Li, T.; Pu, B.; Ma, C.; Ji, X.; Ba, F.; Xiong, C.; et al. Permeability-Engineered Compartmentalization Enables In Vitro Reconstitution of Sustained Synthetic Biology Systems. Adv. Sci. 2022, 9, 2203652. [Google Scholar] [CrossRef] [PubMed]
- Okauchi, H.; Ichihashi, N. Continuous Cell-Free Replication and Evolution of Artificial Genomic DNA in a Compartmentalized Gene Expression System. ACS Synth. Biol. 2021, 10, 3507–3517. [Google Scholar] [CrossRef] [PubMed]
- Sakatani, Y.; Yomo, T.; Ichihashi, N. Self-replication of circular DNA by a self-encoded DNA polymerase through rolling-circle replication and recombination. Sci. Rep. 2018, 8, 13089. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).