GPCRs of Diverse Physiologic and Pathologic Effects with Fingerprints in COVID-19 †
Abstract
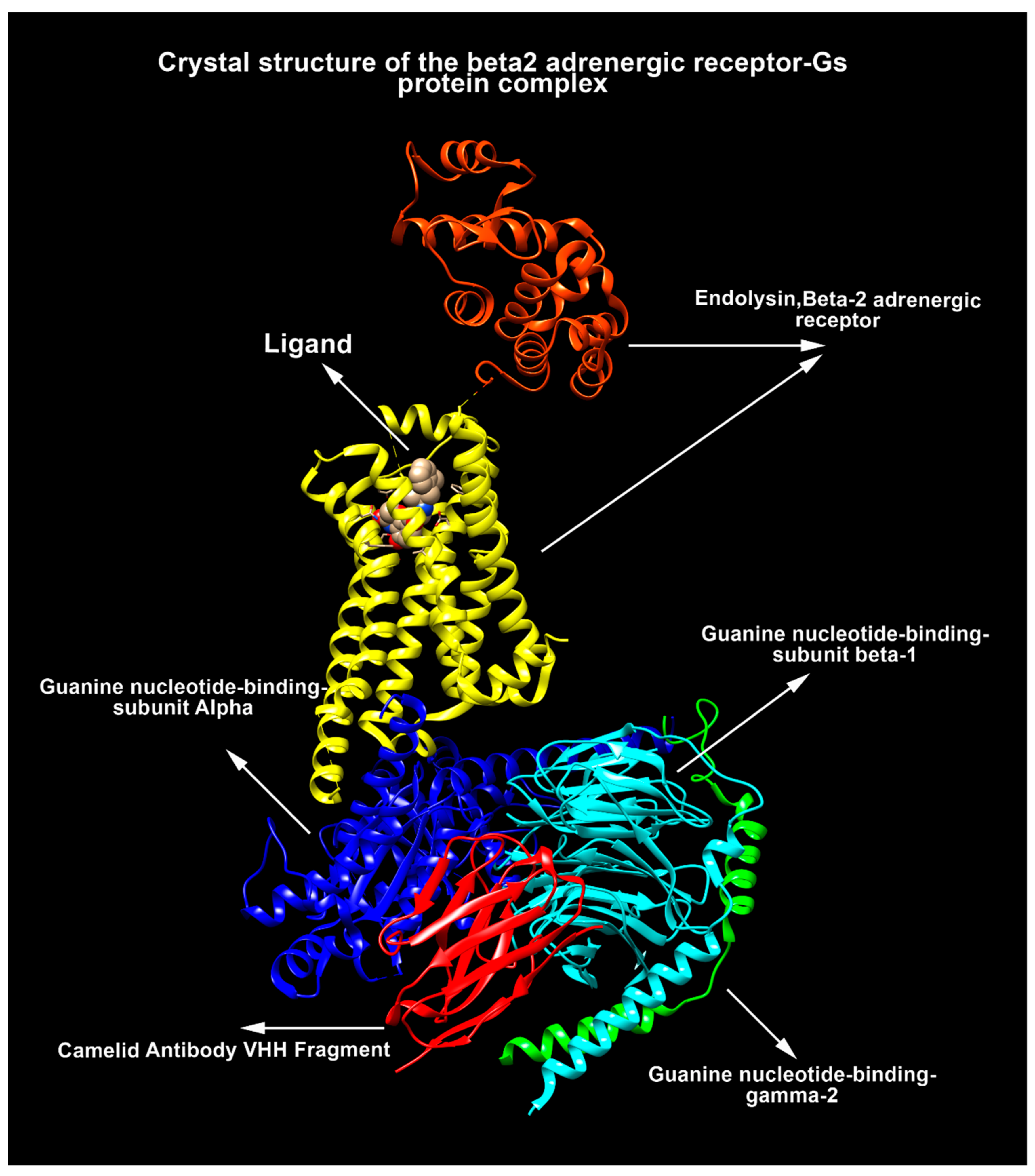
:1. Introduction
2. COVID-19 and GPCRs
3. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ACE | angiotensin converting enzyme |
| ACE2 | angiotensin converting enzyme2 |
| Ang II | angiotensin II |
| AT1R | type I angiotensin II receptor |
| AT2R | type II angiotensin II receptor |
| CFTR | cystic fibrosis transmembrane conductance regulator |
| ENaC | epithelial Na Channel |
| GPCR | G-protein-coupled receptor |
| NOS | nitric oxide synthase |
| RAS | renin angiotensin system |
References
- Cooper, G.M. Cell Signaling. In The Cell—A Molecular Approach; Oxford University Press.: New York, NY, USA, 2019; p. 565. [Google Scholar]
- Quitterer, U.; AbdAlla, S. Discovery of pathologic GPCR aggregation. Front. Med. 2019, 6, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zalewska, M.; Siara, M.; Sajewicz, W. G protein-coupled receptors: Abnormalities in signal transmission, disease states and pharmacotherapy. Acta Pol. Pharm. 2014, 71, 229–243. [Google Scholar]
- Hu, G.-M.; Mai, T.-L.; Chen, C.-M. Visualizing the GPCR network: Classification and evolution. Sci. Rep. 2017, 7, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Alexander, S.P.; Christopoulos, A.; Davenport, A.P.; Kelly, E.; Mathie, A.; Peters, J.A.; Veale, E.L.; Armstrong, J.F.; Faccenda, E.; Harding, S.D. The Concise Guide to PHARMACOLOGY 2019/20: G protein-coupled receptors. Br. J. Pharmacol. 2019, 176, S21–S141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davies, M.N.; Gloriam, D.E.; Secker, A.; Freitas, A.A.; Mendao, M.; Timmis, J.; Flower, D.R. Proteomic applications of automated GPCR classification. Proteomics 2007, 7, 2800–2814. [Google Scholar] [CrossRef] [PubMed]
- Shrivastava, S.; Pardasani, K.; Malik, M. SVM Model for Identification of human GPCRs. arXiv 2010, arXiv:1002.3983. [Google Scholar]
- Lapinsh, M.; Gutcaits, A.; Prusis, P.; Post, C.; Lundstedt, T.; Wikberg, J.E. Classification of G-protein coupled receptors by alignment-independent extraction of principal chemical properties of primary amino acid sequences. Protein Sci. 2002, 11, 795–805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Zhou, X.; Dai, Z.; Zou, X. Classification of G-protein coupled receptors based on support vector machine with maximum relevance minimum redundancy and genetic algorithm. BMC Bioinform. 2010, 11, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Ling, C.; Gao, J. An efficient CNN-based classification on G-protein Coupled Receptors using TF-IDF and N-gram. In Proceedings of the 2017 IEEE Symposium on Computers and Communications (ISCC), Heraklion, Greece, 3–6 July 2017; pp. 924–931. [Google Scholar]
- Kooistra, A.J.; Mordalski, S.; Pándy-Szekeres, G.; Esguerra, M.; Mamyrbekov, A.; Munk, C.; Keserű, G.M.; Gloriam, D.E. GPCRdb in 2021: Integrating GPCR sequence, structure and function. Nucleic Acids Res. 2021, 49, D335–D343. [Google Scholar] [CrossRef]
- Davies, M.N.; Secker, A.; Halling-Brown, M.; Moss, D.S.; Freitas, A.A.; Timmis, J.; Clark, E.; Flower, D.R. GPCRTree: Online hierarchical classification of GPCR function. BMC Res. Notes 2008, 1, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Begum, K.; Mohl, J.E.; Ayivor, F.; Perez, E.E.; Leung, M.-Y. GPCR-PEnDB: A database of protein sequences and derived features to facilitate prediction and classification of G protein-coupled receptors. Database 2020, 2020, baaa087. [Google Scholar]
- Davies, M.N.; Secker, A.; Freitas, A.A.; Mendao, M.; Timmis, J.; Flower, D.R. On the hierarchical classification of G protein-coupled receptors. Bioinformatics 2007, 23, 3113–3118. [Google Scholar] [CrossRef] [Green Version]
- Wootten, D.; Christopoulos, A.; Marti-Solano, M.; Babu, M.M.; Sexton, P.M. Mechanisms of signalling and biased agonism in G protein-coupled receptors. Nat. Rev. Mol. Cell Biol. 2018, 19, 638–653. [Google Scholar] [CrossRef]
- Basith, S.; Cui, M.; Macalino, S.J.; Park, J.; Clavio, N.A.; Kang, S.; Choi, S. Exploring G protein-coupled receptors (GPCRs) ligand space via cheminformatics approaches: Impact on rational drug design. Front. Pharmacol. 2018, 9, 128. [Google Scholar] [CrossRef]
- De Francesco, E.M.; Sotgia, F.; Clarke, R.B.; Lisanti, M.P.; Maggiolini, M. G protein-coupled receptors at the crossroad between physiologic and pathologic angiogenesis: Old paradigms and emerging concepts. Int. J. Mol. Sci. 2017, 18, 2713. [Google Scholar] [CrossRef] [Green Version]
- Schiöth, H.B.; Fredriksson, R. The GRAFS classification system of G-protein coupled receptors in comparative perspective. Gen. Comp. Endocrinol. 2005, 142, 94–101. [Google Scholar] [CrossRef]
- Southan, C.; Sharman, J.L.; Benson, H.E.; Faccenda, E.; Pawson, A.J.; Alexander, S.P.; Buneman, O.P.; Davenport, A.P.; McGrath, J.C.; Peters, J.A.; et al. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: Towards curated quantitative interactions between 1300 protein targets and 6000 ligands. Nucleic Acids Res. 2016, 44, D1054–D1068. [Google Scholar] [CrossRef] [Green Version]
- Beigi, M.; Zell, A. A novel method for classifying subfamilies and sub-subfamilies of G-protein coupled receptors. In Proceedings of the International Symposium on Biological and Medical Data Analysis, Thessaloniki, Greece, 7–8 December 2006; pp. 25–36. [Google Scholar]
- Wittlake, A.; Prömel, S.; Schöneberg, T. The Evolutionary History of Vertebrate Adhesion GPCRs and Its Implication on Their Classification. Int. J. Mol. Sci. 2021, 22, 11803. [Google Scholar] [CrossRef]
- Rajagopal, S.; Rajagopal, K.; Lefkowitz, R.J. Teaching old receptors new tricks: Biasing seven-transmembrane receptors. Nat. Rev. Drug Discov. 2010, 9, 373–386. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.C.; Cao, C.; Zhou, Y.; Zhao, Y. Proton transfer-mediated GPCR activation. Protein Cell 2015, 6, 12–17. [Google Scholar] [CrossRef] [Green Version]
- Ganguly, S.; Saxena, R.; Chattopadhyay, A. Reorganization of the actin cytoskeleton upon G-protein coupled receptor signaling. Biochim. Et Biophys. Acta (BBA) Biomembr. 2011, 1808, 1921–1929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Audet, M.; Bouvier, M. Restructuring G-protein-coupled receptor activation. Cell 2012, 151, 14–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walsh, C.T.; Stupack, D.; Brown, J.H. G Protein–Coupled Receptors Go Extracellular: RhoA Integrates the Integrins. Mol. Interv. 2008, 8, 165. [Google Scholar] [CrossRef] [PubMed]
- Otto, J.C.; Kelly, P.; Chiou, S.-T.; York, J.D. Alterations in an inositol phosphate code through synergistic activation of a G protein and inositol phosphate kinases. Proc. Natl. Acad. Sci. USA 2007, 104, 15653–15658. [Google Scholar] [CrossRef] [Green Version]
- Kamato, D.; Thach, L.; Bernard, R.; Chan, V.; Zheng, W.; Kaur, H.; Brimble, M.; Osman, N.; Little, P.J. Structure, function, pharmacology, and therapeutic potential of the G protein, Gα/q, 11. Front. Cardiovasc. Med. 2015, 2, 14. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.R.; Xu, J.; Maeda, S.; Duc, N.M.; Ahn, D.; Du, Y.; Chung, K.Y. Structural mechanism underlying primary and secondary coupling between GPCRs and the Gi/o family. Nat. Commun. 2020, 11, 1–12. [Google Scholar] [CrossRef]
- Li, Z.; Zhou, X.; Dai, Z.; Zou, X. Classification of G proteins and prediction of GPCRs-G proteins coupling specificity using continuous wavelet transform and information theory. Amino Acids 2012, 43, 793–804. [Google Scholar] [CrossRef]
- Rosenbaum, D.M.; Rasmussen, S.G.; Kobilka, B.K. The structure and function of G-protein-coupled receptors. Nature 2009, 459, 356–363. [Google Scholar] [CrossRef] [Green Version]
- Keshelava, A.; Solis, G.P.; Hersch, M.; Koval, A.; Kryuchkov, M.; Bergmann, S.; Katanaev, V.L. High capacity in G protein-coupled receptor signaling. Nat. Commun. 2018, 9, 1–8. [Google Scholar] [CrossRef]
- Tilley, D.G. G protein–dependent and G protein–independent signaling pathways and their impact on cardiac function. Circ. Res. 2011, 109, 217–230. [Google Scholar] [CrossRef] [Green Version]
- Peterson, Y.K.; Luttrell, L.M. The diverse roles of arrestin scaffolds in G protein–coupled receptor signaling. Pharmacol. Rev. 2017, 69, 256–297. [Google Scholar] [CrossRef] [Green Version]
- Turu, G.; Balla, A.; Hunyady, L. The role of β-arrestin proteins in organization of signaling and regulation of the AT1 angiotensin receptor. Front. Endocrinol. 2019, 10, 519. [Google Scholar] [CrossRef]
- Jong, Y.J.I.; Harmon, S.K.; O’Malley, K.L. GPCR signalling from within the cell. Br. J. Pharmacol. 2018, 175, 4026–4035. [Google Scholar] [CrossRef] [Green Version]
- Eichel, K.; von Zastrow, M. Subcellular organization of GPCR signaling. Trends Pharmacol. Sci. 2018, 39, 200–208. [Google Scholar] [CrossRef]
- Andronico, A.; Kiem, C.T.; Paireau, J.; Succo, T.; Bosetti, P.; Lefrancq, N.; Nacher, M.; Djossou, F.; Sanna, A.; Flamand, C.; et al. Evaluating the impact of curfews and other measures on SARS-CoV-2 transmission in French Guiana. Nat. Commun. 2021, 12, 1–8. [Google Scholar] [CrossRef]
- Molaei, S.; Dadkhah, M.; Asghariazar, V.; Karami, C.; Safarzadeh, E. The immune response and immune evasion characteristics in SARS-CoV, MERS-CoV, and SARS-CoV-2: Vaccine design strategies. Int. Immunopharmacol. 2020, 92, 107051. [Google Scholar] [CrossRef]
- Nejat, R.; Sadr, A.S. SARS virus papain-like protease: A mysterious weapon. J. Biostat. Epidemiol. 2019, 5, 288–295. [Google Scholar] [CrossRef]
- Tufet, M. T cells calm the storm. Nat. Rev. Immunol. 2007, 7, 834–835. [Google Scholar] [CrossRef]
- Henderson, L.A.; Canna, S.W.; Schulert, G.S.; Volpi, S.; Lee, P.Y.; Kernan, K.F.; Caricchio, R.; Mahmud, S.; Hazen, M.M.; Halyabar, O.; et al. On the alert for cytokine storm: Immunopathology in COVID-19. Arthritis Rheumatol. 2020, 72, 1059–1063. [Google Scholar] [CrossRef] [Green Version]
- Barnes, P.J. Receptor heterodimerization: A new level of cross-talk. J. Clin. Investig. 2006, 116, 1210–1212. [Google Scholar] [CrossRef]
- Nejat, R.; Sadr, A.S. Are Losartan and Imatinib Effective Against SARS-CoV2 Pathogenesis? A Pathophysiologic-Based In Silico Study. Silico Pharmacol. 2021, 9, 1. [Google Scholar] [CrossRef] [PubMed]
- Hamming, I.; Cooper, M.E.; Haagmans, B.L.; Hooper, N.M.; Korstanje, R.; Osterhaus, A.D.; Timens, W.; Turner, A.; Navis, G.; van Goor, H. The emerging role of ACE2 in physiology and disease. J. Pathol. A J. Pathol. Soc. Great Br. Irel. 2007, 212, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Shang, J.; Wan, Y.; Luo, C.; Ye, G.; Geng, Q.; Auerbach, A.; Li, F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 11727–11734. [Google Scholar] [CrossRef]
- Banu, N.; Panikar, S.S.; Leal, L.R.; Leal, A.R. Protective role of ACE2 and its downregulation in SARS-CoV-2 infection leading to Macrophage Activation Syndrome: Therapeutic implications. Life Sci. 2020, 256, 117905. [Google Scholar] [CrossRef] [PubMed]
- Reddy Gaddam, R.; Chambers, S.; Bhatia, M. ACE and ACE2 in inflammation: A tale of two enzymes. Inflamm. Allergy-Drug Targets (Former. Curr. Drug Targets-Inflamm. Allergy) (Discontin.) 2014, 13, 224–234. [Google Scholar]
- Wang, X.; Zhang, H.; Ge, Y.; Cao, L.; He, Y.; Sun, G.; Jia, S.; Ma, A.; Liu, J.; Rong, D.; et al. AT1R regulates macrophage polarization through YAP and regulates aortic dissection incidence. Front. Physiol. 2021, 12, 644903. [Google Scholar] [CrossRef] [PubMed]
- Magnani, F.; Pappas, C.G.; Crook, T.; Magafa, V.; Cordopatis, P.; Ishiguro, S.; Ohta, N.; Selent, J.; Bosnyak, S.; Jones, E.S.; et al. Electronic sculpting of ligand-GPCR subtype selectivity: The case of angiotensin II. ACS Chem. Biol. 2014, 9, 1420–1425. [Google Scholar] [CrossRef] [Green Version]
- Lu, S.; He, X.; Yang, Z.; Chai, Z.; Zhou, S.; Wang, J.; Rehman, A.U.; Ni, D.; Pu, J.; Sun, J.; et al. Activation pathway of a G protein-coupled receptor uncovers conformational intermediates as targets for allosteric drug design. Nat. Commun. 2021, 12, 1–15. [Google Scholar] [CrossRef]
- Burghi, V.; Echeverría, E.B.; Sosa, M.H.; Quiroga, D.T.; Muñoz, M.C.; Davio, C.; Monczor, F.; Fernández, N.C.; Dominici, F.P. Participation of Gαi-Adenylate Cyclase and ERK1/2 in Mas Receptor Signaling Pathways. Front. Pharmacol. 2019, 10, 146. [Google Scholar] [CrossRef]
- Zhang, H.; Han, G.W.; Batyuk, A.; Ishchenko, A.; White, K.L.; Patel, N.; Sadybekov, A.; Zamlynny, B.; Rudd, M.T.; Hollenstein, K.; et al. Structural basis for selectivity and diversity in angiotensin II receptors. Nature 2017, 544, 327–332. [Google Scholar] [CrossRef]
- Sadybekov, A.; Katritch, V. Breaking the Enigma Code of angiotensin II type 2 receptor signaling. Structure 2020, 28, 390–392. [Google Scholar] [CrossRef]
- Tóth, A.D.; Turu, G.; Hunyady, L.; Balla, A. Novel mechanisms of G-protein-coupled receptors functions: AT1 angiotensin receptor acts as a signaling hub and focal point of receptor cross-talk. Best Pract. Res. Clin. Endocrinol. Metab. 2018, 32, 69–82. [Google Scholar] [CrossRef] [Green Version]
- Thillaiappan, N.B.; Chakraborty, P.; Hasan, G.; Taylor, C.W. IP3 receptors and Ca2+ entry. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2019, 1866, 1092–1100. [Google Scholar] [CrossRef]
- Leppänen, T.; Tuominen, R.K.; Moilanen, E. Protein kinase C and its inhibitors in the regulation of inflammation: Inducible nitric oxide synthase as an example. Basic Clin. Pharmacol. Toxicol. 2014, 114, 37–43. [Google Scholar] [CrossRef]
- Kim, H.; Zamel, R.; Bai, X.-H.; Liu, M. PKC activation induces inflammatory response and cell death in human bronchial epithelial cells. PLoS ONE 2013, 8, e64182. [Google Scholar] [CrossRef] [Green Version]
- Gray, R.D.; Lucas, C.D.; MacKellar, A.; Li, F.; Hiersemenzel, K.; Haslett, C.; Davidson, D.J.; Rossi, A.G. Activation of conventional protein kinase C (PKC) is critical in the generation of human neutrophil extracellular traps. J. Inflamm. 2013, 10, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Dalal, P.J.; Muller, W.A.; Sullivan, D.P. Endothelial cell calcium signaling during barrier function and inflammation. Am. J. Pathol. 2020, 190, 535–542. [Google Scholar] [CrossRef] [Green Version]
- Manglik, A.; Wingler, L.M.; Rockman, H.A.; Lefkowitz, R.J. β-Arrestin–Biased Angiotensin II Receptor Agonists for COVID-19. Circulation 2020, 142, 318–320. [Google Scholar] [CrossRef]
- Iwasaki, M.; Saito, J.; Zhao, H.; Sakamoto, A.; Hirota, K.; Ma, D. Inflammation triggered by SARS-CoV-2 and ace2 augment drives multiple organ failure of severe COVID-19: Molecular mechanisms and implications. Inflammation 2021, 44, 13–34. [Google Scholar] [CrossRef]
- Angers, S.; Salahpour, A.; Bouvier, M. Dimerization: An emerging concept for G protein–coupled receptor ontogeny and function. Annu. Rev. Pharmacol. Toxicol. 2002, 42, 409–435. [Google Scholar] [CrossRef] [Green Version]
- Faron-Górecka, A.; Szlachta, M.; Kolasa, M.; Solich, J.; Górecki, A.; Kuśmider, M.; Żurawek, D.; Dziedzicka-Wasylewska, M. Understanding GPCR dimerization. Methods Cell Biol. 2019, 149, 155–178. [Google Scholar]
- Bulenger, S.; Marullo, S.; Bouvier, M. Emerging role of homo-and heterodimerization in G-protein-coupled receptor biosynthesis and maturation. Trends Pharmacol. Sci. 2005, 26, 131–137. [Google Scholar] [CrossRef]
- Gahbauer, S.; Böckmann, R.A. Membrane-mediated oligomerization of G protein coupled receptors and its implications for GPCR function. Front. Physiol. 2016, 7, 494. [Google Scholar] [CrossRef] [Green Version]
- Rukavina Mikusic, N.L.; Silva, M.G.; Pineda, A.M.; Gironacci, M.M. Angiotensin Receptors Heterodimerization and Trafficking: How Much Do They Influence Their Biological Function? Front. Pharmacol. 2020, 11, 1179. [Google Scholar] [CrossRef] [PubMed]
- Abdel Hameid, R.; Cormet-Boyaka, E.; Kuebler, W.M.; Uddin, M.; Berdiev, B.K. SARS-CoV-2 may hijack GPCR signaling pathways to dysregulate lung ion and fluid transport. Am. J. Physiol. Lung Cell. Mol. Physiol. 2021, 320, L430–L435. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Al-Khalili, O.; Duke, B.J.; Stockand, J.D.; Eaton, D.C.; Bao, H.-F. The inhibitory effect of Gβγ and Gβ isoform specificity on ENaC activity. Am. J. Physiol. -Ren. Physiol. 2013, 305, F1365–F1373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saint-Criq, V.; Gray, M.A. Role of CFTR in epithelial physiology. Cell. Mol. Life Sci. 2017, 74, 93–115. [Google Scholar] [CrossRef] [Green Version]
- Berdiev, B.K.; Qadri, Y.J.; Benos, D.J. Assessment of the CFTR and ENaC association. Mol. BioSyst. 2009, 5, 123–127. [Google Scholar] [CrossRef] [Green Version]
- Noreng, S.; Bharadwaj, A.; Posert, R.; Yoshioka, C.; Baconguis, I. Structure of the human epithelial sodium channel by cryo-electron microscopy. Elife 2018, 7, e39340. [Google Scholar] [CrossRef]
- Planès, C.; Randrianarison, N.H.; Charles, R.P.; Frateschi, S.; Cluzeaud, F.; Vuagniaux, G.; Soler, P.; Clerici, C.; Rossier, B.C.; Hummler, E. ENaC-mediated alveolar fluid clearance and lung fluid balance depend on the channel-activating protease 1. EMBO Mol. Med. 2010, 2, 26–37. [Google Scholar] [CrossRef]
- Ji, H.-L.; Song, W.; Gao, Z.; Su, X.-F.; Nie, H.-G.; Jiang, Y.; Peng, J.-B.; He, Y.-X.; Liao, Y.; Zhou, Y.-J.; et al. SARS-CoV proteins decrease levels and activity of human ENaC via activation of distinct PKC isoforms. Am. J. Physiol. Lung Cell. Mol. Physiol. 2009, 296, L372–L383. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.-J.; Seth, S.; Yue, G.; Kamat, P.; Compans, R.W.; Guidot, D.; Brown, L.A.; Eaton, D.C.; Jain, L. Influenza virus inhibits ENaC and lung fluid clearance. Am. J. Physiol. Lung Cell. Mol. Physiol. 2004, 287, L366–L373. [Google Scholar] [CrossRef]
- Bhalla, V.; Hallows, K.R. Mechanisms of ENaC regulation and clinical implications. J. Am. Soc. Nephrol. 2008, 19, 1845–1854. [Google Scholar] [CrossRef] [Green Version]
- Bilbao, P.S.; Katz, S.; Boland, R. Interaction of purinergic receptors with GPCRs, ion channels, tyrosine kinase and steroid hormone receptors orchestrates cell function. Purinergic Signal. 2012, 8, 91–103. [Google Scholar] [CrossRef] [Green Version]
- Pochynyuk, O.; Tong, Q.; Staruschenko, A.; Stockand, J.D. Binding and direct activation of the epithelial Na+ channel (ENaC) by phosphatidylinositides. J. Physiol. 2007, 580, 365–372. [Google Scholar] [CrossRef]
- Carvelli, J.; Demaria, O.; Vély, F.; Batista, L.; Benmansour, N.C.; Fares, J.; Carpentier, S.; Thibult, M.-L.; Morel, A.; Remark, R.; et al. Association of COVID-19 inflammation with activation of the C5a–C5aR1 axis. Nature 2020, 588, 146–150. [Google Scholar] [CrossRef]
- Yang, L.V.; Oppelt, K.A.; Thomassen, M.J.; Marie, M.A.; Nik Akhtar, S.; McCallen, J.D. Can GPR4 Be a Potential Therapeutic Target for COVID-19? Front. Med. 2021, 7, 1150. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nejat, R.; Sadr, A.S.; Torshizi, M.F.; Najafi, D.J. GPCRs of Diverse Physiologic and Pathologic Effects with Fingerprints in COVID-19. Biol. Life Sci. Forum 2021, 7, 19. https://doi.org/10.3390/ECB2021-10261
Nejat R, Sadr AS, Torshizi MF, Najafi DJ. GPCRs of Diverse Physiologic and Pathologic Effects with Fingerprints in COVID-19. Biology and Life Sciences Forum. 2021; 7(1):19. https://doi.org/10.3390/ECB2021-10261
Chicago/Turabian StyleNejat, Reza, Ahmad Shahir Sadr, Maziar Fayaz Torshizi, and David J. Najafi. 2021. "GPCRs of Diverse Physiologic and Pathologic Effects with Fingerprints in COVID-19" Biology and Life Sciences Forum 7, no. 1: 19. https://doi.org/10.3390/ECB2021-10261
APA StyleNejat, R., Sadr, A. S., Torshizi, M. F., & Najafi, D. J. (2021). GPCRs of Diverse Physiologic and Pathologic Effects with Fingerprints in COVID-19. Biology and Life Sciences Forum, 7(1), 19. https://doi.org/10.3390/ECB2021-10261






