Abstract
Plant hormones are master regulators of developmental and genetic mechanisms to deal with diverse environmental cues. Upon phosphate (Pi) limitation, vascular plants modify phytohormone metabolism to coordinate diverse mechanisms to overcome such stress. However, the transcriptional program underlying the hormonal signaling in response to Pi scarcity in early branches of land plant phylogeny remains unclear. Therefore, we explored the transcriptional dynamics of key genes involved in auxin, cytokinin, ethylene, jasmonate, gibberellin, and abscisic acid metabolism in the early divergent land plant Marchantia polymorpha upon Pi starvation. Our RNAseq approach revealed major changes in genes associated with auxin and ethylene biosynthesis. Genes involved in cytokinin synthesis are repressed. Interestingly, genes involved in auxin and ethylene signaling, such as MpARF1 and MpARF2, are upregulated. In contrast, MpARRb is down-regulated. Moreover, genes involved in the synthesis of jasmonates were highly upregulated, but those related to signaling did not change in expression. Our data suggest that auxin and ethylene act as positive regulators of rhizoid development under Pi-limited conditions, whereas cytokinin may act as a negative regulator. The transcriptional behavior of some hormone-related genes in Marchantia is similar to those described in controlling root hair development in arabidopsis, maize, and rice, upon Pi scarcity.
1. Introduction
During the process of land colonization by plants (470 mya), one of the most challenging conditions was the poor nutrient availability of the primitive earth crust [1,2]. In this scenario, physico-chemical properties of phosphate (Pi) reduced the accessible amount for plant uptake [3,4], which allowed for the selection of novel morphological and molecular strategies to satisfy its requirements and colonize novel ecosystems. Those biochemical, genetic, and morphological adaptations include the symbiosis with mycorrhizal fungi, the development of novel plant structures, and the rewiring of gene networks to cope with low Pi availability. However, there is a bias to angiosperms in the characterization of PSR along plant phylogeny and little is known about early divergent plants. Those mechanisms have been deeply explored in the flowering plant Arabidopsis thaliana, showing that phenotypic modifications in root system architecture (RSA), such as primary root growth inhibition, the formation of lateral roots, and root hair elongation, are triggered by the external Pi concentrations, defined as the local response (LR) to Pi starvation [5,6]. In contrast, at the molecular level, the transcription factor PHOSPHATE STARVATION RESPONSE 1 (AtPHR1) activates the expression of multiple target genes involved in Pi uptake, recycling, and scavenging—the so-called systemic response (SR) [7,8]. The AtPHR1 activity is negatively regulated by the proteins encoding SIG1-PHO81-XPR1 (SPX) domain, by a protein–protein interaction that depends on internal inositol polyphosphate (IPP) concentration [9,10].
Among the mechanisms developed to cope with low Pi, the roles of plant phytohormones have been explored at multiple levels, uncovering highly relevant changes in auxin, ethylene, jasmonate, gibberellin, and cytokinin metabolism and signaling [11,12]. Auxin acts as a positive regulator of morphological changes in RSA, in particular promoting lateral root development and primary root growth inhibition [11]. Under limited Pi conditions, AUXIN RESPONSE FACTOR 19 (AtARF19) and TRANSPORT INHIBITOR RESPONSE1 (AtTIR1) are transcriptionally induced, suggesting that TIR1, together with the SCF complex, promotes the ubiquitination of AUXIN/INDOLE-3-ACETIC ACID (AtAUX/IAA) for its degradation in order to facilitate the transcription of AtARF19 targets [13]. On the other hand, the expression of AtPHR1 is driven by both AUXIN RESPONSE FACTOR 7/19 (AtARF7 and AtARF19) throughout three auxin response elements (AREs) present in the promoter region of AtPHR1 [14]. Hence, auxin plays an important role in both local and systemic responses of A. thaliana to Pi starvation [11,14]. In addition, ethylene biosynthetic genes, such as AMINOCYCLOPROPANE-1-CARBOXILATE SYNTHASE 2/4/6 (AtACS2/4/6), were found to be upregulated in Pi-starved Arabidopsis plants [15]. Furthermore, pharmacological and genetic experiments showed that the addition of ethylene precursor mimics the RSA phenotype displayed by low Pi [11]. Another study, using ethylene signaling mutants, showed the role of this hormone in root hair development triggered by Pi starvation [16]. Other experiments revealed that ETHYLENE INSENSITIVE 3 (AtEIN3), a transcription factor involved in ethylene signaling, promotes the transcription of AtPHR1 [17]. In contrast, cytokinin negatively impacts the expression of PSR genes since the exogenous application of kinetin was enough to repress the expression of IPS; however, the changes in RSA induced by Pi scarcity were maintained [18]. In addition, mutations of CYTOKININ RESPONSE 1 (AtCRE1) and ARABIDOPSIS HISTIDINE KINASE 4 (AtAHK4) genes, which code for membrane receptors, impair the transcriptional repression of PSR [19]. In the case of jasmonate, it has been reported that its concentration increases in plants grown under low Pi conditions, which potentiates the tolerance to herbivory attacks [20]. Some genes involved in the biosynthesis and signaling of JA, such as JASMONATE ZIM DOMAIN 10 (AtJAZ10), VEGETATIVE STORAGE PROTEIN 2 (AtVSP2), and LIPOXYGENASE 2 (AtLOX2), are transcriptionally upregulated under low Pi conditions [20]. Moreover, genetic evidence suggests that several JA-responsive genes are also under the control of AtPHR1, some of which participate in the anthocyanin accumulation phenotype observed in plants grown in low Pi [20]. On the other hand, it has been found that the active form of Gibberellin decreases under Pi starvation. The low levels of this hormone allow DELLA transcription factors to participate in the regulation of anthocyanin accumulation and RSA modifications [21]. In contrast, the exogenous addition of gibberellins suppresses the transcription of diverse PSR genes, while mutants in genes involved in GA biosynthesis also impair the expression of genes induced by low Pi, when compared to the Wt [21].
In M. polymorpha, the phenotypic alterations under Pi starvation include changes in thallus pigmentation by auronodin accumulation, reduction of thallus weight, decreased internal Pi accumulation, and changes in rhizoid development [22,23]. Underlying such morpho-physiological responses are transcriptional changes, such as the upregulation of MpPHR1, which codes for the homolog of the major transcriptional regulator of PSR, as well as the upregulation of MpMYB14 gene, a master regulator of auronodin biosynthesis, and phosphate transporter-encoding genes, such as MpPHTs and MpPTBs, among others [23]. However, the transcriptional changes triggered by Pi starvation on genes involved in the biosynthesis and signaling of the diverse hormonal pathways remain poorly understood in the liverwort M. polymorpha. Here, we explore the transcriptional changes of such putative orthologous genes in order to explore the probable role of hormonal fluctuations in the response to Pi starvation in Marchantia. These analyses also allow us to speculate on the conservation or divergence in the response along land plant evolution. The resulting transcriptional patterns of genes involved in auxin, ethylene, cytokinin, gibberellin, abscisic acid, and jasmonate pathways revealed that the most relevant changes occur in genes related to auxin, ethylene, and cytokinin metabolism. Pi scarcity modulates auxin and ethylene biosynthesis, apparently to promote the synthesis of both hormones in these conditions. We speculate that downstream, the transcriptional activation of MpARF1/2 and MpERF promotes the expression of their targets in response to Pi limitation. In contrast, the biosynthesis of cytokinins is impaired at the transcriptional level, in correlation with the downregulation of MpARRb in the same conditions.
2. Methods
2.1. Survey of Hormone-Related Homologs in M. polymorpha
To determine the transcriptional behavior of genes related to hormone biosynthesis and signaling in M. polymorpha, we first searched for those genes annotated in the genome version 3.1 (Bowman et al., 2017) using the Pfam, KEGG, or OrthoKEGG identifiers. Then, using the A. thaliana sequences from TAIR V10 (available online: https://phytozome.jgi.doe.gov/pz/portal.html (accessed on 1 June 2020)) as a query, we searched by sequence homology using the BLAST software. As an additional step, we compared the conserved domains identified on the Marchantia proteins with the online tool Conserved Domains (available online: https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi, (accessed on 1 June 2020)). The sequences that did not show conserved domains with their respective A. thaliana orthologs were discarded for posterior analyses.
2.2. RNA-Seq Conditions and Differential Gene Expression Analysis
We used the RNA-seq data for M. polymorpha in low and high Pi conditions previously reported [23]. Briefly, plants were grown in high Pi conditions for 10 days and then transferred to high (500 μM) and low (10 μM) Pi availability on liquid media. The total RNA was sampled at 12, 24, and 150 h post-transference (HPT). The differential gene expression for each time was determined with the online server of DNAsubway, available on the CyVerse platform. We use the MpIDs to search into the table of differential expressed genes induced or repressed in response to low Pi availability. The criteria to define a gene as differentially expressed were FDR < 0.001 and 0.5 > log2FC < −0.5.
2.3. Determination of P1BS Enrichment
We searched for the consensus sequence P1BS along the promoter regions of each gene, and then performed an enrichment analysis for P1BS. The results were reported previously [23].
3. Results
3.1. Pi Limitation Positively Regulates Expression of Auxin-Related Genes in M. polymorpha
The role of auxin in Pi-deprived plants has been extensively studied in angiosperms—mainly the relationship of this hormone with the changes in Arabidopsis RSA and the expression of AtPHR1 under low Pi conditions in A. thaliana [24,25]. Perturbations in auxin metabolism, perception, and transduction have been associated with primary root growth adjustment, increased development and emergence of lateral roots and induction, and elongation of root hairs, all diverse phenotypes altered in response to Pi availability [11]. These are examples of morphological adaptations that lead plants to improve the nutrient assimilation capacity in the richest soil strata [26]. We explored the transcriptional landscape of genes related to auxin biosynthesis, transport, and signaling in the rootless plant M. polymorpha under low Pi conditions. We hypothesized that changes observed in the rhizoid development, which occur under low Pi conditions, are part of the Marchantia strategies to enhance nutrient uptake, which is underpinned by auxin concentration and signaling. Thus, we searched for putative homologs related to auxin metabolism and found that genes involved in auxin biosynthesis were deregulated under Pi starvation (Figure 1). MpTAA1 was downregulated at early times (12 and 24 HPT) and upregulated after 150 HPT. Conversely, the transcripts of MpYUC6 were upregulated at the three time points sampled, and interestingly, we identified two P1BS motives present in the analyzed 1 kb promoter region of this gene. This points to the hypothesis that MpPHR1 could be involved directly in the positive regulation of auxin biosynthesis under low Pi conditions. In the case of auxin transporters, two loci were differentially expressed, MpPIN2 was downregulated three times, and MpPIN3 was downregulated at early times and then showed a strong induction after 150 HPT. In this case, MpPIN2 possessed two enriched P1BS motifs along its promoter region suggesting that MpPHR1 acts as a negative regulator Moreover, the MpTIR locus displayed changes in its expression; it decreased at 12 HPT, was induced at 24 HPT, and was repressed at 150 HPT. Meanwhile, MpAUX/IAA1 was induced at early times, and downregulated after 150 HPT. Downstream, we found MpARF1/2 was induced at early times and downregulated at 150 HPT, which suggests the activation of its direct targets in order to modify the rhizoid development under low Pi. Considering the transcriptional induction of AtPHR1 by the AtARF7/19 under low Pi conditions, we hypothesized that MpARF1/2 likely promotes the expression of MpPHR1 under low Pi conditions, similar to what occurs in A. thaliana.
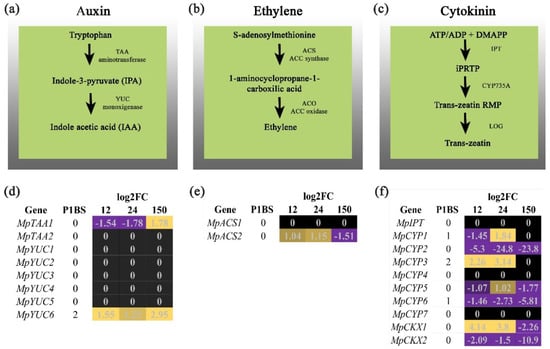
Figure 1.
Transcriptional changes of hormonal biosynthetic pathways. Summary of biosynthetic steps of auxin, ethylene, and cytokinin (a–c panels, respectively). In the heatmaps, purple shows the downregulated genes and yellow shows the upregulated genes for auxin, ethylene, and cytokinin in panels (d–f), respectively. Genes with P1BS present in the 1 Kb promoter region of the respective genes are shown in the heatmap.
3.2. Low Pi-Activated Expression of Ethylene Signaling Genes
It has been reported that Pi starvation impacts ethylene metabolism in A. thaliana. Changes to its biosynthesis and signaling are linked to the regulation of root hair elongation, and AtPHR1 transcriptional levels are modulated by ethylene [16,17]. In the search of homologs related to ethylene metabolism, our results show that putative genes coding for ACC oxidase (ACO) are absent in the M. polymorpha genome, while two loci that code for ACC synthase were identified (Figure 1). We found that MpACS2 was upregulated at early times and was repressed after 150 HPT under low Pi conditions. In the case of ethylene biosynthesis, our results were not conclusive; further experiments are necessary to determine the genes involved in the complete enzymatic pathway for ethylene production in Marchantia. Downstream, the putative Marchantia homologs involved in ethylene signaling show transcriptional changes in the expression of four members of ethylene receptors (Figure 2). MpCTR1 was induced at the three times sampled, while MpETR3 was downregulated at all time points sampled. In addition, MpETR1 and MpCTR3 were upregulated at early times and repressed after 150 HPT. The search for the P1BS revealed that MpETR1 and MpCTR3 have two motives in their respective promoter regions, suggesting that MpPHR1 drives the transcriptional changes of these genes in response to low Pi conditions. Finally, the putative homolog for MpERF1 is upregulated at 12 HPT, downregulated at 24 FPT, and upregulated again at 150 HPT. The transcriptional activation of MpERF1 points to the hypothesis that ethylene biosynthesis and/or perception is promoted by Pi starvation, but how this occurs, and what the phenotypic consequences are of such crosstalk requires additional experiments.
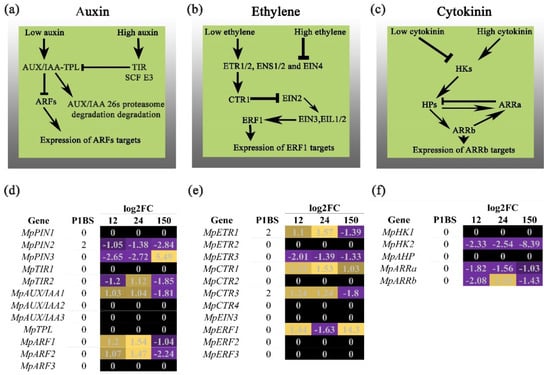
Figure 2.
Expression of hormonal signaling pathways. Summary of the main genes related to auxin, ethylene, and cytokinin signaling (a–c panels, respectively). In the heatmaps, purple shows the downregulated genes and yellow shows the upregulated genes for auxin, ethylene, and cytokinin in (d–f), respectively. The genes that contain the P1BS motif in their promoter regions are also shown.
3.3. Cytokinin Metabolism Negatively Regulates Pi Starvation Responses
Pioneering experiments to elucidate the role of cytokinins in the response of Arabidopsis to cope with low Pi availability revealed that under this nutritional stress, the plant changes its metabolism to decrease the concentration of active cytokinin [18]. The addition of exogenous cytokinin represses the transcription of several genes involved in diverse responses triggered by Pi starvation, supporting the notion that this hormone acts as a negative regulator of such responses [19]. We searched for putative homologs of genes related to CK biosynthesis and signaling and found one locus coding for IPT, seven loci coding for CYP735, and two coding for cytokinin oxidases. In addition, two histidine kinase receptor genes were found, a single gene coding for AHP was identified, and two genes coding for ARR transcription factors. The transcriptional dynamics for these genes show changes induced by Pi starvation in five members of the CYP735 family. MpCYP1 was downregulated at 12 HPT and upregulated after 24 HPT, while MpCYP2/6 was downregulated at the three time points sampled. The transcript levels of MpCYP3 increased at early times, while those of MpCYP5 were downregulated at 12 HPT, upregulated at 24 HPT, and downregulated again at 150 HPT. One and two P1BS motifs were located in the promoter regions of MpCYP1/6 e and MpCYP3, respectively. Moreover, we found that the transcript levels of MpHK2 and MpARRb were downregulated at the three time points sampled, while those of MpARRa were downregulated at 12 HPT, upregulated at 24 HPT, and downregulated after 150 HPT. Altogether, our data show that although some of the Marchantia homologs involved in cytokinin biosynthesis were upregulated, the major transcriptional regulators, such as MpARRa/b, were repressed, suggesting that the negative regulatory effect of CKs in Pi starvation responses could be conserved in M. polymorpha.
3.4. Other Hormonal-Related Genes Regulated by Pi Scarcity
Several other plant hormones explored in the context of Pi starvation response are gibberellins (GA), abscisic acid (ABA), and jasmonates [12]. In A. thaliana, the role of GA and ABA are described as negative regulators of PSR, while the jasmonates are implicated in the induction of ROS and the local modification of RSA [12,27]. Among the biosynthetic genes, we found several of them deregulated, including MpGA20ox, the expression of which was upregulated at the three times sampled. The MpDELLA2 gene was upregulated at 12 HPT and downregulated after 24 and 150 HPT, similar to what was described for A. thaliana. In the case of ABA, we observed that genes involved in the biosynthesis were mainly downregulated under low Pi conditions. Interestingly, MpABI4 was upregulated at 12 HPT and became downregulated after 24 and 150 HPT. In contrast, the genes involved in the biosynthesis of jasmonates were strongly induced in Pi-deprived plants, but those involved in the perception and transcriptional response to this hormone, which include the MYB TFs, did not show differential expression. Only the MpJAZ gene, which codes for a transcriptional repressor, was induced at the three time points of our transcriptomic approach.
4. Discussion
The signaling cascades and transcriptional programs that rely on the morphological and physiological adaptations upstream to deal with low Pi availability have been widely studied in the flowering plant A. thaliana [24,25]. In this field, the potential role of hormones as master coordinators of the Pi starvation response is well-known. Metabolic changes in hormones regulate the modifications in RSA and affect the expressions of genes that are responsive to this nutritional stress [11,28]. However, how this rescue mechanism evolved is poorly understood. In order to gain insight into how the early divergent land plants responded to this nutrient restriction, morphophysiological and transcriptional characterization was performed on the evolutionary model M. polymorpha [23]. The phenotypical changes correlated with the transcriptional dynamic, underpinning the accumulation of auronidins, changes in thallus size, diminution of internal Pi concentration, and developmental modifications of rhizoids [23]. The local and systemic responses were dissected at the transcriptional level, but genes related to the metabolism of hormones remain unexplored. We searched by sequence homology and the conserved domains of the putative homologs related to hormone metabolism and investigated their transcriptional behavior under low Pi.
Our observations indicate that Pi limitation modulates the expression of genes related to auxin biosynthesis and transport. Apparently, MpPHR1 promotes the induction of MpYUC6 through two enriched P1BS motifs present in its promoter region. The presumable induction of this monooxygenase enzyme, via MpPHR1, links auxin synthesis directly to the PSR. These changes in biosynthetic genes are associated with the induction and elongation of rhizoids; an example of this is the effect of exogenous application of auxin on root hairs, which phenocopies the effect of low Pi on trychoblasts [29,30]. In agreement with this, Marchantia auxin-insensitive mutants did not show rhizoid alterations in media supplemented with this hormone [29,30]. In addition, we found transcriptional changes in the auxin transporters. It is worth noting the repression of MpPIN2, which the promoter region possesses in two cis-regulatory P1BS elements made it a potential target of MpPHR1, the master regulator of PSR. This is in agreement with a previously established hypothesis, which suggests that MpPHR1 displays a dual role in inducing and repressing the subsets of genes via its binding to P1BS in a selective manner, and depending on the fluctuations of environmental factors, similar to what occurs in A. thaliana [23,31]. Another notable finding is the induction of MpARF1/2 at early times (12 and 24 HPT), suggesting that Pi starvation (1) increases auxin levels and (2) induces the expression of targets involved in rhizoid development. These observations lead us to suggest that MpARFs could be involved in the transcriptional regulation of MpPHR1, as reported for A. thaliana upon Pi starvation [13]. The notion of a link between auxin signaling and rhizoid development under low Pi conditions is supported by the fact that the loss of function mutants mparf1-1/2 are impaired in the developmental responses of rhizoids after the exogenous application of auxin [29,30]. Hence, the alteration of auxin metabolism under Pi limitation probably allows for the rhizoid response via the induction of MpARF targets, including MpPHR1.
In the case of ethylene’s role in the response of A. thaliana to Pi scarcity, changes in the metabolism of this hormone were linked to the elongation of root hairs in Pi-starved plants [16]. The inhibition or promotion of ethylene synthesis by the addition of silver nitrate or ACC on media, respectively, impacts the development of these structures in the roots [11]. In this study, we were not able to identify a complete biosynthetic pathway in M. polymorpha, and a putative homolog to AtACO was not found according to our search criteria. This suggests two probable scenarios—(1) the ACC oxidase enzyme in Marchantia may possess non-canonical domains that are not conserved in angiosperms, and (2) there is no ACO coding gene in the M. polymorpha genome. Transcriptional changes in other genes, related to ethylene biosynthesis and signaling, suggest that the plant may respond to crosstalk between Pi starvation and this hormone; however, further experiments are necessary to decipher and understand such molecular interactions. The activation of MpERF1 suggests that some players of the ethylene signaling pathway respond to low Pi availability, perhaps to modulate the elongation of rhizoids similarly to what occurs with A. thaliana root hairs [16].
In the case of the cytokinins, the antagonic role of this hormone in the low Pi response has been reported [27]. Based on our results, we hypothesize that such antagonism is partially conserved in M. polymorpha. This is in agreement with the downregulation of key genes, such as MpCYP2/6 and some signaling genes. Moreover, we observed that MpCKX1 was upregulated at 12 and 24 HPT, suggesting that with low Pi, cytokinins are inactivated. Previous reports show that the overexpression of this enzyme decreases the active cytokinin amount [32]. Interestingly, the characterization of the loss of function mutants for MpARRb and the overexpression of MpARRa are involved in the regulation of rhizoid development [32]. We found that under low Pi conditions, both MpARRa/b are downregulated; this correlates with rhizoid modifications observed in response to Pi starvation [23].
Altogether, our results point to major participation of hormone metabolism and signaling in rhizoid developmental changes in response to low Pi (Figure 3). Our transcriptional analyses suggest that under Pi scarcity, auxin and ethylene levels may increase, acting as positive regulators of rhizoid development, while the cytokinin levels, which act as negative regulators of root hair development in other species, may decrease (Figure 3).
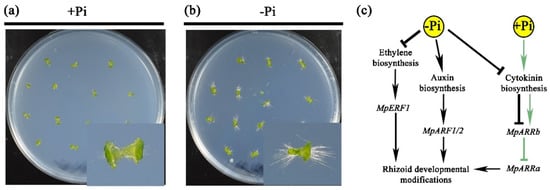
Figure 3.
Changes of rhizoid development and the putative model of phytohormone pathways under low Pi. The phenotypes of plants 7 days after being sown in high and low Pi conditions (panels a,b, respectively). The putative model of rhizoid development under Pi deprivation shows the induction of auxin biosynthesis and represses the synthesis of ethylene and cytokinin. MpERF1 and MpARF1/2 are induced in low Pi conditions and promote the elongation of rhizoids, while MpARRb is downregulated, allowing the activity of MpARRa to promote the rhizoid modifications (c).
5. Conclusions
- Low Pi availability modifies the expression of diverse genes involved in the biosynthesis of auxin, ethylene, cytokinin, gibberellins, abscisic acid, and jasmonates.
- Auxin and ethylene probably act as positive regulators of changes observed in rhizoids under Pi scarcity.
- Cytokinin probably acts as a negative regulator of the PSR and rhizoid developmental modifications.
- The induction of MpARF1/2 and MpERF1 transcription may result in the modification of hormonal levels, promoting the expression of several targets that allow rhizoid elongation.
- The downregulation of MpARRa/b could participate in the phenotypical changes observed in rhizoids.
Author Contributions
F.R.-R., M.A.A.-V., L.H.-E., and A.C.-R. conceived the project. F.R.-R., L.H.-E., and A.C.-R. designed the experiments. F.R.-R., Z.H.U.D.-S., and M.D.-A. performed the experiments. F.R.-R., Z.H.U.D.-S., M.D.-A., K.I., M.A.A.-V., and A.C.-R. analyzed the data. K.I., A.C.-H., J.L.B., M.A.A.-V., L.H.-E., and A.C.-R. contributed key bioinformatic resources, biological materials, and reagents. F.R.-R. and A.C.-R. wrote the manuscript with input from all co-authors. All authors have read and agreed to the published version of the manuscript.
Acknowledgments
A.C.-R wish to thank Kochi from the Graduate School of Biostudies at Kyoto University in Japan for sharing key biological materials. F.R.-R. is indebted to Consejo Nacional de Ciencia y Tecnología (CONACyT) for the PhD scholarship (No. 629985). CONACyT provided the PhD scholarship to MDA (No. 487660). This work was funded by the JSPS KAKENHI grant (19H03247) awarded to K.I. M.A.A.-V. was funded by UCMEXUS (No. 19941-44), CONACYT Ciencia Básica (No. 158550 & A1-S-38383), and the Royal Society Newton Advanced Fellowship (NA150181).
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| Pi | Phosphate |
| RSA | Root system architecture |
| SR | Systemic response |
| IPP | Inositol poly-phosphate |
| PSR | Phosphate starvation response |
| Wt | Wild type |
| µM | Micro molar |
| HPT | Hours post-transference |
| FDR | False discovery rate |
| P1BS | PHR1 binding site |
| Kb | Kilo base |
References
- Sanderson, M.J. Molecular data from 27 proteins do not support a Precambrian origin of land plants. Am. J. Bot. 2003, 90, 954–956. [Google Scholar] [CrossRef] [Green Version]
- Delwiche, C.F.; Cooper, E.D. The evolutionary origin of a terrestrial flora. Curr. Biol. 2015, 25, R899–R910. [Google Scholar] [CrossRef] [Green Version]
- Bieleski, R.L. Phosphate pools, phosphate transport, and phosphate availability. Annu. Rev. Plant Physiol. 1973, 24, 225–252. [Google Scholar] [CrossRef]
- Marschner, H. Marschner’s Mineral Nutrition of Higher Plants; Academic Press: Amsterdam, The Netherlands, 2011. [Google Scholar]
- Sánchez-Calderón, L.; López-Bucio, J.; Chacón-López, A.; Cruz-Ramírez, A.; Nieto-Jacobo, F.; Dubrovsky, J.G.; Herrera-Estrella, L. Phosphate starvation induces a determinate developmental program in the roots of Arabidopsis thaliana. Plant Cell Physiol. 2005, 46, 174–184. [Google Scholar] [CrossRef]
- Svistoonoff, S.; Creff, A.; Reymond, M.; Sigoillot-Claude, C.; Ricaud, L.; Blanchet, A.; Desnos, T. Root tip contact with low-phosphate media reprograms plant root architecture. Nat. Genet. 2007, 39, 792. [Google Scholar] [CrossRef] [Green Version]
- Rubio, V.; Linhares, F.; Solano, R.; Martín, A.C.; Iglesias, J.; Leyva, A.; Paz-Ares, J. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 2001, 15, 2122–2133. [Google Scholar] [CrossRef] [Green Version]
- Bustos, R.; Castrillo, G.; Linhares, F.; Puga, M.I.; Rubio, V.; Pérez-Pérez, J.; Paz-Ares, J. A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet. 2010, 6, e1001102. [Google Scholar] [CrossRef] [Green Version]
- Puga, M.I.; Mateos, I.; Charukesi, R.; Wang, Z.; Franco-Zorrilla, J.M.; de Lorenzo, L.; Leyva, A. SPX1 is a phosphate-dependent inhibitor of Phosphate Starvation Response 1 in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 14947–14952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wild, R.; Gerasimaite, R.; Jung, J.Y.; Truffault, V.; Pavlovic, I.; Schmidt, A.; Mayer, A. Control of eukaryotic phosphate homeostasis by inositol polyphosphate sensor domains. Science 2016, 352, 986–990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- López-Bucio, J.; Hernández-Abreu, E.; Sánchez-Calderón, L.; Nieto-Jacobo, M.F.; Simpson, J.; Herrera-Estrella, L. Phosphate availability alters architecture and causes changes in hormone sensitivity in the Arabidopsis root system. Plant Physiol. 2002, 129, 244–256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rubio, V.; Bustos, R.; Irigoyen, M.L.; Cardona-López, X.; Rojas-Triana, M.; Paz-Ares, J. Plant hormones and nutrient signaling. Plant Mol. Biol. 2009, 69, 361. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Torres, C.A.; López-Bucio, J.; Cruz-Ramírez, A.; Ibarra-Laclette, E.; Dharmasiri, S.; Estelle, M.; Herrera-Estrella, L. Phosphate availability alters lateral root development in Arabidopsis by modulating auxin sensitivity via a mechanism involving the TIR1 auxin receptor. Plant Cell 2007, 20, 3258–3272. [Google Scholar] [CrossRef] [Green Version]
- Huang, K.L.; Ma, G.J.; Zhang, M.L.; Xiong, H.; Wu, H.; Zhao, C.Z.; Li, X.B. The ARF7 and ARF19 transcription factors positively regulate phosphate starvation response1 in Arabidopsis roots. Plant Physiol. 2018, 178, 413–427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lei, M.; Zhu, C.; Liu, Y.; Karthikeyan, A.S.; Bressan, R.A.; Raghothama, K.G.; Liu, D. Ethylene signalling is involved in regulation of phosphate starvation-induced gene expression and production of acid phosphatases and anthocyanin in Arabidopsis. New Phytol. 2011, 189, 1084–1095. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Yu, H.; Dong, J.; Che, X.; Jiao, Y.; Liu, D. The molecular mechanism of ethylene-mediated root hair development induced by phosphate starvation. PLoS Genet. 2016, 12, e1006194. [Google Scholar] [CrossRef]
- Liu, Y.; Xie, Y.; Wang, H.; Ma, X.; Yao, W.; Wang, H. Light and ethylene coordinately regulate the phosphate starvation response through transcriptional regulation of PHOSPHATE STARVATION RESPONSE1. Plant Cell 2017, 29, 2269–2284. [Google Scholar] [CrossRef] [Green Version]
- Martin, A.C.; Del Pozo, J.C.; Iglesias, J.; Rubio, V.; Solano, R.; De La Pena, A.; Leyva, A.; Paz-Ares, J. Influence of cytokinins on the expression of phosphate starvation responsive genes in Arabidopsis. Plant J. 2000, 24, 559–567. [Google Scholar] [CrossRef]
- Franco-Zorrilla, J.M.; Martin, A.C.; Solano, R.; Rubio, V.; Leyva, A.; Paz-Ares, J. Mutations at CRE1 impair cytokinin-induced repression of phosphate starvation responses in Arabidopsis. Plant J. 2002, 32, 353–360. [Google Scholar] [CrossRef] [PubMed]
- Khan, G.A.; Vogiatzaki, E.; Glauser, G.; Poirier, Y. Phosphate deficiency induces the jasmonate pathway and enhances resistance to insect herbivory. Plant Physiol. 2016, 171, 632–644. [Google Scholar] [CrossRef] [Green Version]
- Jiang, C.; Gao, X.; Liao, L.; Harberd, N.P.; Fu, X. Phosphate starvation root architecture and anthocyanin accumulation responses are modulated by the gibberellin-DELLA signaling pathway in Arabidopsis. Plant Physiol. 2007, 145, 1460–1470. [Google Scholar] [CrossRef] [Green Version]
- Voth, P.D.; Hamner, K.C. Responses of Marchantia polymorpha to nutrient supply and photoperiod. Bot. Gaz. 1940, 102, 169–205. [Google Scholar] [CrossRef]
- Rico-Reséndiz, F.; Cervantes-Pérez, S.A.; Espinal-Centeno, A.; Dipp-Álvarez, M.; Oropeza-Aburto, A.; Hurtado-Bautista, E.; Cruz-Hernández, A.; Bowman, J.L.; Ishizaki, K.; Arteaga-Vázquez, M.A.; et al. Transcriptional and Morpho-Physiological Responses of Marchantia polymorpha upon Phosphate Starvation. Int. J. Mol. Sci. 2020, 21, 8354. [Google Scholar] [CrossRef]
- Puga, M.I.; Rojas-Triana, M.; de Lorenzo, L.; Leyva, A.; Rubio, V.; Paz-Ares, J. Novel signals in the regulation of Pi starvation responses in plants: Facts and promises. Curr. Opin. Plant Biol. 2017, 39, 40–49. [Google Scholar] [CrossRef]
- Gutiérrez-Alanís, D.; Ojeda-Rivera, J.O.; Yong-Villalobos, L.; Cárdenas-Torres, L.; Herrera-Estrella, L. Adaptation to phosphate scarcity: Tips from arabidopsis roots. Trends Plant Sci. 2018, 23, 721–730. [Google Scholar] [CrossRef]
- Péret, B.; Clément, M.; Nussaume, L.; Desnos, T. Root developmental adaptation to phosphate starvation: Better safe than sorry. Trends Plant Sci. 2011, 16, 442–450. [Google Scholar] [CrossRef] [PubMed]
- Chacón-López, A.; Ibarra-Laclette, E.; Sánchez-Calderón, L.; Gutiérrez-Alanís, D.; Herrera-Estrella, L. Global expression pattern comparison between low phosphorus insensitive 4 and WT Arabidopsis reveals an important role of reactive oxygen species and jasmonic acid in the root tip response to phosphate starvation. Plant Signal. Behav. 2011, 6, 382–392. [Google Scholar] [CrossRef] [Green Version]
- Franco-Zorrilla, J.M.; Martín, A.C.; Leyva, A.; Paz-Ares, J. Interaction between phosphate-starvation, sugar, and cytokinin signaling in Arabidopsis and the roles of cytokinin receptors CRE1/AHK4 and AHK3. Plant Physiol. 2005, 138, 847–857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kato, H.; Ishizaki, K.; Kouno, M.; Shirakawa, M.; Bowman, J.L.; Nishihama, R.; Kohchi, T. Auxin-mediated transcriptional system with a minimal set of components is critical for morphogenesis through the life cycle in Marchantia polymorpha. PLoS Genet. 2015, 11, e1005084. [Google Scholar] [CrossRef] [Green Version]
- Flores-Sandoval, E.; Eklund, D.M.; Bowman, J.L. A simple auxin transcriptional response system regulates multiple morphogenetic processes in the liverwort Marchantia polymorpha. PLoS Genet. 2015, 11, e1005207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castrillo, G.; Teixeira, P.J.P.L.; Paredes, S.H.; Law, T.F.; de Lorenzo, L.; Feltcher, M.E.; Paz-Ares, J. Root microbiota drive direct integration of phosphate stress and immunity. Nature 2017, 543, 513–518. [Google Scholar] [CrossRef] [PubMed]
- Aki, S.S.; Mikami, T.; Naramoto, S.; Nishihama, R.; Ishizaki, K.; Kojima, M.; Takebayashi, Y.; Sakakibara, H.; Kyozuka, J.; Kohchi, T.; et al. Cytokinin signaling is essential for organ formation in Marchantia polymorpha. Plant Cell Physiol. 2019, 60, 1842–1854. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).