Dissecting Plant Specific Insert Interaction Networks †
Abstract
:1. Introduction
2. Experiments
2.1. Plant Material and Growth Conditions
2.2. Protein Extraction under Non-Denaturing Conditions
2.3. Co-Immunoprecipitation Assay
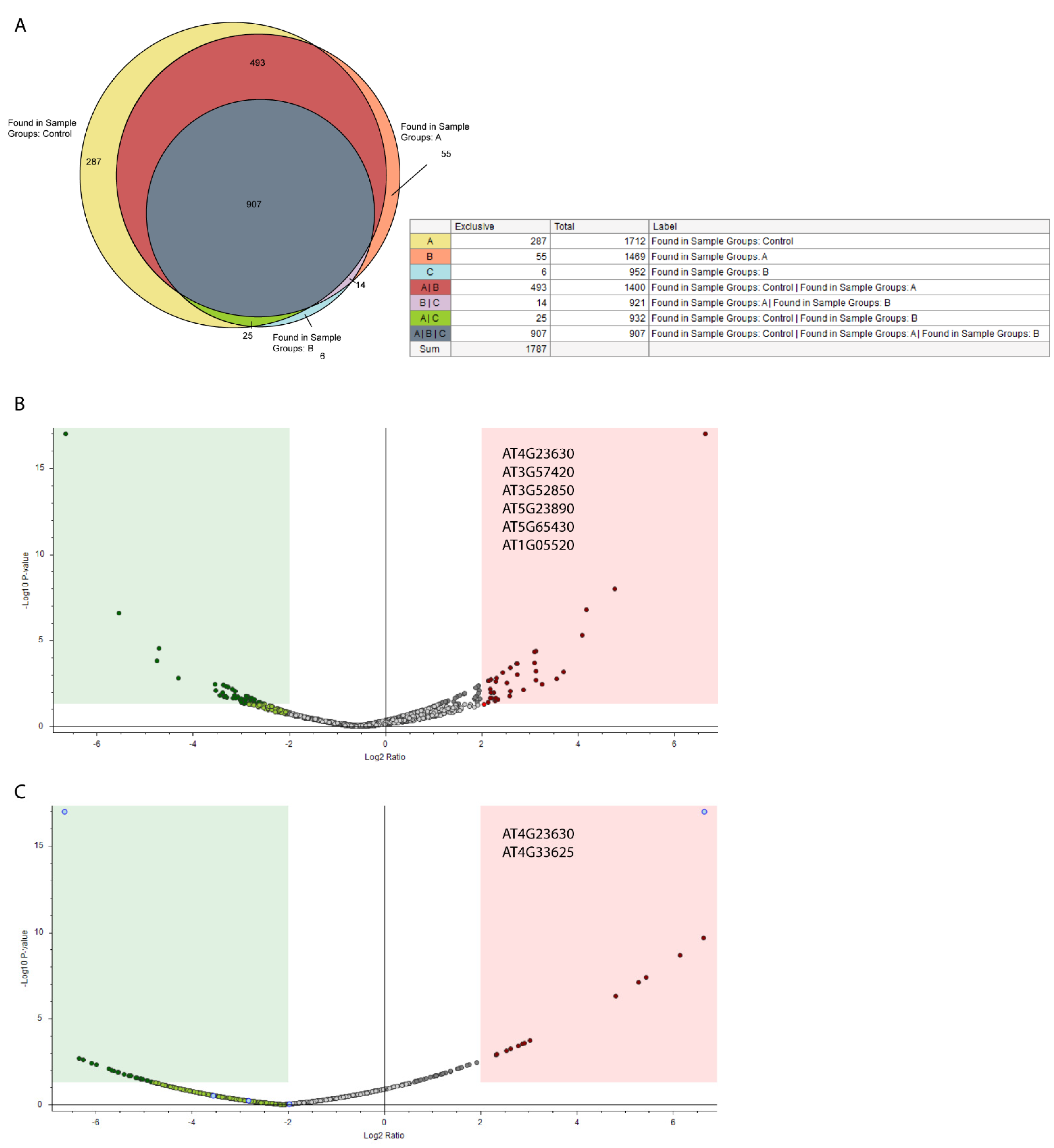
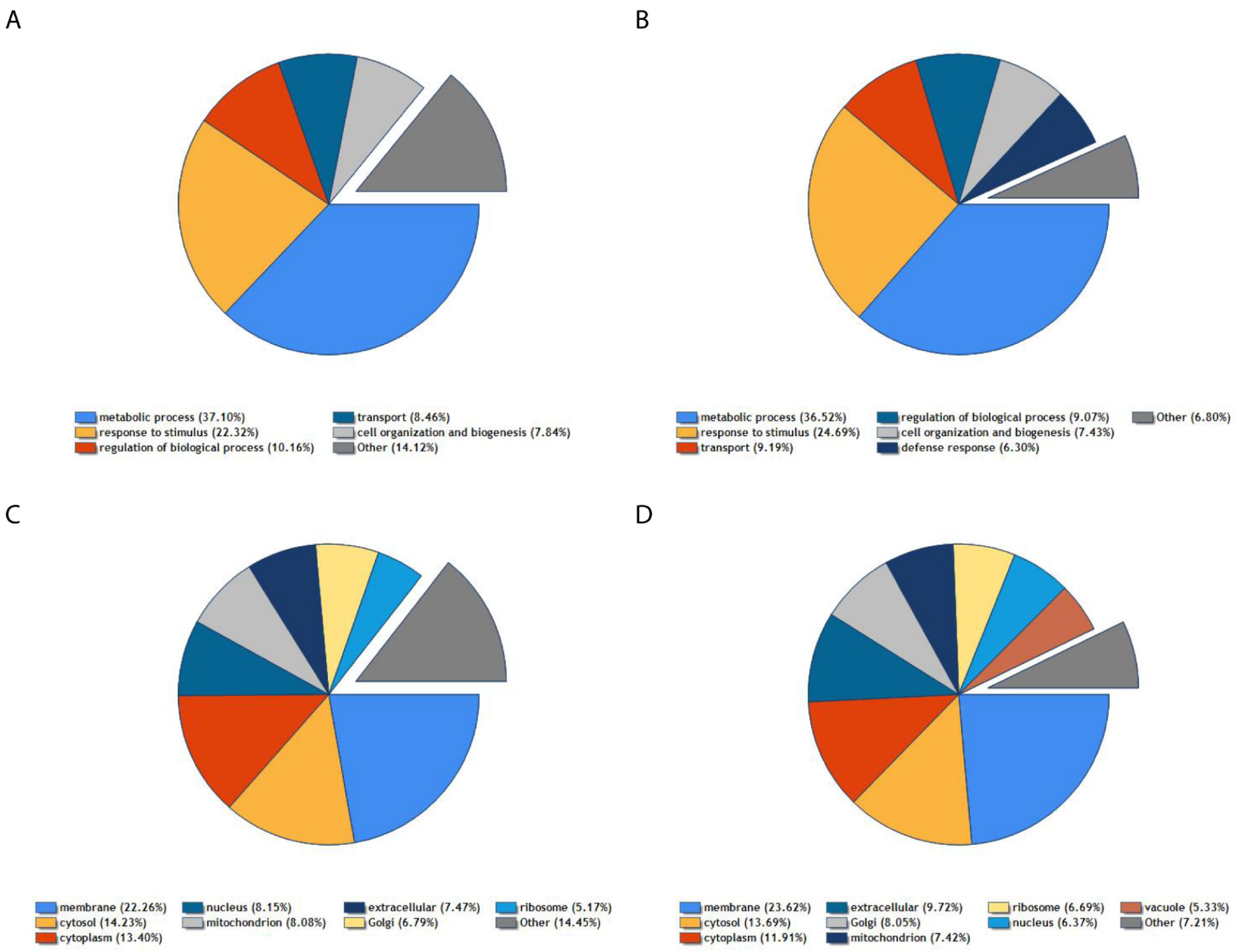
3. Results
4. Discussion
5. Conclusions
Author Contributions
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Rojo, E.; Denecke, J. What is moving in the secretory pathway of plants? Plant Physiol. 2008, 147, 1493–1503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiang, L.; Etxeberria, E.; Van Den Ende, W. Vacuolar protein sorting mechanisms in plants. FEBS J. 2013, 280, 979–993. [Google Scholar] [CrossRef] [Green Version]
- Pereira, C.; Pereira, S.; Satiat-Jeunemaitre, B.; Pissarra, J. Cardosin A contains two vacuolar sorting signals using different vacuolar routes in tobacco epidermal cells. Plant J. 2013, 76, 87–100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sansebastiano, D.; Pietro, G.; Barozzi, F.; Piro, G.; Denecke, J.; Lousa, C.d. Trafficking routes to the plant vacuole: Connecting alternative and classical pathways. J. Exp. Bot. 2018, 69, 79–90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- da Costa, D.S.; Pereira, S.; Moore, I.; Pissarra, J. Dissecting cardosin B trafficking pathways in heterologous systems. Planta 2010, 232, 1517–1530. [Google Scholar] [CrossRef]
- Oliveira, A.; Fidalgo, F.; Teixeira, J.; Teixeira, J.; Oliveira, A.; Pereira, C.; da Costa, D.S.; Pereira, M.; de Oliveira, A. Characterization of aspartic proteinases in C. cardunculus L. callus tissue for its prospective transformation. Plant Sci. 2010, 178, 140–146. [Google Scholar] [CrossRef]
- Egas, C.; Lavoura, N.; Resende, R.; Brito, R.M.M.; Pires, E.; De Lima, M.C.P.; Faro, C. The Saposin-like Domain of the Plant Aspartic Proteinase Precursor Is a Potent Inducer of Vesicle Leakage. J. Biol. Chem. 2002, 275, 38190–38196. [Google Scholar]
- Terauchi, K.; Asakura, T.; Ueda, H.; Tamura, T.; Tamura, K.; Matsumoto, I.; Misaka, T.; Hara-Nishimura, I.; Abe, K. Plant-specific insertions in the soybean aspartic proteinases, soyAP1 and soyAP2, perform different functions of vacuolar targeting. J. Plant Physiol. 2006, 163, 856–862. [Google Scholar] [CrossRef]
- Muñoz, F.F.; Mendieta, J.R.; Pagano, M.R.; Paggi, R.A.; Daleo, G.R.; Guevara, M.G. The swaposin-like domain of potato aspartic protease (StAsp-PSI) exerts antimicrobial activity on plant and human pathogens. Peptides 2010, 31, 777–785. [Google Scholar] [CrossRef]
- Curto, P.; Lufrano, D.; Pinto, C.; Custódio, V.; Gomes, A.C.; Trejo, S.A.; Bakás, L.; Vairo-Cavalli, S.; Faro, C.; Simões, I. Establishing the yeast kluyveromyces lactis as an expression host for production of the saposin-like domain of the aspartic protease cirsin. Appl. Environ. Microbiol. 2014, 80, 86–96. [Google Scholar] [CrossRef] [Green Version]
- De Moura, D.C.; Bryksa, B.C.; Yada, R.Y. In silico insights into protein-protein interactions and folding dynamics of the saposin-like domain of Solanum tuberosum aspartic protease. PLoS ONE 2014, 9, 18–22. [Google Scholar] [CrossRef]
- Muñoz, F.; Palomares-Jerez, M.F.; Daleo, G.; Villalaín, J.; Guevara, M.G. Possible mechanism of structural transformations induced by StAsp-PSI in lipid membranes. Biochim. Biophys. Acta Biomembr. 2014, 1838, 339–347. [Google Scholar] [CrossRef] [Green Version]
- Frey, M.E.; D’Ippolito, S.; Pepe, A.; Daleo, G.R.; Guevara, M.G. Transgenic expression of plant-specific insert of potato aspartic proteases (StAP-PSI) confers enhanced resistance to Botrytis cinerea in Arabidopsis thaliana. Phytochemistry 2018, 149, 1–11. [Google Scholar] [CrossRef]
- De Caroli, M.; Lenucci, M.S.; Di Sansebastiano, G.-P.; Dalessandro, G.; De Lorenzo, G.; Piro, G. Protein trafficking to the cell wall occurs through mechanisms distinguishable from default sorting in tobacco. Plant J. 2011, 65, 295–308. [Google Scholar] [CrossRef]
- De Marchis, F.; Bellucci, M.; Pompa, A. Unconventional pathways of secretory plant proteins from the endoplasmic reticulum to the vacuole bypassing the Golgi complex. Plant Signal. Behav. 2013, 8, e25129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stigliano, E.; Faraco, M.; Neuhaus, J.-M.; Montefusco, A.; Dalessandro, G.; Piro, G.; Di Sansebastiano, G.-P. Two glycosylated vacuolar GFPs are new markers for ER-to-vacuole sorting. Plant Physiol. Biochem. 2013, 73, 337–343. [Google Scholar] [CrossRef] [PubMed]
- Nishizawa, K.; Maruyama, N.; Utsumi, S. The C-terminal region of α′ subunit of soybean β-conglycinin contains two types of vacuolar sorting determinants. Plant Mol. Biol. 2006, 62, 111–125. [Google Scholar] [CrossRef] [PubMed]
- Vieira, V.; Peixoto, B.; Costa, M.; Pereira, S.; Pissarra, J.; Pereira, C. N-linked glycosylation modulates Golgi-independent vacuolar sorting mediated by the plant specific insert. Plants 2019, 8, 312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Occhialini, A.; Gouzerh, G.; Di Sansebastiano, G.P.; Neuhaus, J.M. Dimerization of the vacuolar receptors AtRMR1 and -2 from Arabidopsis thaliana contributes to their localization in the trans-Golgi network. Int. J. Mol. Sci. 2016, 17, 1661. [Google Scholar] [CrossRef] [Green Version]
- Pompa, A.; de Marchis, F.; Pallotta, M.T.; Benitez-Alfonso, Y.; Jones, A.; Schipper, K.; Moreau, K.; Žárský, V.; di Sansebastiano, G.P.; Bellucci1, M. Unconventional transport routes of soluble and membrane proteins and their role in developmental biology. Int. J. Mol. Sci. 2017, 18, 703. [Google Scholar] [CrossRef] [Green Version]
- Hwang, H.H.; Gelvin, S.B. Plant proteins that interact with VirB2, the Agrobacterium tumefaciens pilin protein, mediate plant transformation. Plant Cell 2004, 16, 3148–3167. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.Y.; Bowen, C.H.; Popescu, G.V.; Kang, H.-G.; Kato, N.; Ma, S.; Dinesh-Kumar, S.; Snyder, M.; Popescu, S.C. Arabidopsis RTNLB1 and RTNLB2 reticulon-like proteins regulate intracellular trafficking and activity of the FLS2 immune receptor. Plant Cell 2011, 23, 3374–3391. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Nikolovski, N.; Sorieul, M.; Vellosillo, T.; McFarlane, H.E.; Dupree, R.; Kesten, C.; Schneider, R.; Driemeier, C.; Lathe, R.; et al. Golgi-localized STELLO proteins regulate the assembly and trafficking of cellulose synthase complexes in Arabidopsis. Nat. Commun. 2016, 7, 11656. [Google Scholar] [CrossRef] [Green Version]
- Sanderfoot, A.A.; Ahmed, S.U.; Marty-Mazars, D.; Rapoport, I.; Kirchhausen, T.; Marty, F.; Raikhel, N.V. A putative vacuolar cargo receptor partially colocalizes with AtPEP12p on a prevacuolar compartment in Arabidopsis roots. Proc. Natl. Acad. Sci. USA 1998, 95, 9920–9925. [Google Scholar] [CrossRef] [Green Version]
- Shimada, T.; Fuji, K.; Tamura, K.; Kondo, M.; Nishimura, M.; Hara-Nishimura, I. Vacuolar sorting receptor for seed storage proteins in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2003, 100, 16095–16100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hinz, G.; Colanesi, S.; Hillmer, S.; Rogers, J.C.; Robinson, D.G. Localization of Vacuolar Transport Receptors and Cargo Proteins in the Golgi Apparatus of Developing Arabidopsis Embryos. Traffic 2007, 8, 1452–1464. [Google Scholar] [CrossRef]
- Craddock, C.P.; Hunter, P.R.; Szakacs, E.; Hinz, G.; Robinson, D.G.; Frigerio, L. Lack of a vacuolar sorting receptor leads to non-specific missorting of soluble vacuolar proteins in arabidopsis seeds. Traffic 2008, 9, 408–416. [Google Scholar] [CrossRef] [PubMed]
- Park, M.; Lee, D.; Lee, G.J.; Hwang, I. AtRMR1 functions as a cargo receptor for protein trafficking to the protein storage vacuole. J. Cell Biol. 2005, 170, 757–767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.H.; Oufattole, M.; Rogers, J.C. Golgi-mediated vacuolar sorting in plant cells: RMR proteins are sorting receptors for the protein aggregation/membrane internalization pathway. Plant Sci. 2007, 172, 728–745. [Google Scholar] [CrossRef]
- Kinoshita, T. Glycosylphosphatidylinositol (GPI) anchors: Biochemistry and cell biology: Introduction to a thematic review series. J. Lipid Res. 2016, 57, 4–5. [Google Scholar] [CrossRef] [Green Version]
- Fu, H.; Subramanian, R.R.; Masters, S.C. 14-3-3 Proteins: Structure, function, and regulation. Annu. Rev. Pharmacol. Toxicol. 2000, 40, 617–647. [Google Scholar] [CrossRef]
- Aducci, P.; Camoni, L.; Marra, M.; Visconti, S. From cytosol to organelles: 14-3-3 Proteins as multifunctional regulators of plant cell. IUBMB Life 2002, 53, 49–55. [Google Scholar] [CrossRef]
- Schoonheim, P.J.; Costa Pereira, D.D.; De Boer, A.H. Dual role for 14-3-3 proteins and ABF transcription factors in gibberellic acid and abscisic acid signalling in barley (Hordeum vulgare) aleurone cells. Plant. Cell Environ. 2009, 32, 439–447. [Google Scholar] [CrossRef]
- Denison, F.C.; Paul, A.L.; Zupanska, A.K.; Ferl, R.J. 14-3-3 proteins in plant physiology. Semin. Cell Dev. Biol. 2011, 22, 720–727. [Google Scholar] [CrossRef] [PubMed]
- Camoni, L.; Di Lucente, C.; Pallucca, R.; Visconti, S.; Aducci, P. Binding of phosphatidic acid to 14-3-3 proteins hampers their ability to activate the plant plasma membrane H+-ATPase. IUBMB Life 2012, 64, 710–716. [Google Scholar] [CrossRef]
- Camoni, L.; Visconti, S.; Aducci, P.; Marra, M. 14-3-3 Proteins in Plant Hormone Signaling: Doing Several Things at Once. Front. Plant Sci. 2018, 9, 297. [Google Scholar] [CrossRef]
- Gökirmak, T.; Paul, A.L.; Ferl, R.J. Plant phosphopeptide-binding proteins as signaling mediators. Curr. Opin. Plant Biol. 2010, 13, 527–532. [Google Scholar] [CrossRef]
- Chang, I.-F.; Curran, A.; Woolsey, R.; Quilici, D.; Cushman, J.C.; Mittler, R.; Harmon, A.; Harper, J.F. Proteomic profiling of tandem affinity purified 14-3-3 protein complexes in Arabidopsis thaliana. Proteomics 2009, 9, 2967–2985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Q.; Zhang, S.; Liu, B. 14-3-3 proteins: Macro-regulators with great potential for improving abiotic stress tolerance in plants. Biochem. Biophys. Res. Commun. 2016, 477, 9–13. [Google Scholar] [CrossRef] [PubMed]
- Camoni, L.; Harper, J.F.; Palmgren, M.G. 14-3-3 proteins activate a plant calcium-dependent protein kinase (CDPK). FEBS Lett. 1998, 430, 381–384. [Google Scholar] [CrossRef] [Green Version]
- Fromme, J.C.; Orci, L.; Schekman, R. Coordination of COPII vesicle trafficking by Sec23. Trends Cell Biol. 2008, 18, 330–336. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sampaio, M.; Neves, J.; Pereira, S.; Pissarra, J.; Pereira, C. Dissecting Plant Specific Insert Interaction Networks. Biol. Life Sci. Forum 2021, 4, 65. https://doi.org/10.3390/IECPS2020-08870
Sampaio M, Neves J, Pereira S, Pissarra J, Pereira C. Dissecting Plant Specific Insert Interaction Networks. Biology and Life Sciences Forum. 2021; 4(1):65. https://doi.org/10.3390/IECPS2020-08870
Chicago/Turabian StyleSampaio, Miguel, João Neves, Susana Pereira, José Pissarra, and Cláudia Pereira. 2021. "Dissecting Plant Specific Insert Interaction Networks" Biology and Life Sciences Forum 4, no. 1: 65. https://doi.org/10.3390/IECPS2020-08870
APA StyleSampaio, M., Neves, J., Pereira, S., Pissarra, J., & Pereira, C. (2021). Dissecting Plant Specific Insert Interaction Networks. Biology and Life Sciences Forum, 4(1), 65. https://doi.org/10.3390/IECPS2020-08870






