Abstract
Bacteria actively degrade polymeric materials due to high biochemical activity and the ability to form biofilms. In order to expand our knowledge about soil bacterial biodiversity on the surface of artificial materials from the soil of Chernihiv (Ukraine), in this study, the bacterial strain PET1 was isolated from the biofilm formed on the poly(ethylene terephthalate) bottle material taken from soil. The strain was isolated and purified on Postgate’s “C” medium and modified by us (using poly(ethylene terephthalate) as the only source of carbon). The PET1 strain was identified as Achromobacter xylosoxidans based on a complex of microbiological and molecular genetic features. This study is the first report of the isolation of an A. xylosoxidans strain from a biofilm on the surface of poly(ethylene terephthalate) collected from the soil. Considering the growing interest of researchers in the problem of biodegradation of plastics, attention should be paid to the potential of A. xylosoxidans as biodegradation agents. The involvement of bacteria of the genus Achromobacter in the biodegradation of artificial polymers (polyvinyl chloride, high-density polyethylene, nylon) opens up prospects for the isolated strain PET1 to be studied and, possibly, applied as a biodegrader of poly(ethylene terephthalate).
1. Introduction
Bacteria take an active part in the degradation of polymeric materials due to their high biochemical activity and ability to form biofilms [1,2,3]. Among the microorganisms isolated from the surface of plastic, there are representatives of various species that belong to various ecological and trophic groups [2,4,5]. Poly(ethylene terephthalate) (PET) is a polymer material whose production is growing every year [6]. In Ukraine in 2018, 203.7 thousand tons of PET bottles became waste [7]. Of this amount, about 25% (50,000 tons) went to waste processing enterprises, and 75% was buried in landfills [7]. PET is very resistant to microbial degradation because it has a linear structure and a high proportion of aromatic components [8]. Joo et al. [9] note that according to a report by the US National Park Service, PET bottles take approximately 450 years to decompose. Currently, the accumulation of PET in the environment is considered a global environmental problem [10].
A small number of species of microorganisms that have the ability to biodegrade PET are known [8,10,11]. It is known that some species of microscopic fungi can enzymatically degrade PET [8]. Yoshida et al. [8] isolated a new bacterium Ideonella sakaiensis 201-F6 with the ability to use PET as the main source of energy and carbon, which produces two enzymes capable of hydrolyzing PET and converting it into environmentally harmless monomers. It is known that the ability to destroy highly crystallized PET is inherent in the bacterial enzyme PETase [10]. It was shown that due to the low enzymatic activity of native PETase, a whole-cell biocatalyst was developed to improve its degradation efficiency, in which PETase was contained on the surface of a yeast cell (Pichia pastoris) [10]. Enzymes associated with PET biodegradation are currently known: cutinase-like hydrolase TfH; PET hydrolase; enzymes, surface-modifying PET; LC-cutinase hydrolase; cutinase Cut190; IsPETase; hydrolase TfCut2 [12]. Vague et al. [11] established the presence of bacteria capable of PET degradation in oil-contaminated soil. Three species of lipase-positive pseudomonads and Bacillus cereus were identified as part of two consortia capable of biofilm formation and plastic degradation. At the same time, bacteria formed a biofilm on PET and caused molecular changes on the surface of this material. Currently, the participation of lipase-positive Pseudomonas spp. in the degradation of PET has been little studied, which emphasizes the importance of the conducted research in discovering the potential of using consortia of common soil bacteria for the destruction of plastic waste [11]. The bacterium Gordonia sp. CN2K demonstrated a high ability (40.43% of PET in 45 days of incubation) to degrade PET [13]. Bacteria of the genus Bacillus are known for their ability to biodegradation/biodeterioration plastics [14]. Enzymatic degradation of PET (100% of PET at 360 g/L) by the bacterium Bacillus thermoamylovorans JQ3 as part of the thermophilic microbe-enzyme system with leaf-branch compost cutinase variant was shown [15]. In the study, with bacterium Bacillus safensis YX8 was demonstrated high biodegradation of PET [16].
The genus Achromobacter is also attracting researchers’ attention as a biodegrader of plastics. Recent scientific publications have reported the ability of bacteria of the genus Achromobacter to biodegrade a number of plastics: high-density polyethylene [17], nylon [18], and polyvinyl chloride [19]. However, the isolation of representatives of this genus from biofilms on plastic surfaces has not been reported. To date, the following species of soil bacteria have been isolated and identified from the soil in the Chernihiv city (Ukraine) from the surface of artificial materials (steel): Desulfovibrio oryzae strains NUChC SRB1 and NUChC SRB2, Peribacillus (Bacillus) simplex strain ChNPU F1, Streptomyces gardneri strain ChNPU F3, Streptomyces canus strain NUChC F2, Fictibacillus sp. strain ZVB1, and Anaerotignum (Clostridium) propionicum strain Sat1 [20,21,22]. The ability to form a biofilm has been established for them: P. simplex, S. gardneri, S. canus, and Fictibacillus sp. on glass surfaces [23,24] and D. oryzae and A. propionicum on the surface of polypropylene [25] and on the surface of poly(ethylene terephthalate) [22,26]. In order to expand knowledge about soil bacterial biodiversity on the surface of artificial materials from the soil of Chernihiv city (Ukraine), in this study, a bacterial strain was isolated from the biofilm formed on the PET-bottle material taken from the soil, which was identified by a complex of microbiological and molecular genetic methods. The microbiological and molecular genetic study of the isolated strain for its identification and theoretical justification of its potential as a biodegrader of plastics were the main objectives of the presented study.
2. Materials and Methods
2.1. Isolation of a Bacterial Isolate from a Biofilm Formed on the Surface of Poly(Ethylene Terephthalate)
Bacteria were isolated from the biofilm formed on the surface of the PET bottle, which was located in the soil (Figure 1).

Figure 1.
Research object and its location: (a) poly(ethylene terephthalate) bottle with a formed biofilm; (b) place of sample selection (51°31′ N 31°17′ E, soil, Regional Landscape Park “Yalivshchyna”, Chernihiv, Ukraine), the red circle indicates the location where the PET bottle used to collect the research material was found.
For this purpose, the biofilm was mechanically removed with a sterile scalpel in the sterile medium of Postgate’s “C” [27] modified by us, which contained a sample of PET sterilized in 96% ethyl alcohol (Table 1). After cultivation of microorganisms for 30 days at a temperature of 29 ± 2 °C under aerobic conditions, the biofilm formed on the PET surface was mechanically removed and transferred to test tubes with sterile modified Postgate’s “C” medium with PET samples. On the 5th day of cultivation, the biofilm was removed from the PET sample with a sterile bacteriological loop and cultured on meat-peptone agar (MPA) in a Petri dish by the exhausting stroke method [28]. Incubation took place under aerobic conditions and a temperature of 29 ± 2 °C for 5 days. The material of two isolated colonies [isolates NUChC PET1 (hereafter PET1) and NUChC PET2 (hereafter PET2)] was transferred to a liquid modified medium of Postgate’s “C” with PET samples and incubated under aerobic conditions. Next, we worked with the PET1 isolate, taking into account its ability to grow quickly in the liquid Postgate’s “C” medium modified by us with PET samples: PET1 on the 2nd day of cultivation and PET2 on the 7th day. After five passages on liquid modified Postgate’s “C” medium with PET samples and MPA, a pure culture of isolate PET1 was obtained, which was used in further studies.

Table 1.
The composition of the Postgate’s “C” medium.
2.2. Study of Cultural-Morphological and Some Physiological-Biochemical Properties of the Selected Isolate
Culture purity was checked by microscopy. Light microscopy at magnification (×400 and ×1000) was used to study the morphology of bacteria.
Preparations were made, which were stained according to Gram in Kalina’s modification (to determine Gram affiliation) and according to Hansen’s method (for staining spores) [28,29]. Morphological analysis of colonies on MPA was carried out according to a conventional scheme [28].
Research on the presence of catalase and oxidase was carried out by conventional methods [28,29,30].
2.3. Molecular Genetic Studies of the Selected Isolate
Establishing the systematic position of bacteria was carried out according to the nucleotide sequence of the 16S rRNA gene fragment. For this, a molecular genetic study was carried out, which included the following stages: isolation of DNA from bacterial cells, polymerase chain reaction (PCR), electrophoresis of amplification products, purification of the PCR product, and sequencing.
Bacterial DNA was isolated from the daily culture using the “GeneJET Genomic DNA Purification Kit” (Thermo Fisher Scientific, Waltham, MA, USA), according to the manufacturer’s instructions. Amplification of the 16S rRNA gene was performed with primers 27f (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492r (5′-CGGTTACCTTGTTACGA CTT-3′) [31] under the following temperature regime: initial denaturation 2 min, 95 °C; 30 cycles—30 s, 95 °C; 45 s, 56 °C; 90 s, 72 °C; final elongation 7 min, 72 °C. A 25 μL PCR mix contained 12.5 μL 2× DreamTaq PCR Master Mix (ThermoScientific), 30 pmol of each primer, and 50 ng of DNA. PCR was performed on a Mastercycler Personal 5332 amplifier (Eppendorf, Germany). PCR products were separated in a 1.7% agarose gel containing 0.01% ethidium bromide. The results were visualized in UV light. The resulting amplicon (~1500 bp in size) was excised from the gel and purified using the GeneJet PCR Purification Kit (ThermoScientific). The concentration of DNA was determined on the device “BioPhotometer” (Eppendorf, Germany). The purified PCR product was sequenced in two directions on the device “Genetic Analyzer 3130” using the set of reagents “BigDye Terminator v 3.1 Cycle Sequencing Kit”. The resulting nucleotide sequence was compared with those entered in the GenBank database using the Blast program (http://www.ncbi.nlm.nih.gov/blast (accessed on 3 January 2025). Phylogenetic analysis and alignment of 16S rDNA nucleotide sequences of representatives of different species of the genus Achromobacter were carried out using the MEGA 11 program [32]. A dendrogram of phylogenetic relationships was constructed using the Neighbor Joining method using Kimura’s two-parameter model based on 1000 bootstrap analysis replicates using the MEGA 11 program [32]. The sequences of the 16S rRNA gene of typical cultures of bacteria of the genus Achromobacter were taken by us from the GenBank database.
3. Results and Discussion
3.1. Microbiological Properties of PET1 and PET2 Isolates
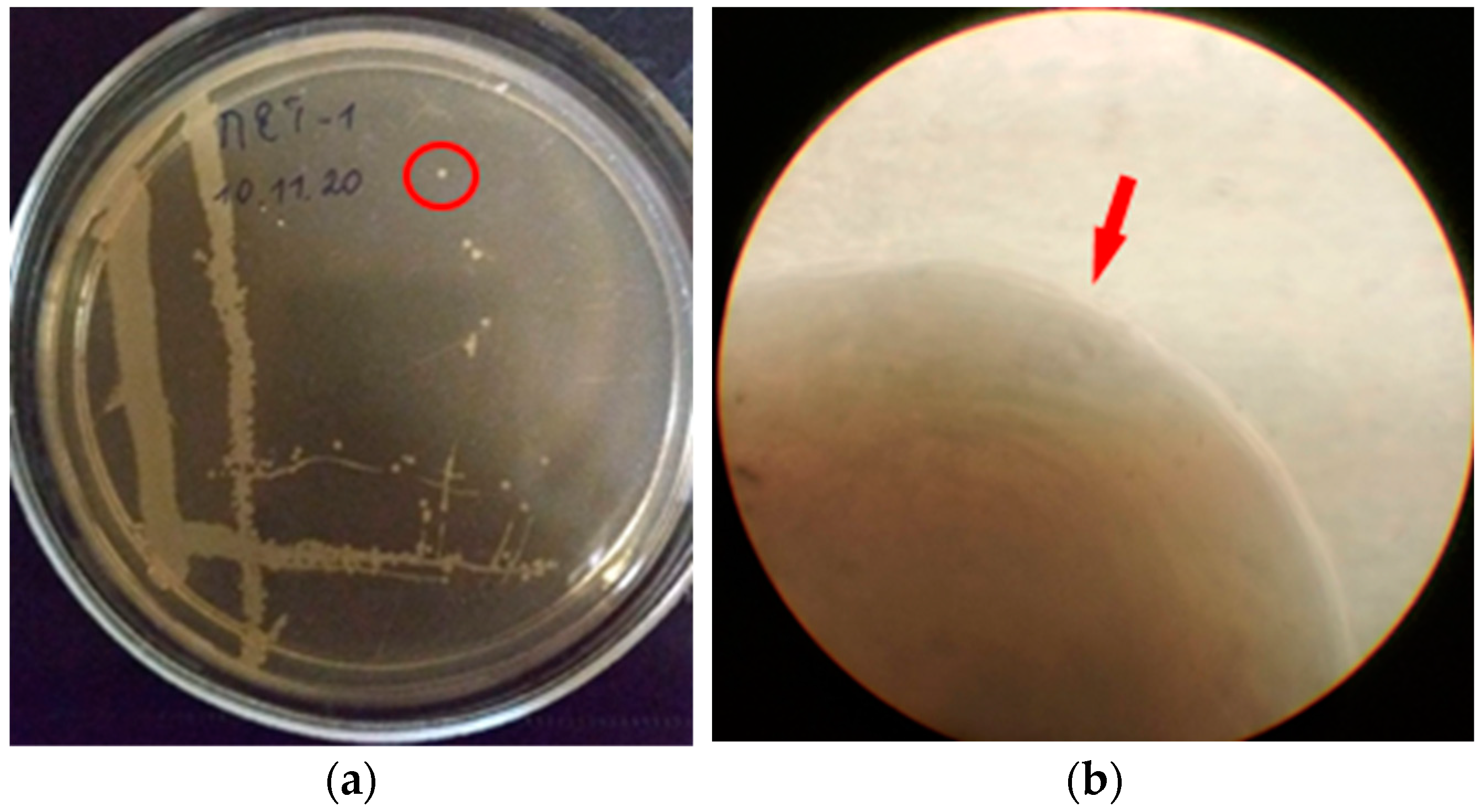
Colonies of isolate PET1 on MPA (2nd day) under aerobic conditions are superficial, whitish-beige in color, rounded in shape, soft in consistency, 1 mm in size. The edges of the colony are even, the structure is homogeneous, the surface is smooth (Figure 2).
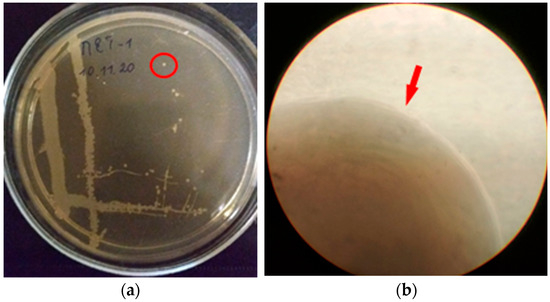
Figure 2.
Colonies of isolate PET1: (a) growth on MPA (2nd day), the red circle indicates the colony that was selected for further study; (b) the edge of the colony (light microscopy, magnification ×135), the red arrow indicates the edge of the colony.
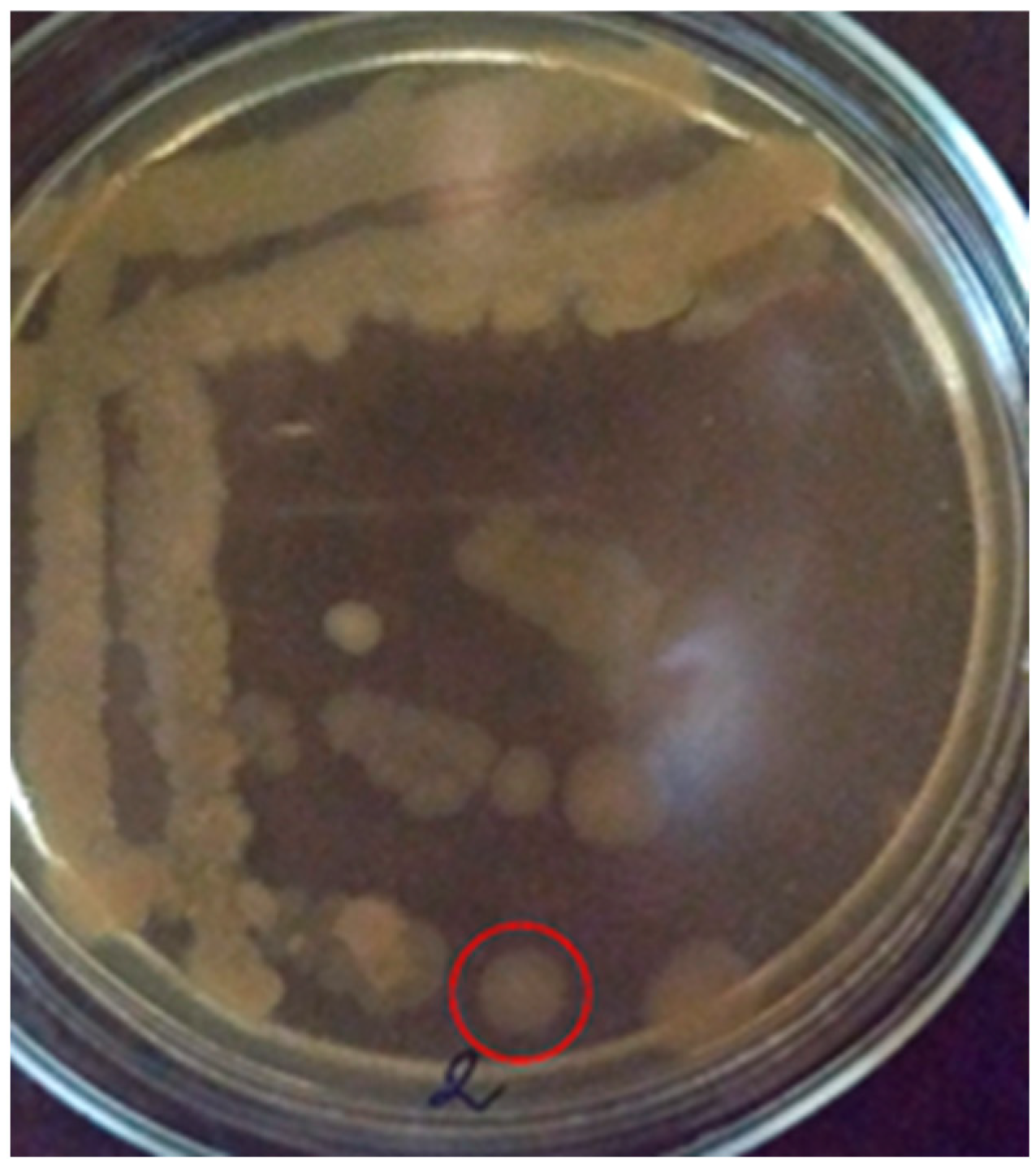
Colonies of the PET2 isolate on MPA (10th day) under aerobic conditions are superficial, beige in color, rounded in shape, soft in consistency, 8 mm in size. The edges of the colony are wavy, the structure is homogeneous, the surface is smooth (Figure 3). Bacteria are catalase-positive.
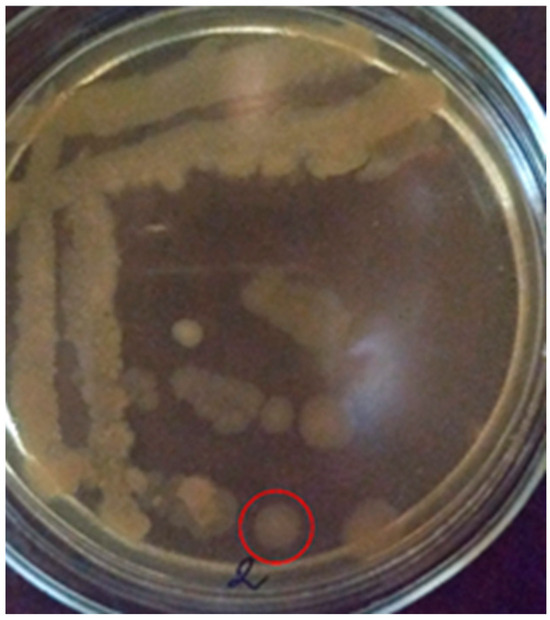
Figure 3.
Colonies of the PET2 isolate on MPA (10th day) (the studied colony is surrounded by a red circle).
Since the PET1 isolate is capable of rapid growth in liquid Postgate’s “C” medium modified by us with PET samples (PET1 on the 2nd day of cultivation, and PET2 on the 7th day), it was further investigated.
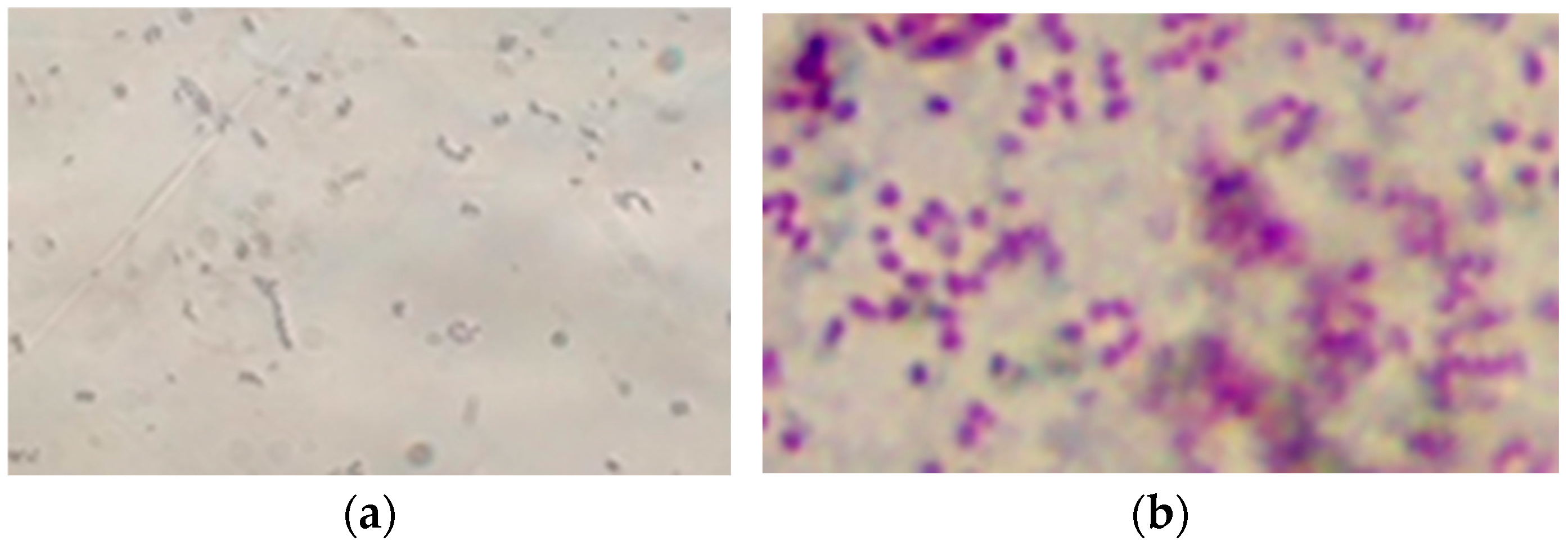
Bacterial cells of isolate PET1 are Gram-negative short rods with rounded ends, single or in pairs (Figure 4). Do not form spores. They are mobile, have a fast helical movement. Bacteria are catalase- and oxidase-positive.
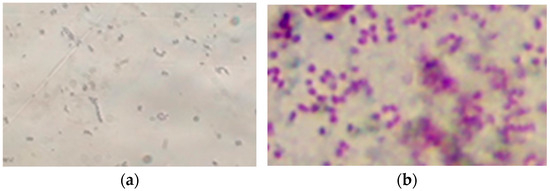
Figure 4.
Bacterial cells of isolate PET1 (light microscopy, immersion, magnification ×1000): (a) preparation-smear (crystal violet staining), bacterial cells are short rods with rounded ends; (b) Gram staining in Kalina’s modification, the cells stain red-purple, indicating that they are Gram-negative.
3.2. Molecular and Genetic Properties of the PET1 Isolate
As a result of the molecular genetic analysis of the genome of the PET1 strain, 16S rRNA gene fragments of 562 and 565 bp were amplified and sequenced. The initial comparison of the obtained sequence using the BLAST program showed 99.82% similarity with representatives of the genus Achromobacter: A. ruhlandii, A. denitificans, A. insolitus, A. animicus, A. xylosoxidans.
Based on the nucleotide sequences of the 16S rRNA gene of typical strains of the most similar species and the studied strain, a dendrogram of genetic similarity was constructed using the nearest neighbor algorithm and the two-parameter Kimura model (Figure 5). As can be seen from the figure, strain PET1 entered the same group as Achromobacter xylosoxidans species, which confirms its belonging to this species.
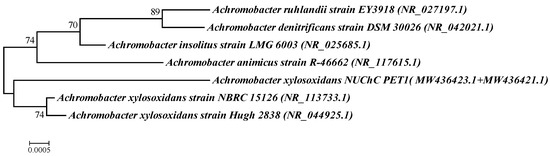
Figure 5.
Results of phylogenetic analysis of strain PET1 and other representatives of the genus Achromobacter (strain PET1 entered the same group as Achromobacter xylosoxidans species, which confirms its belonging to this species).
The nucleotide sequences were deposited in GenBank as Achromobacter xylosoxidans with accession numbers MW436423.1 and MW436421.1.
Therefore, the PET1 strain was identified as Achromobacter xylosoxidans based on a complex of microbiological and molecular genetic features.
According by the biosafety level bacterium A. xylosoxidans belong to the 2nd risk group according to the German Technical Regulations for Biological Agents [33]. Microorganisms of risk group 2 are characterized by a moderate individual risk and low social risk. Risk group 2 includes pathogens that can cause disease in humans or animals but are unlikely to pose a serious risk to laboratory workers, society, livestock, or the environment. Available methods of prevention and treatment effectively prevent the risk of such infections spreading in the population [34,35,36]. Working with microorganisms of risk group 2 is allowed in bacteriological laboratories of medical institutions, diagnostic and research laboratories. Features of laboratory protection are the use of “correct microbiological techniques” in combination with protective clothing and biosafety labeling. Among the protective equipment should be a biological safety box [36]. Therefore, the isolated bacteria cannot be used for educational purposes but can be used in scientific research.
The bacterium A. xylosoxidans is an emerging and opportunistic pathogen in patients with various underlying diseases, such as Cystic Fibrosis and cancers [37]. Its genome is actively studied, in particular, as an example for the genomic study of emerging pathogens [37].
At the same time, the ability of A. xylosoxidans to participate in biogeochemical cycles is known. The heterotrophic bacterial strain Achromobacter xylosoxidans CF-S36, isolated from domestic wastewater, was involved in the study of biogeochemical cycles of carbon and nitrogen in heterotrophic bacteria with the ability for simultaneous nitrification and denitrification [38]. The studied strain effectively removed ammonia, nitrates, and nitrites using different carbon sources. According to the authors, the research conducted by them can be useful to biogeochemical engineers in order to understand the participation of heterotrophic bacteria in the global biogeochemical cycle and the prospects for further diverse applications of these microorganisms.
Achromobacter xylosoxidans strain A8, which can use 2-chlorobenzoate and 2,5-dichlorobenzoate as sole sources of carbon and energy, was isolated from soil contaminated with polychlorinated biphenyls [39]. The authors consider this strain as a microorganism for further development towards a bioremediation tool [39]. Schemes of metabolic pathways in A. xylosoxidans strain A8 are summarized as follows: nitrogen [40], sulfur [41], and 2-oxocarboxylic acid [42].
There are reports on the participation of A. xylosoxidans strain ACM-1 in nitrification processes [43]. The authors suggest that this microorganism can be used to remove ammonium from aquaculture farms [43]. The results of the full-genome sequence of the bacterium A. xylosoxidans strain NH44784-1996 showed the presence of both genes of anaerobic growth via denitrification and genes encoding the biofilm adhesin poly-β-1,6-N-acetyl-D-glucosamin in the genome of the studied bacterium [44]. At the same time, the ability of the studied strain to form a biofilm was confirmed experimentally in in vitro studies [44].
Felgate et al. [45] experimentally established the ability to denitrification in A. xylosoxidans. The ability for simultaneous aerobic denitrification and phosphorus removal, which is promising for the creation of new single-stage wastewater treatment systems, was shown for the Achromobacter sp. strain GAD3 obtained by high-throughput genome sequencing [46].
There is a report that among representatives of the genus Achromobacter Achromobacter denitrificans strain Ebl13 provided degradation of polyvinyl chloride (12.3%) and low-density polyethylene (6.5%) within 6 weeks [47]. At the same time, the processes of biodegradation of plastics with the participation of the bacterium A. xylosoxidans have not been sufficiently studied. Currently, the involvement of A. xylosoxidans strain PE1, isolated from soil, in the process of biodegradation of high-density polyethylene was first established by Kowalczyk et al. [17]. Another strain of A. xylosoxidans, also isolated from the soil, provided a decrease in microplastic’s dry weight by 26.7% (polyethylene microplastics) and 21.3% (nylon 6,6 microplastics) within 40 days, which was somewhat higher compared to the results obtained for mixed culture species in dominance of Pulmonis sp. [18]. The authors showed that laccase and peroxidase enzymes provided cleavage of the C-C bond, reducing the length of the polymer chain [18]. Achromobacter xylosoxidans strain Sb2 isolated from the soil of the plastic waste environment demonstrated after 4 weeks of the experiment the degradation of polyethylene (32.2%) and polyvinyl chloride (17%) samples [19]. Our study did not investigate the weight loss of PET film samples, which is a further prospect for evaluating the biodegradation potential of A. xylosoxidans strain NUChC PET1.
Polycyclic aromatic hydrocarbons are toxic environmental pollutants [48] and they can be found in plastics as a result of the addition of additives at various stages of production and manufacture [49]. The utilization of pyrene as the sole source of carbon by A. xylosoxidans strain PY4 was demonstrated [48]. Achromobacter xylosoxidans strain BP1 was also isolated from contaminated soil at a coal chemical site in northern China by Zhang et al. [50]. For this strain, the property for the degradation of phenanthrene and benzo[a]pyrene is shown [50].
The strain Achromobacter sp. was isolated from the biofilm of the surface of low density polyethylene and demonstrated the ability to degrade low density polyethylene beads (significant weight loss of the samples was about 8%) [51]. However, there are no reports on the isolation of Achromobacter xylosoxidans strains from the biofilm of plastics in the scientific and methodical database available to us. Therefore, the information on the isolation of the A. xylosoxidans strain NUChC PET1 from the biofilm of the surface of poly(ethylene terephthalate) taken from the soil, indicated in the presented study, is the first such report.
Biodegradation of PET with the participation of enzymes (enzymatic degradation) is currently considered the most promising [52,53]. Lipases and cutinases have been widely studied for the purpose of PET biodegradation [52]. A detailed review of the enzymatic mechanisms of PET biodegradation was presented by Kushwaha et al. [54], describing enzyme engineering and modification via mutagenesis, enzyme cooperativity, multiple enzyme utilization, enzyme-substrate interactions, expanded substrate specificity, and improvement of reaction conditions. The authors see future prospects in the development of approaches to PET biodegradation in the application of computational biology and automation, with attention to Industry 4.0, deep learning, and IoT.
A further perspective of our research is the assessment of the biochemical properties of the strain Achromobacter xylosoxidans PET1 with attention to the lipolytic properties, as well as the biofilm-forming properties of the strain on polymeric materials, in particular PET. Also, an important and necessary direction of research should be to determine the ability of the strain to degrade polymeric materials, which will allow us to finally answer the question of its involvement in the processes of cleaning the environment from plastic (in particular PET) and the possibility of classifying it as a bacterium involved in the biodeteriration of plastic pipes and coatings.
4. Conclusions
Thus, the strain NUChC PET1 was isolated from the biofilm that formed on the surface of the PET-bottle, which was located in the soil, on the modified Postgate’s “C” medium with PET as the only carbon source. Based on a complex of microbiological and molecular genetic features, the strain NUChC PET1 was identified as Achromobacter xylosoxidans (MW436423.1 and MW436421.1 in the GenBank). Bacteria of the species Achromobacter xylosoxidans are participants in the biogeochemical cycles of carbon, nitrogen, sulfur, and phosphorus, are known as biodegraders of a number of plastics, belong to risk group 2 (according to the German Technical Regulations for biological agents) and can be used during scientific research on the biodegradation of PET. This study is the first report of the isolation of an A. xylosoxidans strain from a biofilm on the surface of poly(ethylene terephthalate) collected from the soil. Considering the growing interest of researchers in the problem of biodegradation of plastics, attention should be paid to the potential of A. xylosoxidans as biodegradation agents. The perspective of further research may be the study of biochemical properties, the intensity of biofilm formation by this strain on the surface of artificial materials (in particular, plastics) and participation in the processes of their biodegradation.
Author Contributions
Conceptualization, N.T. and L.Z.; methodology, N.T. and L.Z.; formal analysis, N.T. and L.Z.; investigation, N.T. and L.Z.; resources, N.T. and L.Z.; writing—original draft preparation, N.T. and L.Z.; writing—review and editing, N.T. and L.Z.; visualization, N.T. and L.Z.; supervision, N.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data used in this research are available upon request.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Saveliev, Y.V.; Yanovich, I.V.; Akhranovych, O.R.; Markovska, L.A.; Veselov, V.Y. Creation and application of hydrocarbon-based polyurethanes that degrade under environmental conditions. Polymer. J. 2011, 33, 205–217. (In Ukrainian) [Google Scholar]
- Rogers, K.L.; Carreres-Calabuig, J.A.; Gorokhova, E.; Posth, N.R. Micro-by-micro interactions: How microorganisms influence the fate of marine microplastics. Limnol. Oceanogr. Lett. 2020, 5, 18–36. [Google Scholar] [CrossRef]
- Zhang, Y.; Ma, J.; Song, Y.-Q.; Li, G.; O’Connor, P. Stronger deterministic processes shape the plastisphere microbiota of biodegradable microplastics compared to non-biodegradable microplastics in farmland soil. Appl. Soil Ecol. 2024, 196, 105312. [Google Scholar] [CrossRef]
- Ya, H.; Xing, Y.; Zhang, T.; Lv, M.; Jiang, B. LDPE microplastics affect soil microbial community and form a unique plastisphere on microplastics. Appl. Soil Ecol. 2022, 180, 104623. [Google Scholar] [CrossRef]
- Liu, X.; Li, Y.; Yu, Y.; Yao, H. Effect of nonbiodegradable microplastics on soil respiration and enzyme activity: A meta-analysis. Appl. Soil Ecol. 2023, 184, 104770. [Google Scholar] [CrossRef]
- Andrushkiv, B.; Vovk, I.; Pogaidak, O. Improvement of the economic toolkit of finding new resources in the conditions of the post-Soviet society. Galician Econ. Herald. 2012, 3, 80–85. (In Ukrainian) [Google Scholar]
- Burak, O.M.; Kirichenko, A.Y. Creation of PET bottle collection points to improve the waste management process in Ukraine. In Proceedings of the International Science and Practice Internet Conference “Entrepreneurship and Business Administration”, Kharkiv, Ukraine, 10 February 2020. (In Ukrainian). [Google Scholar]
- Yoshida, S.; Hiraga, K.; Takehana, T.; Taniguchi, I.; Yamaji, H.; Maeda, Y.; Toyohara, K.; Miyamoto, K.; Kimura, Y.; Oda, K. A bacterium that degrades and assimilates poly(ethylene terephthalate). Science 2016, 351, 1196–1199. [Google Scholar] [CrossRef] [PubMed]
- Joo, S.; Cho, I.J.; Seo, H.; Son, H.F.; Sagong, H.Y.; Shin, T.J.; Choi, S.Y.; Lee, S.Y.; Kim, K.J. Structural insight into molecular mechanism of poly(ethylene terephthalate) degradation. Nat. Commun. 2018, 9, 382. [Google Scholar] [CrossRef]
- Chen, Z.; Wang, Y.; Cheng, Y.; Wang, X.; Tong, S.; Yang, H.; Wang, Z. Efficient biodegradation of highly crystallized polyethylene terephthalate through cell surface display of bacterial PETase. Sci. Total Environ. 2020, 709, 136138. [Google Scholar] [CrossRef]
- Vague, M.; Chan, G.; Roberts, C.; Swartz, N.A.; Mellies, J.L. Pseudomonas isolates degrade and form biofilms on polyethylene terephthalate (PET) plastic. bioRxiv 2019, 647321, 1–35. [Google Scholar] [CrossRef]
- Ru, J.; Huo, Y.; Yang, Y. Microbial Degradation and Valorization of Plastic Wastes. Front. Microbiol. 2020, 11, 442. [Google Scholar] [CrossRef] [PubMed]
- Chandramouli Swamy, T.M.; Nagarathna, S.V.; Reddy, P.V.; Nayak, A.S. Efficient biodegradation of Polyethylene terephthalate (PET) plastic by Gordonia sp. CN2K isolated from plastic contaminated environment. Ecotoxicol. Environ. Saf. 2024, 281, 116635. [Google Scholar] [CrossRef]
- Tkachuk, N.; Zelena, L. The Impact of Bacteria of the Genus Bacillus upon the Biodamage/Biodegradation of Some Metals and Extensively Used Petroleum-Based Plastics. Corros. Mater. Degrad. 2021, 2, 531–553. [Google Scholar] [CrossRef]
- Yan, Z.F.; Feng, C.Q.; Zhou, J.Q.; Huang, Q.S.; Chen, X.Q.; Xia, W.; Wu, J. Complete degradation of PET waste using a thermophilic microbe-enzyme system. Int. J. Biol. Macromol. 2024, 260 Pt 2, 129538. [Google Scholar] [CrossRef] [PubMed]
- Zeng, C.; Ding, F.; Zhou, J.; Dong, W.; Cui, Z.; Yan, X. Biodegradation of Poly(ethylene terephthalate) by Bacillus safensis YX8. Int. J. Mol. Sci. 2023, 24, 16434. [Google Scholar] [CrossRef]
- Kowalczyk, A.; Chyc, M.; Ryszka, P.; Latowski, D. Achromobacter xylosoxidans as a new microorganism strain colonizing high-density polyethylene as a key step to its biodegradation. Environ. Sci. Pollut. Res. 2016, 23, 11349–11356. [Google Scholar] [CrossRef]
- Tiwari, N.; Santhiya, D.; Sharma, J.G. Significance of landfill microbial communities in biodegradation of polyethylene and nylon 6,6 microplastics. J. Hazard Mater. 2024, 462, 132786. [Google Scholar] [CrossRef] [PubMed]
- Saeed, S.; Iqbal, A.; Deeba, F. Biodegradation study of Polyethylene and PVC using naturally occurring plastic degrading microbes. Arch. Microbiol. 2022, 204, 497. [Google Scholar] [CrossRef]
- Tkachuk, N.; Zelena, L.; Mazur, P.; Lukash, O. Genotypic, physiological and biochemical features of Desulfovibrio strains in a sulfidogenic microbial community isolated from the soil of ferrosphere. Ecol. Quest. 2020, 31, 79–88. [Google Scholar] [CrossRef]
- Tkachuk, N.; Zelena, L. Some corrosive bacteria isolated from the technogenic soil ecosystem in Chernihiv city (Ukraine). Stud. Quat. 2021, 38, 101–108. [Google Scholar] [CrossRef]
- Tkachuk, N.; Zelena, L. Bacterial sulfidogenic community from the surface of technogenic materials in vitro: Composition and biofilm formation. Biofouling 2023, 39, 327–338. [Google Scholar] [CrossRef]
- Tkachuk, N.; Zelena, L. Inhibition of heterotrophic bacterial biofilm in the soil ferrosphere by Streptomyces spp. and Bacillus velezensis. Biofouling 2022, 38, 916–925. [Google Scholar] [CrossRef]
- Tkachuk, N.; Zelena, L. The intensity of biofilm formation by heterotrophic bacteria isolated from soil ferrosphere. Ecol. Quest. 2023, 34, 37–41. [Google Scholar] [CrossRef]
- Tkachuk, N.; Zelena, L.; Mazur, P. Properties of anaerobic bacteria from ferrosphere crucial for biofilm development. Ecol. Quest. 2021, 32, 107–112. [Google Scholar]
- Tkachuk, N.; Zelena, L.; Lukash, O.; Mazur, P. Microbiological and genetic characteristics of Bacillus velezensis bacillibactin-producing strains and their effect on the sulfate-reducing bacteria biofilms on the poly(-ethylene terephthalate) surface. Ecol. Quest. 2021, 32, 119–129. [Google Scholar] [CrossRef]
- Postgate, J.R. The Sulfate-Reducing Bacteria; Cambridge University Press: Cambridge, UK, 1984; 224p. [Google Scholar]
- Furzikova, T.M.; Serhiychuk, M.G.; Vlasenko, V.V.; Shvets, Y.V.; Pozur, V.K. Microbiology. Practicum; Phytosocial Center: Kyiv, Ukraine, 2006; 210p. (In Ukrainian) [Google Scholar]
- Dikiy, I.L.; Holupyak, I.Y.; Sidorchuk, I.I. Microbiology. Laboratory Manual; Publishing house of NUPh “Zolotyie stranitsyi”: Harkov, Ukraine, 2002; 444p. (In Russian) [Google Scholar]
- Gerhardt, P. Methods for General and Molecular Bacteriology, 2nd ed.; Gerhardt, P., Murray, R.G.E., Krieg, N.R., Wood, W.A., Eds.; American Society for Microbiology: Washington, DC, USA, 1994; 791p. [Google Scholar]
- Lane, D.J. 16S/23S rRNA Sequencing. In Nucleic Acid Techniques in Bacterial Systematic; Stackebrandt, E., Goodfellow, M., Eds.; John Wiley and Sons: New York, NY, USA, 1991; pp. 115–175. [Google Scholar]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Bio. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- TRBA 466 “Einstufung von Prokaryonten (Bacteria und Archaea) in Risikogruppen”. Available online: https://www.baua.de/DE/Angebote/Rechtstexte-und-Technische-Regeln/Regelwerk/TRBA/pdf/TRBA-466.pdf?__blob=publicationFile&v=4 (accessed on 28 September 2024).
- Directive 2000/54/EC of the European Parliament and of the Council of 18 September 2000 on the Protection of Workers from Risks Related to Exposure to Biological Agents at Work, 2000. Official Journal of the European Communities. 17 October 2000, pp. 22–45. Available online: https://osha.europa.eu/en/legislation/directives/exposure-to-biological-agents/77 (accessed on 29 September 2024).
- Zlenko, V.V.; Piryatinska, N.E.; Lytvynenko, M.I.; Yavorska, O.M.; Denyshchuk, G.P.; Nechvoglod, T.O.; Slabodchykova, S.Y. (Eds.) Organization of Work and Provision of Sanitary and Anti-Epidemic Regime in Laboratory-Diagnostic Institutions of Various Profiles: Teaching. Manual; Under the editorship Zalyubovska, O.I.; KhNMU: Kharkiv, Ukraine, 2015; 56p, Available online: https://repo.knmu.edu.ua/server/api/core/bitstreams/49027a42-4e81-4cd7-b61b-7893a0b05abd/content (accessed on 29 September 2024). (In Ukrainian)
- Golubnycha, V.M.; Pogorelov, M.V.; Kornienko, V.V. Biosafety and Bioprotection in Biological Laboratories of the 1st and 2nd Levels of Biosafety: Monograph; Sumy State University: Sumy, Ukraine, 2016; 123p, Available online: https://core.ac.uk/download/pdf/141450992.pdf (accessed on 29 September 2024). (In Ukrainian)
- Pongchaikul, P. Comparative Genomic Approaches to Understanding Achromobacter xylosoxidans. Ph.D. Thesis, University of Liverpool, Liverpool, UK, September 2015. Available online: https://livrepository.liverpool.ac.uk/2050219/1/PongchaikulPis_Sep2015_2050219.pdf (accessed on 11 February 2024). [CrossRef]
- Padhi, S.K.; Maiti, N.K. Molecular insight into the dynamic central metabolic pathways of Achromobacter xylosoxidans CF-S36 during heterotrophic nitrogen removal processes. J. Biosci. Bioeng. 2017, 123, 46–55. [Google Scholar] [CrossRef] [PubMed]
- Strnad, H.; Ridl, J.; Paces, J.; Kolar, M.; Vlcek, C.; Paces, V. Complete genome sequence of the haloaromatic acid-degrading bacterium Achromobacter xylosoxidans A8. J. Bacteriol. 2011, 193, 791–792. [Google Scholar] [CrossRef] [PubMed]
- KEEG. Nitrogen Metabolism—Achromobacter xylosoxidans A8. Available online: https://www.genome.jp/kegg-bin/show_pathway?axy00910 (accessed on 11 February 2024).
- KEEG. Sulfur Metabolism—Achromobacter xylosoxidans A8. Available online: https://www.genome.jp/kegg-bin/show_pathway?axy00920 (accessed on 11 February 2024).
- KEEG. 2-Oxocarboxylic Acid Metabolism—Achromobacter xylosoxidans A8. Available online: https://www.kegg.jp/pathway/axy01210+AXYL_02631 (accessed on 11 February 2024).
- Basha, K.A.; Joseph, T.; Lalitha, K.V.; Vineetha, D.; Rathore, G.; Tripati, G.; Prasad, K. Nitrification Potential of Achromobacter xylosoxidans Isolated from Fresh Water Finfish Farms of Kerala, India. Int. J. Curr. Microbiol. App. Sci. 2018, 7, 2645–2654. [Google Scholar] [CrossRef]
- Jakobsen, T.H.; Hansen, M.A.; Jensen, P.Ø.; Hansen, L.; Riber, L.; Cockburn, A.; Kolpen, M.; Rønne Hansen, C.; Ridderberg, W.; Eickhardt, S.; et al. Complete genome sequence of the cystic fibrosis pathogen Achromobacter xylosoxidans NH44784-1996 complies with important pathogenic phenotypes. PLoS ONE 2013, 8, e68484. [Google Scholar] [CrossRef]
- Felgate, H.; Giannopoulos, G.; Sullivan, M.J.; Gates, A.J.; Clarke, T.A.; Baggs, E.; Rowley, G.; Richardson, D.J. The impact of copper, nitrate and carbon status on the emission of nitrous oxide by two species of bacteria with biochemically distinct denitrification pathways. Environ. Microbiol. 2012, 14, 1788–1800. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Chen, Q.; Ma, T.; Wang, M.; Ni, J. Genomic insights into metabolic potentials of two simultaneous aerobic denitrification and phosphorus removal bacteria, Achromobacter sp. GAD3 and Agrobacterium sp. LAD9. FEMS Microbiol. Ecol. 2018, 94, fiy020. [Google Scholar] [CrossRef] [PubMed]
- Rad, M.M.; Moghimi, H.; Azin, E. Biodegradation of thermo-oxidative pretreated low-density polyethylene (LDPE) and polyvinyl chloride (PVC) microplastics by Achromobacter denitrificans Ebl13. Mar. Pollut. Bull. 2022, 181, 113830. [Google Scholar] [CrossRef]
- Nzila, A.; Ramirez, C.O.; Musa, M.M.; Sankara, S.; Basheer, C.; Li, Q.X. Pyrene biodegradation and proteomic analysis in Achromobacter xylosoxidans, PY4 strain. Int. Biodeter. Biodegrad. 2018, 130, 40–47. [Google Scholar] [CrossRef]
- Alassali, A.; Calmano, W.; Gidarakos, E.; Kuchta, K. The degree and source of plastic recycles contamination with polycyclic aromatic hydrocarbons. RSC Adv. 2020, 10, 44989. [Google Scholar] [CrossRef]
- Zhang, B.; Xu, W.; Ma, Y.; Gao, X.; Ming, H.; Jia, J. Effects of bioaugmentation by isolated Achromobacter xylosoxidans BP1 on PAHs degradation and microbial community in contaminated soil. J. Environ. Manag. 2023, 334, 117491. [Google Scholar] [CrossRef]
- Dey, A.S.; Bose, H.; Mohapatra, B.; Sar, P. Biodegradation of Unpretreated Low-Density Polyethylene (LDPE) by Stenotrophomonas sp. and Achromobacter sp., Isolated From Waste Dumpsite and Drilling Fluid. Front. Microbiol. 2020, 11, 603210. [Google Scholar] [CrossRef] [PubMed]
- Sui, B.; Wang, T.; Fang, J.; Hou, Z.; Shu, T.; Lu, Z.; Liu, F.; Zhu, Y. Recent advances in the biodegradation of polyethylene terephthalate with cutinase-like enzymes. Front. Microbiol. 2023, 14, 1265139. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Chen, Y.; Zhang, L.; Wu, J.; Zeng, X.; Shi, X.; Liu, L.; Chen, J. A comprehensive review on enzymatic biodegradation of polyethylene terephthalate. Environ. Res. 2024, 240 Pt 2, 117427. [Google Scholar] [CrossRef]
- Kushwaha, A.; Goswami, L.; Singhvi, M.; Kim, B.S. Biodegradation of poly(ethylene terephthalate): Mechanistic insights, advances, and future innovative strategies. Chem. Eng. J. 2023, 457, 141230. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).