Dynamics Insight of Dodonaea viscosa Phytochemicals as a Potent Inhibitor Targeting Dengue Virus NS5 Methyltransferase †
Abstract
1. Introduction
2. Materials and Methods
2.1. Target and Ligands Preparation
2.2. Grid Generation and Docking Investigation
2.3. Molecular Dynamics Simulation
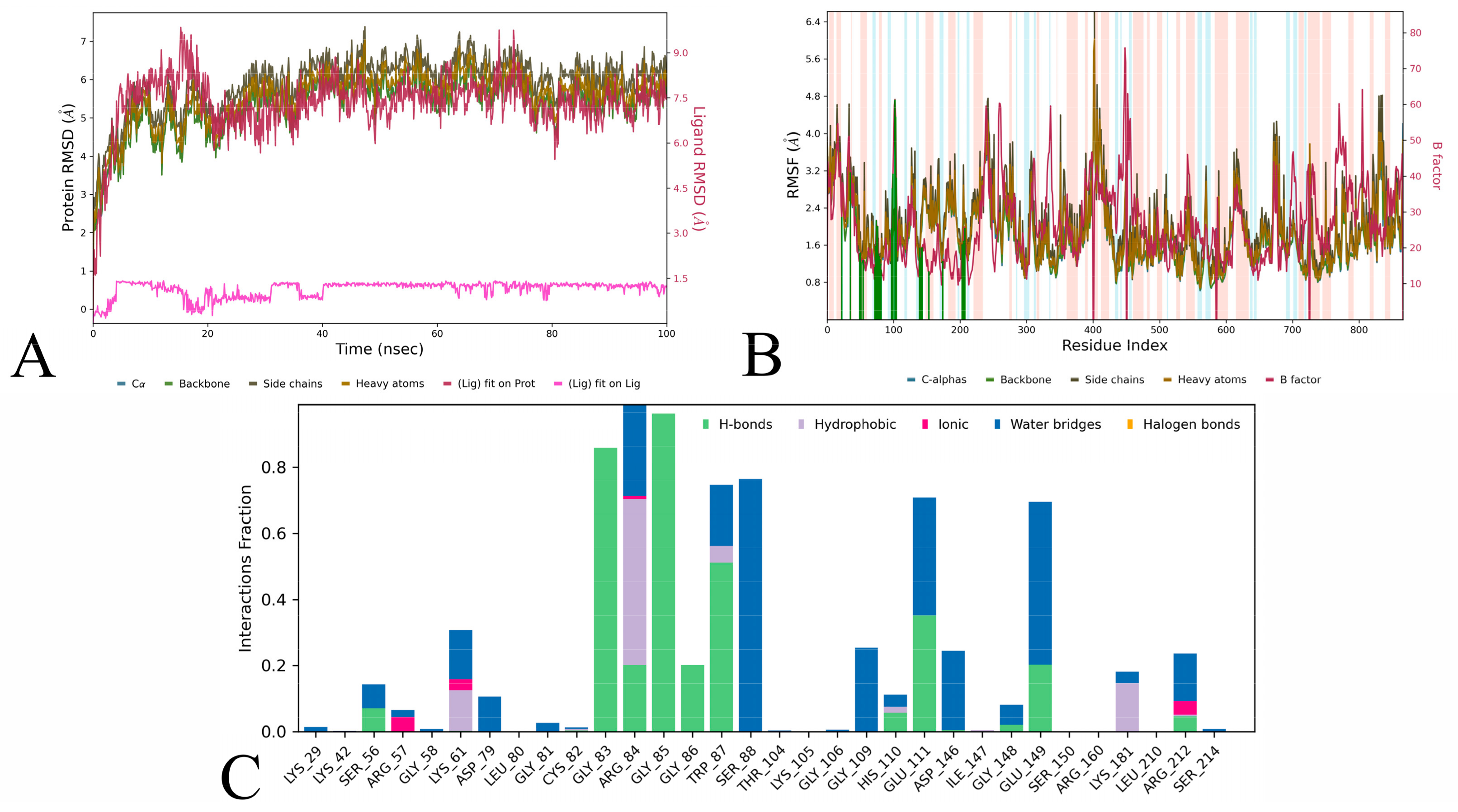
3. Results and Discussion
3.1. Target and Ligand Retrieval and Their Preparation
3.2. Grid Generation and Docking Investigation
3.3. Molecular Dynamics Simulation
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ali, S.; Ali, U.; Safi, K.; Naz, F.; Jan, M.I.; Iqbal, Z.; Ali, T.; Ullah, R.; Bari, A. In silico homology modeling of dengue virus non-structural 4B (NS4B) protein and its molecular docking studies using triterpenoids. BMC Infect. Dis. 2024, 24, 688. [Google Scholar] [CrossRef] [PubMed]
- Abduljalil, J.M.; Abd Al Galil, F.M. Molecular pathogenesis of dengue virus infection in Aedes mosquitoes. J. Insect Physiol. 2022, 138, 104367. [Google Scholar] [CrossRef] [PubMed]
- Leta, S.; Beyene, T.J.; De Clercq, E.M.; Amenu, K.; Kraemer, M.U.; Revie, C.W. Global risk mapping for major diseases transmitted by Aedes aegypti and Aedes albopictus. Int. J. Infect. Dis. 2018, 67, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Mazri, R.; Ouassaf, M.; Kerassa, A.; Alhatlani, B.Y. Exploring potential therapeutics: Targeting dengue virus NS5 through molecular docking, ADMET profiling, and DFT analysis. Chem. Phys. Impact 2024, 8, 100468. [Google Scholar] [CrossRef]
- Kularatne, S.A.; Dalugama, C. Dengue infection: Global importance, immunopathology and management. Clin. Med. 2022, 22, 9–13. [Google Scholar] [CrossRef]
- Martina, B.E.; Koraka, P.; Osterhaus, A.D. Dengue virus pathogenesis: An integrated view. Clin. Microbiol. Rev. 2009, 22, 564–581. [Google Scholar] [CrossRef]
- El Sahili, A.; Lescar, J. Dengue Virus Non-Structural Protein 5. Viruses 2017, 9, 91. [Google Scholar] [CrossRef]
- Ullah, A.; Gong, P.; Khan, A.M.; Choudhary, M.I. Identification of new inhibitors of NS5 from dengue virus using saturation transfer difference (STD-NMR) and molecular docking studies. RSC Adv. 2022, 13, 355–369. [Google Scholar] [CrossRef]
- Kachhadiya, D.K.; Pooja, K.; Mishra, S.K.; George, J.J. In Silico Based Identification Of Novel Inhibitors For Selected MDR Protein From Shigella Species: A Validation Through Molecular Docking Analysis. Educ. Adm. Theory Pract. 2024, 30, 309–316. [Google Scholar] [CrossRef]
- Mishra, S.K.; Priya, P.; Rai, G.P.; Haque, R.; Shanker, A. Coevolution based immunoinformatics approach considering variability of epitopes to combat different strains: A case study using spike protein of SARS-CoV-2. Comput. Biol. Med. 2023, 163, 107233. [Google Scholar] [CrossRef]
- Vaghasia, V.V.; Sharma, K.; Mishra, S.K.; Georrge, J.J. In silico identification of natural product inhibitor for multidrug resistance proteins from selected gram-positive bacteria. In Nanotechnology and In Silico Tools; Elsevier: Amsterdam, The Netherlands, 2024; pp. 309–317. [Google Scholar]
- Vakhariya Sakina, S.; Mishra, S.K.; Sharma, K.; Georrge, J.J. Designing of a novel curcumin analogue to inhibit mitogen-activated protein kinase: A cheminformatics approach. J. Phytonanotechnol. Pharm. Sci. 2023, 3, 37–47. [Google Scholar]
- Vinjoda, P.; Mishra, S.K.; Sharma, K.; Georrge, J.J. In silico identification of novel drug target and its natural product inhibitors for herpes simplex virus. In Nanotechnology and In Silico Tools; Elsevier: Amsterdam, The Netherlands, 2024; pp. 377–383. [Google Scholar]
- Mishra, S.K.; Praba, J.J.; Georrge, J.J. An emerging trends of bioinformatics and big data analytics in healthcare. In Digital Transformation in Healthcare 5.0: Volume 2: Metaverse, Nanorobots and Machine Learning; Walter de Gruyter GmbH & Co KG.: Berlin, Germany, 2024; p. 159. [Google Scholar]
- Al-Snafi, A.E. A review on Dodonaea viscosa: A potential medicinal plant. IOSR J. Pharm. 2017, 7, 10–21. [Google Scholar] [CrossRef]
- Madhavi Sastry, G.; Adzhigirey, M.; Day, T.; Annabhimoju, R.; Sherman, W. Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 2013, 27, 221–234. [Google Scholar] [CrossRef] [PubMed]
- Kim, S. Exploring Chemical Information in PubChem. Curr. Protoc. 2021, 1, e217. [Google Scholar] [CrossRef]
- Mishra, S.K.; Roy, S.; Chhetri, T.; Georrge, J.J. Computer-assisted Methods and Tools for Structure-and Ligand-based Drug Design. In Computational Methods for Rational Drug Design; John Wiley & Sons: Hoboken, NJ, USA, 2025; pp. 69–95. [Google Scholar]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [CrossRef]
- Sayyed, S.K.; Quraishi, M.; Jobby, R.; Rameshkumar, N.; Kayalvizhi, N.; Krishnan, M.; Sonawane, T. A computational overview of integrase strand transfer inhibitors (INSTIs) against emerging and evolving drug-resistant HIV-1 integrase mutants. Arch. Microbiol. 2023, 205, 142. [Google Scholar] [CrossRef]
- Bhachoo, J.; Beuming, T. Investigating protein–peptide interactions using the Schrödinger computational suite. In Modeling Peptide-Protein Interactions: Methods and Protocols; Humana: New York, NY, USA, 2017; pp. 235–254. [Google Scholar]
- Saini, R.; Kumar, V.; Patel, C.N.; Sourirajan, A.; Dev, K. Synergistic antibacterial activity of Phyllanthus emblica fruits and its phytocompounds with ampicillin: A computational and experimental study. Naunyn Schmiedebergs Arch. Pharmacol. 2024, 397, 857–871. [Google Scholar] [CrossRef]
- Bouzidi, A.; Azizi, A.; Messaoudi, O.; Abderrezzak, K.; Vidari, G.; Hellal, A.N.; Patel, C.N. Phytochemical analysis, biological activities of methanolic extracts and an isolated flavonoid from Tunisian Limoniastrum monopetalum (L.) Boiss: An in vitro and in silico investigations. Sci. Rep. 2023, 13, 19144. [Google Scholar] [CrossRef]
- Abouelwafa, M.; Ibrahim, T.M.; El-Hadidi, M.S.; Mahnashi, M.H.; Owaidah, A.Y.; Saeedi, N.H.; Attia, H.G.; Georrge, J.J.; Mostafa, A. Using CADD tools to inhibit the overexpressed genes FAP, FN1, and MMP1 by repurposing ginsenoside C and Rg1 as a treatment for oral cancer. Front. Mol. Biosci. 2023, 10, 1248885. [Google Scholar] [CrossRef]
- Salamat, A.; Kosar, N.; Mohyuddin, A.; Imran, M.; Zahid, M.N.; Mahmood, T. SAR, Molecular Docking and Molecular Dynamic Simulation of Natural Inhibitors against SARS-CoV-2 Mpro Spike Protein. Molecules 2024, 29, 1144. [Google Scholar] [CrossRef]
- Mohanraj, K.; Karthikeyan, B.S.; Vivek-Ananth, R.P.; Chand, R.B.; Aparna, S.R.; Mangalapandi, P.; Samal, A. IMPPAT: A curated database of Indian Medicinal Plants, Phytochemistry And Therapeutics. Sci. Rep. 2018, 8, 4329. [Google Scholar] [CrossRef]
- Duke, J.A. Database of Biologically Active Phytochemicals & Their Activity; CRC Press: Boca Raton, FL, USA, 2020. [Google Scholar]
- Shinbo, Y.; Nakamura, Y.; Altaf-Ul-Amin, M.; Asahi, H.; Kurokawa, K.; Arita, M.; Saito, K.; Ohta, D.; Shibata, D.; Kanaya, S. KNApSAcK: A comprehensive species-metabolite relationship database. In Plant Metabolomics. Biotechnology in Agriculture and Forestry; Springer: Berlin/Heidelberg, Germany, 2006; pp. 165–181. [Google Scholar]


| Sl. No. | PubChem ID | Name | Docking Score (Kcal/Mol) |
|---|---|---|---|
| 1 | 5280343 | Quercetin | −7.164 |
| 2 | 440833 | Leucocianidol | −6.839 |
| 3 | 5281654 | Isorhamnetin | −6.392 |
| 4 | 5320462 | Penduletin | −5.850 |
| 5 | 5280863 | kaempferol | −5.837 |
| 3034034 | Quinine (Control) | −3.050 |
| Name | #Stars | CNS | mol MW | RuleOfFive |
|---|---|---|---|---|
| Quercetin | 0 | −2 | 302.24 | 0 |
| Leucocianidol | 0 | −2 | 306.271 | 1 |
| Isorhamnetin | 0 | −2 | 316.267 | 0 |
| Penduletin | 0 | −1 | 344.32 | 0 |
| Kaempferol | 0 | −2 | 286.24 | 0 |
| Quinine (Control) | 0 | −1 | 324.422 | 0 |
| Range | 0 to 5 | −2 (inactive), +2 (active) | 130.0 to 725.0 | Maximum 4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mishra, S.K.; Roy, S.; Chhetri, T.; Patel, C.; Georrge, J.J. Dynamics Insight of Dodonaea viscosa Phytochemicals as a Potent Inhibitor Targeting Dengue Virus NS5 Methyltransferase. Biol. Life Sci. Forum 2024, 35, 12. https://doi.org/10.3390/blsf2024035012
Mishra SK, Roy S, Chhetri T, Patel C, Georrge JJ. Dynamics Insight of Dodonaea viscosa Phytochemicals as a Potent Inhibitor Targeting Dengue Virus NS5 Methyltransferase. Biology and Life Sciences Forum. 2024; 35(1):12. https://doi.org/10.3390/blsf2024035012
Chicago/Turabian StyleMishra, Saurav Kumar, Sneha Roy, Tabsum Chhetri, Chirag Patel, and John J. Georrge. 2024. "Dynamics Insight of Dodonaea viscosa Phytochemicals as a Potent Inhibitor Targeting Dengue Virus NS5 Methyltransferase" Biology and Life Sciences Forum 35, no. 1: 12. https://doi.org/10.3390/blsf2024035012
APA StyleMishra, S. K., Roy, S., Chhetri, T., Patel, C., & Georrge, J. J. (2024). Dynamics Insight of Dodonaea viscosa Phytochemicals as a Potent Inhibitor Targeting Dengue Virus NS5 Methyltransferase. Biology and Life Sciences Forum, 35(1), 12. https://doi.org/10.3390/blsf2024035012






