Abstract
Halophiles are microorganisms that inhabit saline and hypersaline environments, requiring salinity to survive in such extreme conditions. These microorganisms are mainly researched for their biotechnological potential. This study aims to investigate the phenology of the studied strain, Idiomarina loihiensis, and to demonstrate its extracellular proteolytic activity, as well as the production of a protease via batch fermentation in halophilic microorganisms. Macroscopic studies revealed small colonies (≤5 mm) with a convex spherical structure, regular outline, smooth surface, and color ranging from beige to opaque cream. Protease production was investigated in high-salinity conditions with a moderately halophilic bacterium using basal media with varying nitrogen sources. This study found that the highest proteolytic activity occurred in media with tryptone and casein peptone as nitrogen sources, at pH 10, a temperature of 70 °C, and 22.5% salt concentration. The results also demonstrated that the studied protease was a thermostable enzyme.
1. Introduction
Extremophile microorganisms dwelling in harsh environmental habitats have evolved unique properties that can be of biotechnological and commercial significance. These non-conventional microorganisms living in such extremely hostile environmental habitats possess special adaptation strategies that make them interesting [1], and have developed the capacity to produce extremophile enzymes such as proteases with biotechnological potential [2].
Besides being intrinsically stable and active at high salt concentrations, halophilic enzymes offer important opportunities in biotechnological applications, such as food processing, environmental bioremediation, and biosynthetic processes [3].
It is important to highlight that the use of enzymes from halophiles in industrial applications is not limited to their stability at high salt concentrations, since these extremozymes are usually also tolerant to high temperatures and they are stable in the presence of organic solvents [2,3].
Halophilic microbes isolated from different saline soils have been found to produce proteases with potential industrial biotechnological value [4]. They are most significant as their properties can be easily modified through genetic manipulations to suit their various applications [5].
The industrial demand of proteases with special properties extends to stimulating a search for new enzymes. The genus Bacillus is the main producer of industrial alkaline proteases; however, other genus such as Pseudomonas and Streptomyces isolated from various environments have also been reported as producers of alkaline and thermostable proteases with industrial importance. Likewise, Idiomarina could represent an important source of novel proteases with commercial applications [2].
The aim of this study was to characterize the phenology and demonstrate the extracellular proteolytic activity of the studied strain Idiomarina loihiensis and to produce a protease via batch fermentation in halophilic microorganisms.
2. Materials and Methods
2.1. Samples and Strain Isolation and Identification
A moderately halophilic bacterium was isolated from an evaporation table in Sebkhet Ez-Zemoul (Ain M’lila, eastern Algeria, https://maps.app.goo.gl/JfXTqcLnfk7jFwAk8) and its proteolytic potential was confirmed using agar with 1% casein/gelatin added (MGM solid medium—0.3% peptone and 0.1% yeast extract with 12.5% salt) and via an enzymatic test on liquid medium in the presence of azocasein [6]. Molecular identification was carried out using the 16S rRNA gene [7].
2.2. Morphology and Physiology
The strain was examined for morphology analysis using cells from exponentially growing cultures. The aspect of the colonies was examined. Cell morphology and Gram staining were observed. Its proteolytic activity was tested on MGM (modified growth medium) solid media (0.3% peptone and 0.1% yeast extract) supplemented with 1% (w/v) casein [8] or 1% (w/v) gelatin [9]; positive results were detected after 5 days at 37 °C due to the presence of a precipitate around the colonies for casein and/or a translucent halo (after an addition of Frazier’s reagent) for gelatin. Growth rates were estimated on solid MGM medium at different salt concentrations (0–27%), pH levels (5–10) when incubated at 37 °C, and temperatures (12–55 °C) under aerobic conditions for 24–72 h.
2.3. Inoculum Preparation and Halophilic Protease Production
The inoculum was prepared from a 24 h culture on Petri dishes, inoculated into tubes containing 5 mL MGM medium with 10% salt, and incubated at 37 °C for 48 h. The enzyme extract was prepared by inoculating 10% v/v bacterial suspension into 250 mL flasks containing 45 mL production medium (MGM 0.05% peptone and 0.01% yeast extract) supplemented with different nitrogen sources: glycine, peptone, gelatin, casein, casein-peptone, tryptone, cream, and gelatin-tryptone. The pH, salt concentration, and temperature were adjusted according to the optimal growth conditions for the culture [10].
The media were incubated in a shaking bath at 37 °C for 6 days. The cultures were then centrifuged (4000× g for 10 min) and the supernatant was cooled to 0 °C. To compare the proteolytic activity across the different nitrogen sources, a two-factor analysis of variance was performed using Minitab 18 Statistical software.
2.4. Enzymatic Assay
The enzymatic assay was performed using two methods to confirm proteolytic activity at 0% and 10% salinity. A positive control with proteinase K and a blank assay (with an addition of TCA prior to reaction) were performed each time.
2.4.1. Method 1
Proteolytic activity was determined according to the method of Tsuchida et al. (1986) [11]. The reaction mixture contained 1 mL of 2% casein (prepared in phosphate buffer at pH 7) and 1 mL of the enzyme extract. The mixture was incubated in a water bath for 30 min at 40 °C (standard conditions). The enzymatic reaction was stopped with an addition of 2 mL of 0.4 M TCA (Trichloroacetic acid) and centrifuged at 4000× g for 10 min. Then, 1 mL of the supernatant was transferred to another tube, to which 5 mL of 0.4 M Na2CO3 and 1 mL of 10% v/v Folin’s calcium reagent were added. The reaction was incubated for 20 min at 40 °C in the dark. The absorbance was measured at a wavelength of 750 nm. One unit of proteolytic activity was defined as the amount of enzyme capable of releasing 1 µg of tyrosine per ml/min [11].
2.4.2. Method 2
Halophilic protease activity was determined according to the method of Brock et al. (1982) modified by Chuprom et al. (2016) [10], using azocasein (sulfanilamide azocasein) as the substrate. For this method, 1 mL of a reaction mixture consisting of 1/1 crude enzyme (cell-free supernatant) and 0.8% (w/v) azocasein (in 0.1 M Tris-HCl buffer (pH 8.0) at 10% (w/v) salinity) was incubated at 40 °C for 2 h in a bain-marie. The reaction was stopped by adding 1 mL of 10% (w/v) TCA solution and the mixture was allowed to stand for 30 min at room temperature. After centrifugation at 4000× g for 10 min, 600 µL of the supernatant was transferred to a microcentrifuge tube and 700 µL of 1.0 N NaOH was added. Absorbance was measured at 440 nm using a UV-vis spectrophotometer (one unit (U) of halophilic protease activity was defined as the amount of enzymatic activity that produced a 0.01 change in absorbance at 440 nm in 2 h at 40 C under standard assay conditions). Halophilic protease activity was calculated using the following equation:
where A and B are the optical densities of the crude enzyme and the control, respectively; Vt is the total reaction volume; and Ve is the volume of crude enzyme [10].
2.5. Partial Physico-Chemical Characterization of the Enzyme Extract
In order to determine the parameters required for optimum proteolytic activity, the enzymatic reaction was carried out in different buffer solutions from pH 4 to 12, at different temperatures (30, 40, 50, 60, 70, 80, and 90 °C) for 30 min, in the presence of 0–30% salt. Finally, to study thermostability, the reaction mixture was heated to 70 °C for 4 periods (30, 60, 90, and 120 min) and the residual activity was compared with the optimum activity. Proteolytic activity was determined in duplicate via the Tsuchida method [12].
3. Results and Discussion
A moderately halophilic bacterium strain producing a halophilic extracellular protease was isolated from a hypersaline environment. The bacteria hydrolyzed casein and gelatin, demonstrating their proteolytic potential. The halophilic strain was identified as Idiomarina loihiensis.
3.1. Strain Isolation and Identification
The studied strain showed small colonies (≤5 mm) with a domed spherical shape, regular outline, and smooth surface, beige to opaque cream in color. Gram staining using the technique of Dussault, 1955 [13] showed that the strain comprised Gram-negative small rods with cells arranged in chains or pairs, or dispersed, as is the case with most isolates of moderately halophilic, halotolerant, and halophilic bacteria isolated from different saline habitats [14,15,16]. The halophilic bacterium was identified as Idiomarina loihiensis through molecular characterization.
3.2. Nitrogen Source
Different nitrogen sources affecting the production of halophilic protease by the isolate induce differences in activity depending on the dosing method and the salinity of the medium, as shown in Figure 1. In the absence of salt, the most cost-effective nitrogen sources are peptone for the azocasein method and casein peptone for the Tsuchida method, yielding higher activity than the positive control. However, in the presence of 10% (w/v) salt, the best nitrogen source for both methods is tryptone, which results in greater proteolysis compared to proteinase K (the control). Our analysis of variance confirms that tryptone is the best carbon source (Table 1).
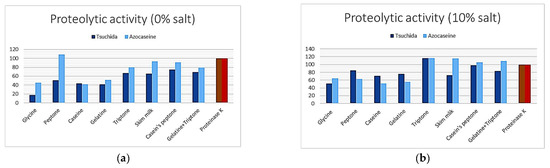
Figure 1.
Proteolytic activity of enzymatic curd from Idiomarina loihiensis against various nitrogen sources, (a) at 0% salt and (b) at 10% salt concentrations.

Table 1.
Variance analysis (Tukey’s test): Tukey’s method and 95% confidence level for grouping information.
3.3. Partial Physico-Chemical Characterization
A characteristic curve of the effect of the pH on enzymatic activity is shown in Figure 2a. Maximum activity is observed at pH 10; before and after this value, it decreases until reaching a minimum at pH 7, leading to the conclusion that our extract contains alkaline proteases. Similar results have been obtained with proteases produced by the moderate halophilic bacterium Halobacillus karajensis, whose optimum pH is 9 [17]; other proteolytic enzymes of Bacillus luteus H11 have a pH optimum of 10.5 [18]. Other similar results have been obtained with proteases produced by Thermoactinomyces sp. HS682, whose optimum pH is 11 [19].
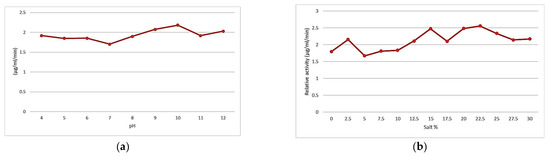
Figure 2.
Physiological effects of pH (a) and salinity (b) on the proteolytic activity of enzyme curd.
Figure 2b shows a characteristic curve of the effect of salinity on enzymatic activity. Indeed, the maximum activity is observed at 22.5% salt, with a maximum activity of 2.55 U/mL/min. Similar results were obtained for the halophilic bacteria Halobacillus sp. SR5-3, which showed optimum activity at 20% salt. The activity of the enzyme increased approximately 2.5-fold after the addition of 20 to 35% salt, and the enzyme was strongly stabilized by the salt.
Regarding the effect of the temperature, it can be seen that the proteolytic activity increases at a temperature of 70 °C, yet sharply declines beyond this temperature threshold (Figure 3a). Consequently, we can infer that the enzyme extract exhibits maximum proteolytic activity at 70 °C. Results similar to ours have been obtained for the strain Chromohalobacter sp. TVSP101, which shows a protease with maximum activity at 75 °C [20]. The strain Thermoactinomyces sp. HS682 also has an active protease at 70 °C [11].
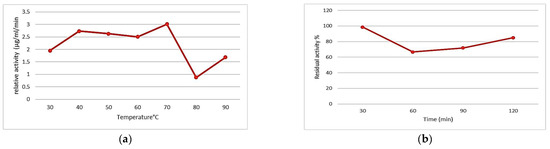
Figure 3.
Effect of temperature on (a) proteolytic activity and (b) the thermal stability of the enzyme curd.
Thermostability analysis of the enzyme extract at the optimum temperature showed the maximum activity to be at 30 min, as shown in Figure 3b, and a 33% loss of activity at 60 min, which remained stable until 90 min and increased to 86% after 120 min. The results obtained reveal that the protease from the Idiomarina loihiensis strain is a thermostable enzyme. We can compare this with other thermostable bacterial enzymes such as thermolysin, a protease produced by Bacillus thermoproteolyticus, which retains half of its activity after one hour at 80 °C [21].
4. Conclusions
The moderately halophilic strain Idiomarina loihiensis secretes a proteolytic enzyme extract that exhibits maximum activity at 22.5% salinity, pH 10, and a temperature of 70 °C, maintaining more than 80% of its proteolytic activity after 120 min at this temperature.
This shows that we are dealing with a halophilic, alkaliphilic, and thermostable protease capable of maintaining a good hydrolytic activity under extreme conditions, which makes it very interesting from a biotechnological point of view.
Author Contributions
Conceptualization, methodology, software, validation, formal analysis, investigation, resources, data curation, writing—original draft preparation, writing—review and editing, H.B. and K.K.; visualization, H.B.; supervision, K.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article.
Acknowledgments
We kindly thank R. Guessoum and F. Z. Boumezouzd for their contribution.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Jothi Basu, M.; Indra, V.; Hemapriya, J.; Vijayanand, S. Bioprocess Optimization of Halophilic Protease Production by a Halophilic Bacterial Strain JS4. Int. J. Curr. Res. Acad. Rev. 2015, 3, 309–315. Available online: www.ijcrar.com (accessed on 26 June 2019).
- Flores-Fernández, C.N.; Chávez-Hidalgo, E.; Santos, M.; Zavaleta, A.I.; Arahal, D.R. Molecular characterization of protease producing Idiomarina species isolated from Peruvian saline environments. Microbiol. Biotechnol. Lett. 2019, 47, 4001–4411. [Google Scholar] [CrossRef]
- De Lourdes Moreno, M.; Pérez, D.; García, M.; Mellado, E. Halophilic bacteria as a source of novel hydrolytic enzymes. Life 2013, 3, 38–51. [Google Scholar] [CrossRef] [PubMed]
- Corral, J.C.C.; De Los Santos Villalobos, S.; Barrgàn, L.A.P.; Figueroa, J.J.B.; Vásquez-Murrieta, M.S.; Alvarado, M.I.E.; Chávez, L.A.C. Isolation of moderately halophilic bacteria in saline environments of sonora state searching for proteolytic hydrolases. Open Agric. 2018, 3, 207–213. [Google Scholar] [CrossRef]
- Asha, B.; Palaniswamy, M. Optimization of alkaline protease production by Bacillus cereus FT 1isolated from soil. J. Appl. Pharm. Sci. 2018, 8, 119–127. [Google Scholar] [CrossRef]
- Gomri, M.A.; Rico-Díaz, A.; Escuder-Rodríguez, J.-J.; Khaldi, T.E.M.; González-Siso, M.-I.; Kharroub, K. Production and Characterization of an Extracellular Acid Protease from Thermophilic Brevibacillus sp. OA30 Isolated from an Algerian Hot Spring. Microorganisms 2018, 6, 31. [Google Scholar] [CrossRef] [PubMed]
- Benmebarek, H.; Escuder-Rodríguez, J.J.; González-Siso, M.I.; Karroub, K. Test for the production and assay of the proteolytic activities of halophilic bacteria and archaea isolated from algerian hypersaline environments. Proceedings 2020, 66, 12. [Google Scholar] [CrossRef]
- Cojoc, R.; Merciu, S.; Popescu, G.; Dumitru, L.; Enache, M.; Kamekura, M.; Enache, M. Extracellular hydrolytic enzymes of halophilic bacteria isolated from a subterranean rock salt crystal. Rom. Biotechnol. Lett. 2009, 14, 4658–4664. Available online: https://www.researchgate.net/publication/228413520_Extracellular_hydrolytic_enzymes_of_halophilic_bacteria_isolated_from_a_subterranean_rock_salt_crystal (accessed on 12 October 2023).
- Gutiérrez, C.; González, C. Method for simultaneous detection of proteinase and esterase activities in extremely halophilic bacteria. Appl. Microbiol. 1972, 24, 516–517. [Google Scholar] [CrossRef] [PubMed]
- Chuprom, J.; Bovornreungroj, P.; Ahmad, M.; Kantachote, D.; Dueramae, S. Approach toward enhancement of halophilic protease production by Halobacterium sp. strain LBU50301 using statistical design response surface methodology. Biotechnol. Rep. (Amst.) 2016, 10, 17–28. [Google Scholar] [CrossRef] [PubMed]
- Tsuchida, O.; Yamagata, Y.; Ishizuka, T.; Arai, T.; Yamada, J.-I.; Takeuchi, M.; Eiji, I. An Alkaline Proteinase of an alkalophilic Bacillus sp. Curr. Microbiol. 1986, 14, 7–12. [Google Scholar] [CrossRef]
- Boughachiche, F.; Rachedi, K.; Duran, R.; Lauga, B.; Karama, S.; Bouyoucef, L.; Boulezaz, S.; Boukrouma, M.; Boutaleb, H.; Boulahrouf, A. Optimization of alkaline protease production by Streptomyces sp. strain isolated from saltpan environment. Afr. J. Biotechnol. 2016, 15, 1401–1412. [Google Scholar] [CrossRef]
- Dussault, H.P. An improved technique for staining red halophiloc bacteria. J. Bacteriol. 1955, 70, 484–485. [Google Scholar] [CrossRef] [PubMed]
- Ventosa, A.; De La Haba, R.R. Chromohalobacter. In Bergey’s Manual of Systematics of Archaea and Bacteria, 1st ed.; Whitman, W.B., Ed.; Wiley: Hoboken, NJ, USA, 2020; pp. 1–16. [Google Scholar] [CrossRef]
- Arahal, D.R.; Ludwig, W.; Schleifer, K.H.; Ventosa, A. Phylogeny of the family Halomonadaceae based on 23S and 165 rDNA sequence analyses. IJSEM 2002, 52, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.; Govind, C.K.; Qiu, H.; Kim, S.J.; Dong, J.; Hinnebusch, A.G. Recruitment of the ArgR/Mcm1p repressor is stimulated by the activator Gcn4p: A self-checking activation mechanism. Proc. Natl. Acad. Sci. USA 2004, 101, 11713–11718. [Google Scholar] [CrossRef] [PubMed]
- Karbalaei-Heidari, H.R.; Amoozegar, M.A.; Hajighasemi, M.; Ziaee, A.A.; Ventosa, A. Production, optimization and purification of a novel extracellular protease from the moderately halophilic bacterium Halobacillus karajensis. J. Ind. Microbiol. Biotechnol. 2009, 36, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Kalwasińska, A.; Jankiewicz, U.; Felföldi, T.; Burkowska-But, A.; Swiontek Brzezinska, M. Alkaline and halophilic protease production by Bacillus luteus h11 and its potential industrial applications. Food Technol. Biotechnol. 2018, 56, 553–561. [Google Scholar] [CrossRef] [PubMed]
- Tsuchiya, K.; Nakamura, Y.; Sakashita, H.; Kimura, T. Purification and characterization of a thermostable alkaline protease from alkalophilic Thermoactinomyces sp. H8682. Biosci. Biotechnol. Biochem. 1992, 56, 246–250. [Google Scholar] [CrossRef] [PubMed]
- Vidyasagar, M.; Prakash, S.; Mahajan, V.; Shouche, Y.S.; Sreeramulu, K. Purification and characterization of an extreme halothermophilic protease from a halophilic bacterium Chromohalobacter sp. TVSP101. Braz. J. Microbiol. 2009, 40, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Majumder, D.R.; Kanekar, P.P.; Gaikwad, S.M. Purification and characterization of a thermolysin like protease from Thermoactinomyces thalpophilus MCMB-380. Protein Pept. Lett. 2013, 20, 918–925. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).