Isolation and Identification of Culturable Gut Microbiota in the Larval Stage of Lesser Mealworm (Alphitobius diaperinus) †
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation and Count of Culturable Bacteria and Yeast from the Gut of A. diaperinus
2.2. Bacterial and Yeast Identification
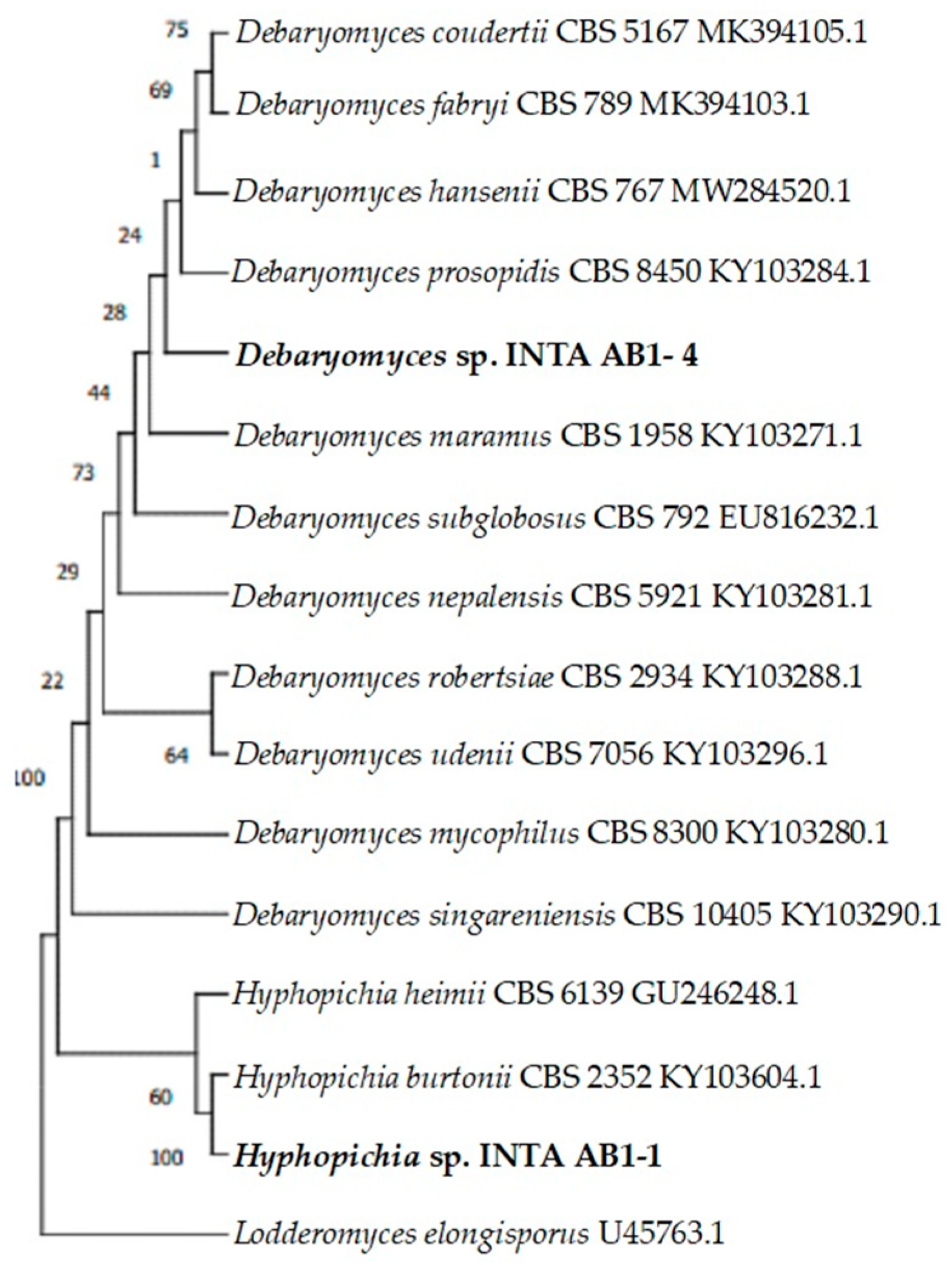
3. Results and Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kurečka, M.; Kulma, M.; Petříčková, D.; Plachy, V.; Kouřimská, L. Larvae and pupae of Alphitobius diaperinus as promising protein alternatives. Eur. Food Res. Technol. 2021, 247, 2527–2532. [Google Scholar] [CrossRef]
- Rumbos, C.I.; Karapanagiotidis, I.T.; Mente, E.; Athanassiou, C.G. The lesser mealworm Alphitobius diaperinus: A noxious pest or a promising nutrient source? Rev. Aquacult. 2019, 11, 1418–1437. [Google Scholar] [CrossRef]
- Cucini, C.; Funari, R.; Mercati, D.; Nardi, F.; Carapelli, A.; Marri, L. Polystyrene shaping effect on the enriched bacterial community from the plastic-eating Alphitobius diaperinus (Insecta: Coleoptera). Symbiosis 2022, 86, 305–313. [Google Scholar] [CrossRef]
- Cucini, C.; Leo, C.; Vitale, M.; Frati, F.; Carapelli, A.; Nardi, F. Bacterial and fungal diversity in the gut of polystyrene-fed Alphitobius diaperinus (Insecta: Coleoptera). Animal Gene 2020, 17–18, 200109. [Google Scholar] [CrossRef]
- Vaughan, J.A.; Turner, E.C.; Ruszler, P.L. Infestation and Damage of Poultry House Insulation by the Lesser Mealworm, Alphitobius diaperinus (Panzer). Poult. Sci. 1984, 63, 1094–1100. [Google Scholar] [CrossRef]
- Lu, F.; Kang, X.; Jiang, C.; Lou, B.; Jiang, M.; Way, M. Isolation and characterization of bacteria from midgut of the rice water weevil (Coleoptera: Curculionidae). Environ. Entomol. 2013, 42, 874–881. [Google Scholar] [CrossRef] [PubMed]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA Genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Martinis, V. Aspetti Microbiologici e Composizione Chimica di larve di Alphitobius diaperinus, Tenebrio molitor e Zophobas morio Destinati al Consumo Umano. Master’s Thesis, Università di Pisa, Pisa, Italy, 2020. [Google Scholar]
- Wynants, E.; Crauwels, S.; Verreth, C.; Gianotten, N.; Lievens, B.; Claes, J.; Van Campenhout, L. Microbial dynamics during production of lesser mealworms (Alphitobius diaperinus) for human consumption at industrial scale. Food Microbiol. 2018, 70, 181–191. [Google Scholar] [CrossRef] [PubMed]


| Isolate | Sequenced Gene/Genes | Nucleotide Length (bp) | GenBank Accession Number |
|---|---|---|---|
| INTA AN 1-1 | 16S rRNA | 1401 | OP339834.1 |
| INTA AN 1-5 | 16S rRNA | 1369 | OP346784.1 |
| INTA AN 1-10 | 16S rRNA | 1396 | OP346981.1 |
| INTA AN 1-15 | 16S rRNA | 1397 | OP347118.1 |
| INTA AC 1-3 | 16S rRNA | 1395 | OP348220.1 |
| INTA AC 1-6 | 16S rRNA | 1396 | OP348874.1 |
| INTA AC 1-9 | 16S rRNA | 1399 | OP348886.1 |
| INTA AC 1-14 | 16S rRNA | 1369 | OP351273.1 |
| INTA AC 1-4 | 16S rRNA | 1417 | OP348929.1 |
| INTA AC 1-8 | 16S rRNA | 967 | OP348932.1 |
| INTA AB 1-1 | rRNA genes ITS region | 446 | OP348991.1 |
| INTA AB 1-4 | rRNA genes ITS region | 612 | OP348992.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Antonuccio, G.I.; Sauka, D.H. Isolation and Identification of Culturable Gut Microbiota in the Larval Stage of Lesser Mealworm (Alphitobius diaperinus). Biol. Life Sci. Forum 2024, 31, 12. https://doi.org/10.3390/ECM2023-16465
Antonuccio GI, Sauka DH. Isolation and Identification of Culturable Gut Microbiota in the Larval Stage of Lesser Mealworm (Alphitobius diaperinus). Biology and Life Sciences Forum. 2024; 31(1):12. https://doi.org/10.3390/ECM2023-16465
Chicago/Turabian StyleAntonuccio, Gisele Ivonne, and Diego Herman Sauka. 2024. "Isolation and Identification of Culturable Gut Microbiota in the Larval Stage of Lesser Mealworm (Alphitobius diaperinus)" Biology and Life Sciences Forum 31, no. 1: 12. https://doi.org/10.3390/ECM2023-16465
APA StyleAntonuccio, G. I., & Sauka, D. H. (2024). Isolation and Identification of Culturable Gut Microbiota in the Larval Stage of Lesser Mealworm (Alphitobius diaperinus). Biology and Life Sciences Forum, 31(1), 12. https://doi.org/10.3390/ECM2023-16465







