Associations between Vitamin D Receptor (VDR) Polymorphisms and Gut Microbiota in a Spanish Population Sample †
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Sample Collection
2.3. DNA Extraction
2.4. VDR Genotyping
2.5. Sequencing and Bioinformatics
2.6. Statistical Analysis
3. Results
3.1. VDR Genotyping
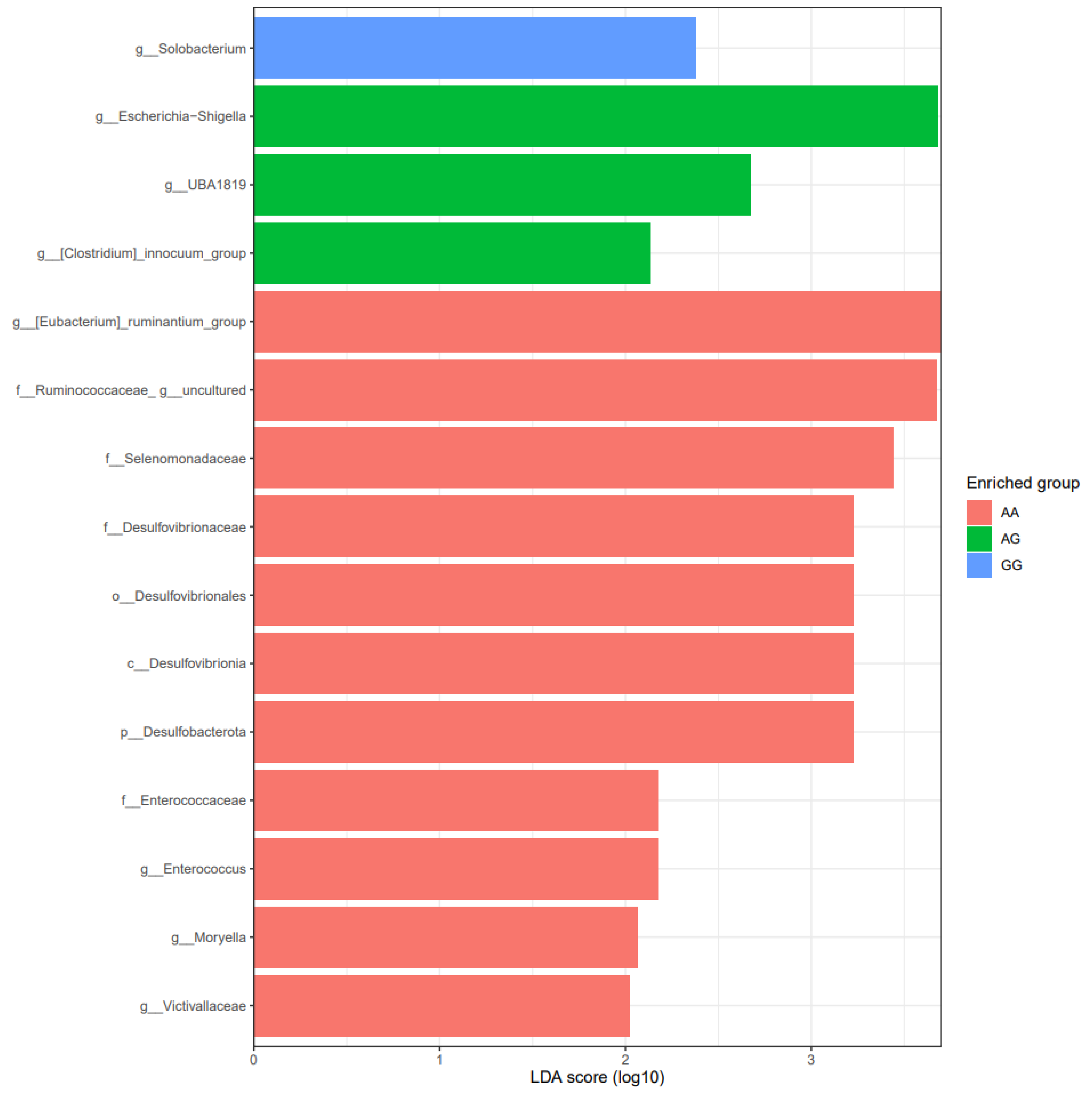
3.2. Bacterial Composition According to Genotypes
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pike, J.W.; Meyer, M.B. The vitamin D receptor: New paradigms for the regulation of gene expression by 1,25-dihydroxyvitamin D3. Endocrinol. Metab. Clin. N. Am. 2010, 39, 255–269. [Google Scholar] [CrossRef] [PubMed]
- Ishizawa, M.; Akagi, D.; Makishima, M. Lithocholic Acid Is a Vitamin D Receptor Ligand That Acts Preferentially in the Ileum. Int. J. Mol. Sci. 2018, 19, 1975. [Google Scholar] [CrossRef] [PubMed]
- Khammissa, R.A.G.; Fourie, J.; Motswaledi, M.H.; Ballyram, R.; Lemmer, J.; Feller, L. The Biological Activities of Vitamin D and Its Receptor in Relation to Calcium and Bone Homeostasis, Cancer, Immune and Cardiovascular Systems, Skin Biology, and Oral Health. BioMed Res. Int. 2018, 2018, 9276380. [Google Scholar] [CrossRef] [PubMed]
- Feldman, D.; Malloy, P.J. Mutations in the vitamin D receptor and hereditary vitamin D-resistant rickets. Bonekey Rep. 2014, 3, 510. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, I.; Lu, R.; Zhang, Y.; Zhang, J.; Dai, Y.; Xia, Y.; Sun, J. Vitamin D receptor promotes healthy microbial metabolites and microbiome. Sci. Rep. 2020, 10, 7340. [Google Scholar] [CrossRef] [PubMed]
- Loughran, G.; Jungreis, I.; Tzani, I.; Power, M.; Dmitriev, R.I.; Ivanov, I.P.; Kellis, M.; Atkins, J.F. Stop codon readthrough generates a C-terminally extended variant of the human vitamin D receptor with reduced calcitriol response. J. Biol. Chem. 2018, 293, 4434–4444. [Google Scholar] [CrossRef] [PubMed]
- Goode, E.L.; Dunning, A.M.; Kuschel, B.; Healey, C.S.; E Day, N.; Ponder, B.A.J.; Easton, D.F.; Pharoah, P.P.D. Effect of germ-line genetic variation on breast cancer survival in a population-based study. Cancer Res. 2002, 62, 3052–3057. [Google Scholar] [PubMed]
- Lundin, A.C.; Söderkvist, P.; Eriksson, B.; Bergman-Jungeström, M.; Wingren, S.; South-East Sweden Breast Cancer Group. Association of breast cancer progression with a vitamin D receptor gene polymorphism. Cancer Res. 1999, 59, 2332–2334. [Google Scholar] [PubMed]
- Vasilopoulos, Y.; Sarafidou, T.; Kotsa, K.; Papadimitriou, M.; Goutzelas, Y.; Stamatis, C.; Bagiatis, V.; Tsekmekidou, X.; Yovos, J.G.; Mamuris, Z. VDR TaqI is associated with obesity in the Greek population. Gene 2013, 512, 237–239. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Thingholm, L.B.; Skiecevičienė, J.; Rausch, P.; Kummen, M.; Hov, J.R.; Degenhardt, F.; Heinsen, F.-A.; Rühlemann, M.C.; Szymczak, S.; et al. Genome-wide association analysis identifies variation in vitamin D receptor and other host factors influencing the gut microbiota. Nat. Genet. 2016, 48, 1396–1406. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Zhang, Y.-G.; Lu, R.; Xia, Y.; Zhou, D.; O Petrof, E.; Claud, E.C.; Chen, D.; Chang, E.B.; Carmeliet, G.; et al. Intestinal epithelial vitamin D receptor deletion leads to defective autophagy in colitis. Gut 2015, 64, 1082–1094. [Google Scholar] [CrossRef] [PubMed]
- Kaehler, B.D.; Bokulich, N.A.; McDonald, D.; Knight, R.; Caporaso, J.G.; Huttley, G.A. Species-level microbial sequence classification is improved by source-environment information. Nat. Commun. 2019, 10, 4643. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Caporaso, J.G. Optimizing taxonomic classification of marker gene sequences. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef] [PubMed]
- Bailén, M.; Tabone, M.; Bressa, C.; Lominchar, M.G.M.; Larrosa, M.; González-Soltero, R. Unraveling Gut Microbiota Signatures Associated with PPARD and PARGC1A Genetic Polymorphisms in a Healthy Population. Genes 2022, 13, 289. [Google Scholar] [CrossRef] [PubMed]
- Sole, X.; Guino, E.; Valls, J.; Iniesta, R.; Moreno, V. SNPStats: A web tool for the analysis of association studies. Bioinformatics 2006, 22, 1928–1929. [Google Scholar] [CrossRef] [PubMed]
- Alauzet, C.; Aujoulat, F.; Lozniewski, A.; Brahim, S.B.; Domenjod, C.; Enault, C.; Lavigne, J.-P.; Marchandin, H. A new look at the genus solobacterium: A retrospective analysis of twenty-seven cases of infection involving s. moorei and a review of sequence databases and the literature. Microorganisms 2021, 9, 1229. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Chi, X.; Zhang, Q.; Chen, F.; Deng, X. Characterization of the salivary microbiome in people with obesity. PeerJ. 2018, 6, e4458. [Google Scholar] [CrossRef] [PubMed]
| rs731236 (TaqI) (C/T) | SNP Allele Frequencies (n = 87) | Genotypes | Frequencies (H-W) |
|---|---|---|---|
| T | 0.39 | TT | 0.43 |
| C | 0.61 | TC | 0.38 |
| CC | 0.20 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
González Soltero, R.; Bressa, C.; Tabone, M.; Clemente, S.; Larrosa, M.; Bailén, M. Associations between Vitamin D Receptor (VDR) Polymorphisms and Gut Microbiota in a Spanish Population Sample. Biol. Life Sci. Forum 2023, 29, 4. https://doi.org/10.3390/IECN2023-15530
González Soltero R, Bressa C, Tabone M, Clemente S, Larrosa M, Bailén M. Associations between Vitamin D Receptor (VDR) Polymorphisms and Gut Microbiota in a Spanish Population Sample. Biology and Life Sciences Forum. 2023; 29(1):4. https://doi.org/10.3390/IECN2023-15530
Chicago/Turabian StyleGonzález Soltero, Rocío, Carlo Bressa, Mariangela Tabone, Sara Clemente, Mar Larrosa, and María Bailén. 2023. "Associations between Vitamin D Receptor (VDR) Polymorphisms and Gut Microbiota in a Spanish Population Sample" Biology and Life Sciences Forum 29, no. 1: 4. https://doi.org/10.3390/IECN2023-15530
APA StyleGonzález Soltero, R., Bressa, C., Tabone, M., Clemente, S., Larrosa, M., & Bailén, M. (2023). Associations between Vitamin D Receptor (VDR) Polymorphisms and Gut Microbiota in a Spanish Population Sample. Biology and Life Sciences Forum, 29(1), 4. https://doi.org/10.3390/IECN2023-15530










